Abstract
Background
The insulin-like growth factor-1 receptor (IGF-1R) pathway is known to play a role in the acquisition of resistance to epidermal growth factor receptor (EGFR)-specific tyrosine kinase inhibitors (TKIs) in non-small cell lung cancer (NSCLC). However, its exact role in TKI resistance has so far remained unclear. Here, we interrogated the hypothesis that the IGF-1R may serve as a biomarker for, and may play a role in, intrinsic resistance to the EGFR-specific TKI gefitinib in NSCLC.
Methods
Total-IGF-1R and phosphorylated (p)-IGF-1R expression levels were related to gefitinib sensitivity in 23 NSCLC cell lines. This sensitivity was re-evaluated after knocking down IGF-1R expression and after IGF-1R up‐regulation through exogenous IGF-1 expression. The utility of IGF-1R expression as a predictive biomarker was also evaluated by immunohistochemistry (IHC) in 98 primary NSCLC samples from patients treated with gefitinib.
Results
Seventeen of the cell lines tested were resistant to gefitinib, whereas 3 cell lines were sensitive. The three remaining cell lines showed intermediate values. Thirteen resistant cell lines were found to be positive for total-IGF-1R expression, while all the sensitive cell lines were negative, resulting in a positive predictive value (PPV) of 81 % for total-IGF-1R to predict resistance. Seven resistant cell lines exhibited high p-IGF-1R levels, whereas all 3 sensitive cell lines were negative for p-IGF-1R, resulting in a PPV of 100 % for p-IGF-1R to predict resistance. Neither a knock-down of IGF-1R expression nor an activation of the IGF1-R pathway through exogenous IGF-1 expression affected gefitinib sensitivity. In primary NSCLC tissues, IGF-1R expression was found to be significantly higher in patients with progressive disease, i.e., showing gefitinib resistance, as compared to those with a complete or partial response.
Conclusions
IGF-1R acts as a predictor for resistance to gefitinib in NSCLC cell lines and NSCLC patients, but does not seem to play a role in the intrinsic resistance to this drug. High total-IGF-1R and p-IGR-1R levels may predict such a resistance. Since the underlying mechanism does not appear to be related to proliferation induction, alternative pathways should be explored.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer is the leading cause of cancer-related mortality with 157,300 deaths in 2010 in the United States alone, and more than 1,300,000 deaths each year globally [1]. The overall 5 year survival rate for lung cancer patients remains extremely low (approximately 16 %), highlighting the need for more effective treatment strategies. One of the most important advances in the treatment of non-small cell lung cancer (NSCLC) over the last decade has been the development of targeted therapies aimed at the epidermal growth factor receptor (EGFR) [2]. The EGFR signaling pathway has been studied extensively and, by doing so, it was found that 40–80 % of NSCLCs exhibit over-expression of the EGFR. In addition, it was found that tumors that carry activating mutations in the EGFR gene are particularly sensitive to EGFR-specific tyrosine kinase inhibitors (TKIs) [2, 3]. Currently, among the existing bio-markers, the EGFR mutation status plays a prominent role in the treatment of advanced NSCLC in 1st line therapies, and EGFR TKIs have been shown to be superior to chemotherapy in EGFR mutation-positive NSCLC patients [2, 4–7]. The TKI gefitinib has been FDA approved for patients harboring EGFR mutations. However, as of yet all NSCLC patients treated with EGFR-specific TKIs develop at some point resistance to these TKIs.
Resistance to EGFR-specific TKIs may result from intrinsic and/or acquired processes. Acquired resistance may occur e.g. through the occurrence of additional mutations in the EGFR gene, such as the T790M mutation, which is associated with >50 % of NSCLC cases with acquired TKI resistance [8–10]], cMET amplification [11–14], epithelial-mesenchymal transition (EMT) [15], or insulin-like growth factor 1 receptor (IGF-1R) pathway activation [16–18]. Currently, there is a growing body of evidence for a role of the latter pathway in the acquisition of resistance to EGFR-specific TKIs. The IGF-1R binds both insulin-like growth factor-1 (IGF-1) and IGF-2. IGF-1 is a potent stimulator of mitogenesis and can promote cellular differentiation and transformation. Furthermore, IGF-1 acts as a major survival factor and protects cells from apoptosis in a wide variety of conditions [18]. It has been reported that lung cancer patients exhibit high levels of circulating IGF-1, which is in line with the notion that the IGF-1R pathway may be involved in the development of this disease [18]. The IGF-1R is expressed in many cancers [19], and its stimulation has been found to interfere with the anti-tumor activity of EGFR inhibitors through apoptosis inhibition by amphiregulin [18]. In addition, TKIs can induce EGFR/IGF-1R heterodimerization, thereby transmitting survival signals through the IGF-1R and its downstream mediators [20]. Ludovini et al. [21] reported that among 125 NSCLC patients, co-expression of IGF-1R and EGFR was associated with a shorter disease-free survival in resected stage I patients, and others reported that high IGF-1R expression was associated with a poor survival in surgically resected NSCLC patients [17, 22]. Additionally, IGF-1R over-expression was found in EGFR TKI-resistant primary human glioblastomas [16] and down-regulation of IGFBP-3 and IGFBP-4 in EGFR TKI-resistant squamous carcinoma cancer cell lines [23]. Intrinsic (primary) resistance to TKIs has been related to e.g. the presence of KRAS mutations [13, 24–28].
The aim of this study was to evaluate the role of the IGF-1R as a potential biomarker for resistance to the EGFR-specific TKI gefitinib. To this end, we assessed in a series of lung cancer cell lines whether (i) total or phosphorylated IGF-1R can predict resistance to gefitinib, (ii) knocking down IGF-1R expression through shRNA can render gefitinib-resistant cell lines sensitive, and (iii) IGF-1 can increase the proliferation rate of gefitinib-resistant or gefitinib-sensitive cell lines. Next, we validated our results in a clinical cohort of 98 Japanese patients with NSCLC who were treated with gefitinib.
2 Materials and methods
2.1 Cell lines and culture conditions
The 23 NSCLC cell lines used in this study were obtained from the American Tissue Culture Collection (ATCC, Manassas, VA, USA) and propagated and maintained in RPMI-1640 medium (Sigma, USA) supplemented with 10 % foetal bovine serum (Sigma, USA). The packaging cell line phoenix 293 T was maintained in Dulbecco’s modified Eagle’s medium (DMEM) (GIBCO, USA) supplemented with 10 % foetal bovine serum. All cells were cultured at 37 °C in a humidified atmosphere (95 %) and mixture of air and CO2 (5 %).
2.2 MTS assay
The sensitivity of cells to gefitinib was determined by using a MTS in vitro cytotoxicity assay according to the manufacturer’s instructions (Promega Corp, USA). In brief, cells were seeded overnight at optimized densities in 96-well plates in RPMI 1640 medium containing 10 % FBS. Next, cells were treated with gefitinib for 72 h at concentrations ranging from 0–30 μM. Cell growth in the presence of only the vehicle (DMSO) was calculated and set at 100 %. IC50 was defined as the drug concentration that leads to 50 % decrease in cell growth.
2.3 Lentiviral transduction and gefitinib sensitivity
Three vectors containing RFP-labeled IGF-1R shRNA sequences (B5, B10 and G6), a vector control (NSC), and two lentiviral packaging plasmids (pCMV-VSV-G and pHR-8.2ΔR) were purchased from Thermo Fisher Open Biosystems. The recombinant and control lentiviruses were produced through transfection of phoenix 293 T cells by the LT1 method. In brief, 7.5 × 106 293 T cells were cultured in DMEM with 10 % foetal bovine serum in a T75 flask. After 24 h and 1 h prior transfection, 8 ml of fresh DMEM growth medium was added. Transfection of 3 vectors (pCMV-VSV-G, pHR-8.2 ΔR, and a recombinant lentiviral vector or control vector) using the LT1 transfection reagent (Thermo Fisher Open Biosystems) was performed and the transfected 293 T cells were maintained at 37 °C. After 48 h, the produced lentiviruses were harvested and NSCLC H226 cells were transduced. These transduced cells were screened in puromycin selective medium for 3 days. For gefitinib sensitivity detection, the transduced cells were seeded in 96-well plates using RPMI-1640 with 10 % FBS. After 24 h the cells were treated with different concentrations of gefitinib and incubated with or without doxycycline. Five days later, the levels of growth inhibition were determined by the MTS assay.
2.4 Preparation of total cell extracts and immunoblot analysis
After growth to 70–80 % confluence, cells were harvested and resuspended in RIPA lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 0.1 % SDS, 0.5 % deoxycholic acid, 0.02 % sodium azide, 1 % NP-40, 2.0 μg/ml aprotinin, 1 mM phenylmethylsulfonylfluoride [PMSF]). To detect the effect of IGF-1R knock-down, the cells were seeded into 10 cm dishes. After 24 h the cells were incubated with doxycycline for varying lengths of time. After lysis, equivalent amounts of proteins were used for total IGF-1R detection. The effects of IGF-1 stimulation were detected by serum starving the cells for 24 h and then stimulating the cells with 5 or 10 ng IGF-1 for 10 min. Equivalent amounts of proteins were used for total-IGF-1R, Erk1/2, phosphorylated-Erk1/2, AKT, and phosphorylated-AKT (Cell Signaling Technology, Beverly, MA) immunoblot detection.
2.5 Total-IGF-1R and p-IGFR-1 expression levels
The expression level of total-IGF-1R in the gefitinib-sensitive H-827 cell line was chosen as the lower limit of total-IGF-1R positivity by immunoblotting (see above). Using this cut-off, 13 of the 17 gefitinib-resistant cell lines were positive for total-IGF-1R, whereas all 3 of the gefitinib-sensitive cell lines were negative. The expression level of phosphorylated (p)-IGF-1R in the gefitinib-resistant H1975 cell line was chosen as the lower limit of p-IGF-1R positivity. Using this cut-off, 7 out of 17 cell lines that were gefitinib-resistant exhibited high levels of p-IGF-1R and 3 of the gefitinib-sensitive cell lines were negative for p–IGF-1R. The immunoblot bands were quantified using ImageJ software.
2.6 Immunoprecipitation of IGF-1 activated p-IGF-1R
To detect basal levels of p-IGF-1R, 48 hours before collection the cells were seeded at optimized densities in 10 cm dishes in RPMI 1640 medium containing 10 % FBS. When the cells were grown to 70–80 % confluence, they were washed with ice-cold 1x PBS and lysed on ice in RIPA lysis buffer (see above). Next, the lysates were centrifuged at 10,000 g for 15 min at 4 °C. Equal amounts of whole cell lysates (1 mg) were used for immunoprecipitation. After pre-clearing with 40 μl of protein G agarose for 30 min, the lysates were incubated with the phosphotyrosine antibody PY20 (Santa Cruz Biotechnology, USA) overnight on a rotating wheel at 4 °C. Enrichment of specifically bound antibody was achieved by the addition of 60 μl protein G agarose for 2 h. After brief centrifugation and washing, the precipitated proteins were released from the bead complexes by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and analyzed directly by SDS-PAGE and immunoblotting. In the IGF-1 stimulation experiments, cells were seeded in 10 cm dishes 24 h before collection. Then the cells in monolayer were washed twice with serum-free medium to remove residual FBS. After overnight serum starvation, the cells were stimulated for 10 min at 37 °C and 5 % CO2 with serum-free medium containing different concentrations of IGF-1. After this, the cells were washed with ice-cold 1x PBS and lysed on ice using lysis buffer, and subjected to immunoprecipitation as described above.
2.7 Cell proliferation assay
Cell proliferation was assessed by measuring 5-bromo-2-deoxyuridine (BrdU) incorporation during DNA synthesis in proliferating cells according to the manufacturer’s instructions (Roche Molecular Biochemicals). Briefly, 100 μl of cell suspension (2 × 105 cells/ml) was seeded into 96-well plates containing RPMI 1640 medium with 0.5 % FBS. Cells were allowed to adhere to the substratum for 24 h. Next, the medium was replaced with fresh RPMI 1640 containing IGF-1 (50 μg/ml or 100 μg/ml) and the cells were incubated for another 24 h. Then 10 μM BrdU was added to incorporate into the newly synthesized DNA. After 2 h of incubation the culture medium was removed, the cells were fixed and permeabilized, and the DNA was denatured to enable antibody binding to the incorporated BrdU. Anti-BrdU antibody was added to the wells for 1 h at room temperature. Unbound antibody was removed and horseradish peroxidase-(HRP)-conjugated goat-anti mouse antibody was added. Next, a substrate solution was added to each well, resulting in a colour change proportional to the amount of DNA synthesized by the cells. Finally, the colour reaction was stopped and the optical density was determined using a Molecular Devices Vmax kinetic microplate reader, set to 450 nm.
2.8 IGF-1R protein expression analysis by immunohistochemistry
Immunohistochemical (IHC) evaluation was performed using the Ventana G11 anti-IGF-1R antibody following the manufacturer’s instructions (CONFIRM™, Ventana Medical Systems, Tucson, AZ). Briefly, 4 μM sections were stained on the Ventana BenchMark XT autostainer via a 16 min primary antibody incubation and evaluated using the ultraView detection kit. Placental tissue was included as a positive control, and as a negative control a non-immune rabbit antibody was used instead of the primary antibody on the positive control tissue. The score was determined according to the ‘hybrid scoring system’ (H-score) criteria, through which the products of the five staining intensity categories (0–4) are multiplied by the percentage of positive cells (0–100 %). Thus, the final IHC scores range from 0 to 400. IHC staining was evaluated at the University of Colorado by a certified pathologist and a trained reader who were blinded to the clinical data.
2.9 Clinical cohorts
2.9.1 Patients
In this study we included 98 Japanese NSCLC patients who were treated with gefitinib as monotherapy (250 mg per day), and who were assessed for recurrent disease after having undergone curative pulmonary resection at the Tokyo Medical University Hospital between May 1995 and March 2008. The clinical characteristics of these patients have been described before in [3] and [29]. Briefly, this NSCLC cohort includes 46 male and 52 female patients of which 50 were smokers, 80 patients with adenocarcinomas (ADC), 9 patients with squamous cell carcinomas (SCC), 5 patients with large cell carcinomas and 4 patients with other NSCLCs, defined according to the World Health Organization criteria [30]. Pathological staging at the time of surgery using the TNM classification of malignant tumors in conjunction with H&E stained sections [31] revealed 14, 17, 10, 9, 36, 7 and 5 patients at stages IA, IB, IIA, IIB, IIIA, IIIB and IV, respectively. The responses to gefitinib therapy were defined according to response evaluation criteria in solid tumors (RECIST), in conjunction with computed tomography scanning [32], as complete response (CR), partial response (PR), stable disease (SD) and progressive disease (PD). At least 4 weeks after the initial determination of therapy response a confirmatory evaluation was repeated. During gefitinib therapy, assessments were performed every 4 weeks for the first 4 months and then every 8 weeks until disease progression. As RECIST recommends that the duration of SD should specify the minimal time interval required between two measurements, disease control (DC = CR + PR + SD) was evaluated at 12 weeks. Of the 98 patients included, 10 (10 %) received gefitinib as 1st systemic anticancer therapy after recurrence, 43 (44 %) received gefitinib as 2nd and 45 (46 %) as ≥3rd line therapy. Objective responses were considered only for patients treated with gefitinib for at least 4 weeks. Responses were available for 94/98 patients. Progression free survival (PFS) was defined as the time from the start of gefitinib therapy to progression or death.
2.9.2 Tissue microarrays
Paraffin-embedded tissue blocks were prepared following fixation with 10 % formalin from the 98 surgical resection specimens included in this study, and subjected to pathological review. Whole tissue mounts were used from 28 of the patients, whereas tissue microarrays (TMAs) were generated from 70 patients. Three replicate core samples (1.2 mm) from the most representative tumor areas were collected and the TMAs were assembled using a tissue-arraying device. Normal liver tissues were used for slide orientation purposes. After sectioning, the slides were stored at 4 °C until use.
3 Results
3.1 Total-IGF-1R and p-IGF-1R expression and its association with gefitinib sensitivity
In order to identify cells with different responses to gefitinib, a panel of 23 NSCLC cell lines was screened using the MTS assay. The sensitivity of each cell line is presented as IC50 (Fig. 1a). A wide range of sensitivity was observed across this panel. The resistant/sensitive phenotype to gefitinib was classified as follows: cell lines with an IC50 above 10 μM were defined as resistant and those with an IC50 below 2 μM as sensitive, whereas those in between were defined as intermediate. Using this classification, the Calu-3, HCC4006 and H827 cell lines were sensitive, the HCC95, H2122 and HCC193 cell lines intermediate and the remaining 17 cell lines resistant to gefitinib (Fig. 1a). The mutations status of each cell line is indicated below the MTS assay in Fig. 1a.
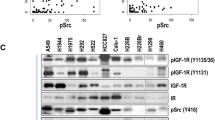
Gefitinib IC50 values for 27 NSCLC cell lines and expression of total-IGF-1R and p-IGF-1R. a Graphic representation of IC50 values for 27 NSCLC cell lines treated with gefitinib and measured using the MTS assay. The EGFR and KRAS mutation status, as well as ALK-fusion status, of the cell lines are shown below. b Basal levels of total-IGF-1R and p-IGF-1R in the 27 NSCLC cell lines. p-IGF-1R was first immunoprecipitated with a p-tyrosine specific antibody followed by immunoblotting with an IGF-1R specific antibody (bands were quantified by ImageJ software). c Graphic representation of gefitinib-resistant and sensitive cell lines (total-IGF-1R lower limit reference is H827; p-IGF-1R lower limit reference is H1975; IC50 cut-off values: Sensitive <2 mM, Resistant ≥10 mM). d Gefitinib sensitivity and specificity of NSCLC cell lines in relation to total-IGF-1R and p-IGF-1R expression (PPV Positive predictive value, NPV Negative predictive value)
To assess whether IGF-1R activation is correlated with gefitinib-resistance, the protein expression levels of both total and activated p-IGF-1R were determined in the 23 NSCLC cell lines using immunoblotting and immunoprecipation followed by immunoblotting, respectively. Endogenous GAPDH was used as a loading control. By doing so, we found that total-IGF-1R was expressed at varying levels in all cell lines, with the exception of the gefitinib-resistant cell line H441, which did not appear to express any total-IGF-1R (Fig. 1b). The expression level of total-IGF-1R in the gefitinib sensitive cell line H-827 was arbitrarily chosen as the lower limit of total-IGF-1R positivity. By using this cut-off level, 13 of the 17 gefitinib-resistant cell lines were positive for total-IGF-1R. Also, all 3 gefitinib-sensitive cell lines were positive. These results provide a specificity of 76 %, a sensitivity of 0 %, a positive predictive value of 81 %, and negative predictive value of 0 % for total-IGF-1R to predict resistance to gefitinib (Fig. 1c, d). The 3 cell lines defined as having an intermediate sensitivity to gefitinib were all positive for total-IGF-1R. Phosphorylated (p)-IGF-1R was also observed in a number of cell lines with varying degrees of expression. The expression level of p-IGF-1R in the H1975 cell line was arbitrarily chosen as the lower limit of positivity. By using this cut-off level, 7 out of the 17 gefitinib-resistant cell lines exhibited high levels of p-IGF-1R, whereas all 3 gefitinib-sensitive cell lines were negative for p-IGF-1R. This results in a specificity of 100 %, a sensitivity of 41 %, a positive predictive value of 100 %, and negative predictive value of 23 % for high p-IGF-1R to predict resistance to gefitinib (Fig. 1c, d). From the 3 cell lines defined as having an intermediate sensitivity to gefitinib, 2 were positive for p-IGF-1R and one was negative. These cell lines were not included in the specificity and sensitivity analyses. Together, these data suggest that a high total-IGF-1R level does discriminate between cell lines resistant or sensitive to gefitinib. However, while high p-IGF-1R can predict resistance to gefitinib (7/7), low p-IGF-1R expression cannot predict resistance, as 10 out of 13 cell lines which expressed low levels of p-IGF-1R were resistant to gefitinib (Fig. 1d).
3.2 IGF-1R expression knock-down and its effect on gefitinib sensitivity
To further investigate the correlation between IGF-1R expression and gefitinib resistance in NSCLC, we assessed whether reducing the expression of IGF-1R in a resistant cell line would affect its sensitivity to gefitinib. To this end, H226, a gefitnib resistant cell line, was transduced with either of 3 IGF-1R shRNA lentiviruses (B5, B10 and G6). The efficiency of the knock-down (KD) was evaluated by immunoblotting of the 3 respective cell lines generated following a time course of doxycycline induction of the shRNAs (Fig. 2a). Compared to the normal control parental H226 cell line (NC), IGF-1R expression was significantly reduced in the B5 and B10 transduced cell lines, with a maximum reduction at day 6. While IGF-1R expression was also reduced in the G6 transduced cell line relative to the normal parental control, the expression at day 6 did not appear to differ from that at day zero. (Fig. 2a). In order to next assess whether a reduction in IGF-1R protein expression does affect gefitinib sensitivity, the viability of the B5 and B10 transduced cell lines was measured using the MTS assay following treatment with increasing concentrations of gefitinib with and without induction of the shRNAs. By doing so, we found that there was no difference in viability between the induced and the non-induced cell lines at any concentration of gefitinib and, thus, that both cell lines remained resistant (Fig. 2b). These results indicate that the presence of IGF-1R is not responsible for the resistance to gefitininb in this cell line and that, in addition, this cell line is not dependent on IGF-1R for growth.
Knock-down of IGF-1R protein expression with shRNAs and its effect on sensitivity to gefitinib. a Immunoblot of total-IGF-1R and GAPDH in the normal control parental H226 cell line (NC) following 6 days of doxycycline treatment, and three H266 cell lines transduced with IGF-1R shRNA lentiviruses (B5, B10, G6) induced for 0, 2, 4 or 6 days with doxycycline. b Graphs showing the survival curves of the B5 and B10 transduced cell lines treated with increasing concentrations of gefitinib and induced or not induced with doxycycline to produce shRNAs
3.3 IGF-1 stimulation of IGF-1R and its downstream signaling
In order to assess whether gefitinib resistant cell lines are capable of activating the IGF-1R pathway and inducing proliferation, 3 gefitinib-resistant cell lines (H1975, A549 and H226), 2 gefitinib-sensitive cell lines (HCC4006 and Calu-3) and 1 intermediate cell line (HCC95) were stimulated with IGF-1 following a 24 h serum starvation. As shown in Fig. 3a, IGF-1 was capable of activating IGF-1R in all the gefitinib-resistant cell lines tested and the gefitinib-sensitive cell line HCC4006, but not the gefitinib-sensitive cell line Calu-3. The downstream mediator AKT was, however, activated in the Calu-3 cell line, as well as in all the other cell lines tested. All cell lines had high basal levels of activated ERK (p-ERK) except the A549 cell line, which had a lower but still detectable level of p-ERK. While all cell lines exhibited some level of responsiveness to IGF-1, none of the cell lines showed any change in proliferation rate, as measured by BrdU incorporation, following IGF-1 stimulation (Fig. 3b). These results indicate that neither the gefitinib-resistant nor the gefitinib-sensitive cell lines appear dependent the IGF-1R pathway as a predominant driver for growth, although they respond to IGF-1 by activating the receptor and/or its downstream pathway.
IGF-1R pathway activation and cell proliferation following IGF-1 stimulation. a Immunoblot analysis of p-IGF-1R, total-IGF-1R, p-AKT, p-ERK1/2, and GAPDH in 3 gefitinib-resistant cell lines (H1975, A549, H226), 2 gefitinib-sensitive cell lines (HCC4006 and Calu-3) and one intermediate (HCC95) cell line following IGF-1 stimulation. b Histogram of BrdU incorporation in newly synthesized DNA in NSCLC cell lines following either IGF-1stimulation at 50 or 100 ng/ml or no stimulation
3.4 IGF-1R protein expression in a clinical cohort treated with gefitinib
3.4.1 IGF-1R protein expression levels and clinical characteristics
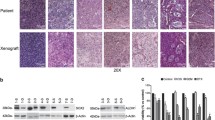
In order to further explore the hypothesis that higher IGF-1R protein expression predicts resistance to EGFR-targeted therapy, we examined total-IGF-1R protein expression levels in a cohort of 98 NSCLC patients who were treated with gefitinib following post-surgical relapse of their disease. In Table 1 the levels of IGF-1R protein expression compared to patients’ age, gender, histology, pathological stage, smoking status and EGFR mutation status are listed. Assessment of IGF-1R on a continuous scale shows relatively higher expression levels in males (mean H-score 170 for males versus 113 for females, p = 0.0037), squamous cell carcinoma (mean 253 for SCC versus 123 for ADC versus 177 for other histologies, p = 0.0002) and patients with wild-type (WT) or non-exon 19 deleted or non-exon 21 L858R point mutated EGFR (mean 173 for WT or non-exon 19 deletion or exon 21 mutation-positive versus 103 for exon 19 deletion or exon 21 mutation-positive, p = 0.0004). No differences were found regarding age, stage or smoking status. In Fig. 4a the IGF-1R distributions for the entire population, as well as by gender, histology and EGFR mutation status, are shown.
IGF-1R protein expression distribution in a clinical NSCLC cohort treated with gefitinib. a Scatter plot showing the distribution of IGF-1R protein expression in the entire cohort by gender, histology, and EGFR mutation status. b Scatter plot showing the distribution of IGF-1R protein expression in relation to the response in the entire cohort and the two EGFR mutation status subgroups
3.4.2 IGF-1R protein expression levels and associations with clinical response
We also examined IGF-1R protein expression levels relative to the best responses that the patients achieved upon gefitinib treatment. We found that IGF-1R expression was significantly higher in patients with progressive disease (PD), i.e. resistance, as compared to those with a complete response (CR) or a partial response (PR) (mean H-score 202 for PD versus 102 for CR/PR, p = 0.0003) (Table 1 and Fig. 4b). The IGF-1R expression was also significantly higher in PD versus disease control, which includes CR, PR and stable disease (SD) (mean 202 for PD versus 119 for DC, p = 0.0006). Assessment of IGF-1R expression relative to response within the EGFR WT (or non-exon 19 deletion or exon 21 mutation-positive) subgroup showed that IGF-1R was higher, but not statistically significant, in those with PD versus CR/PR (mean 201 versus 115, p = 0.1029) and PD versus DC (mean 201 versus 150, p = 0.1366). Within the EGFR exon 19 deletion or exon 21 mutation-positive population, i.e., those predicted to respond based on EGFR mutation status, IGF-1R expression was significantly higher in patients whose best response to gefitinib was PD compared to those with a CR or PR (mean 190 versus 98, p = 0.0283) or DC (mean 190 versus 96, p = 0.0161).
3.4.3 IGF-1R protein expression discriminates between progressive disease and clinical response or disease control
Due to the significantly higher expression levels of IGF-1R in patients with PD, we assessed whether IGF-1R expression can discriminate between PD and either CR/PR or DC using receiver operator characteristics (ROC) curve analyses. By doing so, we indeed found that IGF-1R can discriminate between PD and CR/PR with a significant area under the curve (AUC) of 0.82 (p = 0.0006) and, likewise, that there was a significant discrimination between PD and DC (AUC 0.76, p = 0.0006) (Fig. 5a and Table 1). There was also a trend towards significance in the EGFR WT or non-exon 19 deletion or exon 21 mutation-positive subgroup with an AUC of 0.76 (p = 0.0888) for PD versus CR/PR and of 0.65 (p = 0.1060) for PD versus DC. However, within the EGFR exon 19 or 21 mutation-positive population, those most likely to be responsive to gefitinib, IGF-1R protein expression could significantly discriminate PD from CR/PR (AUC 0.92, p = 0.0254) and PD from DC (AUC 0.90, p = 0.0222).
Receiver operator characteristic (ROC) curves for discrimination of gefitinib response, and Kaplan-Meier progression-free survival curves. a ROC curves showing the ability of IGF-1R protein expression to discriminate between either progressive disease (PD) and complete or partial response (CR/PR), or between PD and disease control (DC = CR + PR + stable disease, SD). Curves are shown for the entire cohort and the two EGFR mutation status populations. b Kaplan-Meier progression-free survival (PFS) curves for high versus low IGF-1R protein expression. Curves are shown for the entire cohort and the two EGFR mutation status subgroups
3.4.4 IGF-1R protein expression and its association with disease outcome
Given the ability of IGF-1R to discriminate between PD and either CR/PR or DC and the significantly higher expression of IGF-1R in the cases with PD, we assessed whether there is a difference in progression-free survival (PFS) after gefitinib for the NSCLC cases high in IGF-1R protein expression as compared to those with a low expression. For this analysis we chose a cut-off point of ≥ 155 for positive, as this was the point on the ROC curve that gave the best discrimination between PD and CR/PR, while maintaining both a sensitivity and specificity above 70 %. Figure 5b shows that within the entire cohort high IGF-1R expression significantly associated with a shorter PFS (p = 0.0175, HR 1.7 (95 % CI, 1.1 to 2.7), which is consistent with its higher expression in patients with disease progression. Within the EGFR WT or non-exon 19 deletion or exon 21 mutation-positive population there was no difference in PFS for those with a high versus a low IGF-1R protein expression (p = 0.1394, HR 1.6 (95 % CI, 0.9 to 2.8). There was, however, a trend for a decreased PFS in the EGFR exon 19 deletion or exon 21 mutation-positive patients that were high for IGF-1R (p = 0.0856, HR 2.1 (95 % CI, 0.9 to 4.9), which is also consistent with the gefitinib response data in this subgroup.
4 Discussion
Activating mutations in the EGFR gene confer sensitivity to the EGFR-specific TKIs gefinitib and erlotinib in patients with NSCLC. Even though many patients show an initial response, they all succumb to relapse due to resistance to this treatment. The goal of this study was to explore, using multiple approaches, the hypothesis that the IGF-1R may serve as a biomarker for, and plays a role in, intrinsic resistance to the EGFR TKI gefitinib in lung cancer cells. We demonstrated that (i) the presence of high total-IGF-1R and p-IGF-1R does discriminate between cell lines that are either resistant or sensitive to gefitinib, (ii) while shRNA was effective in knocking-down IGF-1R protein expression in a gefitinib-resistant cell line, there was no increase in sensitivity to gefitinib after this knock-down, (iii) IGF-1 did not increase the proliferation rate of gefinitib-resistant or gefinitib-sensitive cell lines, and (iv) in a gefitinib treated clinical NSCLC cohort high IGF-1R protein expression was associated with PD and a shorter PFS. Collectively, this study suggests that total- and p-IGR-1R may serve as biomarkers for intrinsic resistance to the EGFR-specific TKI gefitinib in lung cancer cell lines and in NSCLC patients treated with gefitinib. These results may have significant clinical implications, as there is currently no biomarker available that can predict resistance to EGFR-specific TKIs. Known biomarkers can, however, predict responses to EGFR-specific TKIs. For example, NSCLC patients with KRAS mutations have shown poor clinical outcomes when treated with EGFR-specific TKIs and chemotherapy [33], and another study has shown that tumors with KRAS exon 2 mutations were associated with a lack of response to these TKIs [26]. Our in vitro studies suggest that high total-IGF-1R can predict resistance to gefitinib with a specificity of 76 % and a positive predictive value of 81 %, while high p-IGF-1R can predict resistance with a specificity of 100 %, a sensitivity of 41 %, a positive predictive value of 100 % and a negative predictive value of 23 %. Most importantly, the clinical data support a discriminative power within patients with EGFR mutations, i.e., high IGF-1R expression predicts gefitinib resistance even in the presence of EGFR mutations (Fig. 4). However, our current study suggests that IGF-1R signaling does not play a role as a de novo by pass mechanism for EGFR-specific TKI therapy in lung cancer cells in vitro. This notion is supported by (i) the fact that knocking down IGF-1R expression in a resistant cell line (H226, which was selected since this cell line is WT for EGFR and KRAS; Fig. 1), did not affect the resistance to gefitinib (Fig. 2) and (ii) the fact that IGF-1R activation had a similar proliferative effect both on gefitinib-sensitive and gefitinib-resistant cell lines (Fig. 3). One possible explanation for this lack of association may be that we have only examined the IGF-1R downstream signaling pathway. Thus, we cannot exclude the possibility that acquired resistance to EGFR may serve as an escape pathway form EGFR-specific TKI therapy. In addition, it is deemed necessary to screen additional gefitinib-resistant cell lines in order to definitely conclude that knocking down IGF-1R expression in a resistant cell line affects the resistance to gefitinib. Our findings stand in contrast to data available on the role of IGF-1R signaling in the acquisition of resistance to EGFR-specific TKIs. Guix et al. [23] developed a cell line model with an acquired resistance to EGFR-specific TKIs, which maintained its resistance by activating the IGR-1R pathway. Gene expression profiling of the resistant cells revealed that down-regulation of IGF-binding protein-3 (IGFBP-3) was responsible for the increase in IGF-1R activation. In addition, these authors found that treatment of cancer cell lines and mouse xenografts with both EGFR-specific TKIs and an IGF-1R monoclonal antibody prevented tumor recurrence. The latter study suggests that a combined therapy aimed at the inhibition of both the EGFR and the IGF-1R may prohibit acquired drug resistance. Using both EGFR and IGF-1R inhibitors in the clinic could potentially prolong the benefit of anti-EGFR therapy. However, a clinical trial (phase III), aimed at determining whether the addition of the IGF-1R antibody figitumumab (Pfizer Pharmaceuticals) in combination with paclitaxel plus carboplatin prolongs survival in NSCLC patients (Stage IIIB (with pleural effusion) and Stage IV or recurrent) with a non adenocarcinoma histology failed and, consequently, was terminated (ClinicalTrial no. NCT00596830). A possible explanation for this failure may be the lack of patient selection beyond histology.
Within this context, other reports have provided supportive evidence that erlotinib induces survival of NSCLC cells by inducing EGFR and IGF-1R heterodimerization, thereby stimulating IGF-1R and its downstream mediators AKT, p44/42 and MAPK. This activation, in turn, stimulates mTOR-mediated synthesis of survivin, a protein that plays a pivotal role in inhibiting apoptosis [20]. These data suggest that a combined treatment with EGFR-specific TKIs and IGF-1R inhibitors may decrease the proliferation of NSCLCs and induce its apoptosis in vitro and in vivo.
Gong et al. [17] reported that activation of IGF-1R by IGF-1 or inhibition of IGF-1R by R1507 had a minimal effect on ERK activation in either R1507-sensitive or -resistant cell lines. Thus, IGF-1R appears to be the major driving force for AKT activation in drug-sensitive (R1507) cells. The dependence of AKT on IGF-1R signaling was further underscored by a knock-down of IGF-1R using siRNAs. Consistent with these and our current results (Fig. 3), previous studies in rhabdomyosarcoma cells have shown that the phospho-AKT status is predominantly controlled via IGF-1R activation [34]. Insulin receptor substrate 1 (IRS-1) is known to mediate the activation of PI3K by associating with src homology 2 domains of the p85 subunit of PI3K upon IGF-1R pathway activation [35]. In EGFR dependent cells, the activation of the PI3K/AKT pathway is most likely controlled by ERBB3 [18]. This latter option should, however, be further investigated.
Two retrospective analyses of IGF-1R expression in NSCLCs treated with gefitinib showed that this expression was not associated with resistance to gefitinib. Cappuzzo et al. [36] evaluated IGF-1R expression in 77 NSCLCs by immunohistochemistry and failed to find any significant difference in response to gefitinib treatment between high and low IGF-1R expressing cases. Moreover, Fidler et al. [37] examined the expression of IGF-1R in 83 NSCLCs by immunohistochemistry and found no relationship between high IGF-1R expression and gefitinib resistance. The differences between these two studies and the results from our study could be explained by differences in the use of diagnostic antibodies, assessment methods and/or patient cohorts. We carried out our expression experiments in in vitro systems, while Cappuzzo et al. [36] and Fidler et al. [37] used solely clinical specimens. In addition, we measured total-IGF-1R and p-IGF-1R by immunoblotting, while the two clinical studies only used a total IGF-1R antibody for immunohistochemistry. These assays have distinct sensitivity and specificity parameters.
Our study also has some limitations. First, the expression level cut-off values for gefitinib resistant or sensitive cell lines were set based on the 23 NSCLC cell lines studied and second, the suggested predictive value of IGF-1R cannot achieve 100 % as its positive prediction value is 81. Taken together, we conclude that IGF-1R expression may serve as a biomarker to predict de novo resistance to gefitinib in NSCLC.
References
A. Jemal, R. Siegel, J. Xu, E. Ward, Cancer statistics, 2010. CA Cancer J Clin 60, 277–300 (2010)
T.S. Mok, Y.L. Wu, S. Thongprasert, C.H. Yang, D.T. Chu, N. Saijo, P. Sunpaweravong, B. Han, B. Margono, Y. Ichinose, Y. Nishiwaki, Y. Ohe, J.J. Yang, B. Chewaskulyong, H. Jiang, E.L. Duffield, C.L. Watkins, A.A. Armour, M. Fukuoka, Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361, 947–957 (2009)
Y. Kato, N. Peled, M.W. Wynes, K. Yoshida, M. Pardo, C. Mascaux, T. Ohira, M. Tsuboi, J. Matsubayashi, T. Nagao, N. Ikeda, F.R. Hirsch, Novel epidermal growth factor receptor mutation-specific antibodies for non-small cell lung cancer: immunohistochemistry as a possible screening method for epidermal growth factor receptor mutations. J Thorac Oncol 5, 1551–1558 (2010)
F.R. Hirsch, M. Varella-Garcia, P.A. Bunn Jr., W.A. Franklin, R. Dziadziuszko, N. Thatcher, A. Chang, P. Parikh, J.R. Pereira, T. Ciuleanu, J. von Pawel, C. Watkins, A. Flannery, G. Ellison, E. Donald, L. Knight, D. Parums, N. Botwood, B. Holloway, Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol 24, 5034–5042 (2006)
M. Maemondo, A. Inoue, K. Kobayashi, S. Sugawara, S. Oizumi, H. Isobe, A. Gemma, M. Harada, H. Yoshizawa, I. Kinoshita, Y. Fujita, S. Okinaga, H. Hirano, K. Yoshimori, T. Harada, T. Ogura, M. Ando, H. Miyazawa, T. Tanaka, Y. Saijo, K. Hagiwara, S. Morita, T. Nukiwa, Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362, 2380–2388 (2010)
R. Rosell, T. Moran, C. Queralt, R. Porta, F. Cardenal, C. Camps, M. Majem, G. Lopez-Vivanco, D. Isla, M. Provencio, A. Insa, B. Massuti, J.L. Gonzalez-Larriba, L. Paz-Ares, I. Bover, R. Garcia-Campelo, M.A. Moreno, S. Catot, C. Rolfo, N. Reguart, R. Palmero, J.M. Sanchez, R. Bastus, C. Mayo, J. Bertran-Alamillo, M.A. Molina, J.J. Sanchez, M. Taron, Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 361, 958–967 (2009)
K. Tamura, I. Okamoto, T. Kashii, S. Negoro, T. Hirashima, S. Kudoh, Y. Ichinose, N. Ebi, K. Shibata, T. Nishimura, N. Katakami, T. Sawa, E. Shimizu, J. Fukuoka, T. Satoh, M. Fukuoka, Multicentre prospective phase II trial of gefitinib for advanced non-small cell lung cancer with epidermal growth factor receptor mutations: results of the West Japan Thoracic Oncology Group trial (WJTOG0403). Br J Cancer 98, 907–914 (2008)
S. Kobayashi, T.J. Boggon, T. Dayaram, P.A. Janne, O. Kocher, M. Meyerson, B.E. Johnson, M.J. Eck, D.G. Tenen, B. Halmos, EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352, 786–792 (2005)
W. Pao, V.A. Miller, K.A. Politi, G.J. Riely, R. Somwar, M.F. Zakowski, M.G. Kris, H. Varmus, Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS medicine 2, e73 (2005)
J.A. Engelman, P.A. Janne, Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 14, 2895–2899 (2008)
P.C. Ma, R. Jagadeeswaran, S. Jagadeesh, M.S. Tretiakova, V. Nallasura, E.A. Fox, M. Hansen, E. Schaefer, K. Naoki, A. Lader, W. Richards, D. Sugarbaker, A.N. Husain, J.G. Christensen, R. Salgia, Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res 65, 1479–1488 (2005)
G. Maulik, T. Kijima, P.C. Ma, S.K. Ghosh, J. Lin, G.I. Shapiro, E. Schaefer, E. Tibaldi, B.E. Johnson, R. Salgia, Modulation of the c-Met/hepatocyte growth factor pathway in small cell lung cancer. Clin Cancer Res 8, 620–627 (2002)
F. Cappuzzo, P.A. Janne, M. Skokan, G. Finocchiaro, E. Rossi, C. Ligorio, P.A. Zucali, L. Terracciano, L. Toschi, M. Roncalli, A. Destro, M. Incarbone, M. Alloisio, A. Santoro, M. Varella-Garcia, MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol 20, 298–304 (2009)
J.A. Engelman, K. Zejnullahu, T. Mitsudomi, Y. Song, C. Hyland, J.O. Park, N. Lindeman, C.M. Gale, X. Zhao, J. Christensen, T. Kosaka, A.J. Holmes, A.M. Rogers, F. Cappuzzo, T. Mok, C. Lee, B.E. Johnson, L.C. Cantley, P.A. Janne, MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 (2007)
M. Xie, L. Zhang, C.S. He, F. Xu, J.L. Liu, Z.H. Hu, L.P. Zhao, Y. Tian, Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J Cell Biochem 113, 1501–1513 (2012)
A. Chakravarti, J.S. Loeffler, N.J. Dyson, Insulin-like growth factor receptor I mediates resistance to anti-epidermal growth factor receptor therapy in primary human glioblastoma cells through continued activation of phosphoinositide 3-kinase signaling. Cancer Res 62, 200–207 (2002)
Y. Gong, E. Yao, R. Shen, A. Goel, M. Arcila, J. Teruya-Feldstein, M.F. Zakowski, S. Frankel, M. Peifer, R.K. Thomas, M. Ladanyi, W. Pao, High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507). PLoS One 4, e7273 (2009)
A. Hurbin, L. Dubrez, J.L. Coll, M.C. Favrot, Inhibition of apoptosis by amphiregulin via an insulin-like growth factor-1 receptor-dependent pathway in non-small cell lung cancer cell lines. J Biol Chem 277, 49127–49133 (2002)
A. Gualberto, M. Pollak, Emerging role of insulin-like growth factor receptor inhibitors in oncology: early clinical trial results and future directions. Oncogene 28, 3009–3021 (2009)
F. Morgillo, J.K. Woo, E.S. Kim, W.K. Hong, H.Y. Lee, Heterodimerization of insulin-like growth factor receptor/epidermal growth factor receptor and induction of survivin expression counteract the antitumor action of erlotinib. Cancer Res 66, 10100–10111 (2006)
V. Ludovini, G. Bellezza, L. Pistola, F. Bianconi, L. Di Carlo, A. Sidoni, A. Semeraro, R. Del Sordo, F.R. Tofanetti, M.G. Mameli, G. Daddi, A. Cavaliere, M. Tonato, L. Crino, High coexpression of both insulin-like growth factor receptor-1 (IGFR-1) and epidermal growth factor receptor (EGFR) is associated with shorter disease-free survival in resected non-small-cell lung cancer patients. Ann Oncol 20, 842–849 (2009)
R. Dziadziuszko, D.T. Merrick, S.E. Witta, A.D. Mendoza, B. Szostakiewicz, A. Szymanowska, W. Rzyman, K. Dziadziuszko, J. Jassem, P.A. Bunn Jr., M. Varella-Garcia, F.R. Hirsch, Insulin-like growth factor receptor 1 (IGF1R) gene copy number is associated with survival in operable non-small-cell lung cancer: a comparison between IGF1R fluorescent in situ hybridization, protein expression, and mRNA expression. J Clin Oncol 28, 2174–2180 (2010)
M. Guix, A.C. Faber, S.E. Wang, M.G. Olivares, Y. Song, S. Qu, C. Rinehart, B. Seidel, D. Yee, C.L. Arteaga, J.A. Engelman, Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest 118, 2609–2619 (2008)
C.Q. Zhu, G. da Cunha Santos, K. Ding, A. Sakurada, J.C. Cutz, N. Liu, T. Zhang, P. Marrano, M. Whitehead, J.A. Squire, S. Kamel-Reid, L. Seymour, F.A. Shepherd, M.S. Tsao, Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol 26, 4268–4275 (2008)
V.A. Miller, G.J. Riely, M.F. Zakowski, A.R. Li, J.D. Patel, R.T. Heelan, M.G. Kris, A.B. Sandler, D.P. Carbone, A. Tsao, R.S. Herbst, G. Heller, M. Ladanyi, W. Pao, D.H. Johnson, Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol 26, 1472–1478 (2008)
W. Pao, T.Y. Wang, G.J. Riely, V.A. Miller, Q. Pan, M. Ladanyi, M.F. Zakowski, R.T. Heelan, M.G. Kris, H.E. Varmus, KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS medicine 2, e17 (2005)
N.T. W. Brugger, M. Blasinska-Morawiec, S. Curescu, R. Sakalauskas, G. Manikhas, J. Mazieres, R. Whittom, K. Rohr, F. Cappuzzo, on behalf of the SATURN Investigators, Biomarker analyses from the phase III placebo-controlled SATURN study of maintenance erlotinib following first-line chemotherapy for advanced NSCLC. ASCO 2009 (2009)
P.A. Zucali, M.G. Ruiz, E. Giovannetti, A. Destro, M. Varella-Garcia, K. Floor, G.L. Ceresoli, J.A. Rodriguez, I. Garassino, P. Comoglio, M. Roncalli, A. Santoro, G. Giaccone, Role of cMET expression in non-small-cell lung cancer patients treated with EGFR tyrosine kinase inhibitors. Ann Oncol 19, 1605–1612 (2008)
C. Mascaux, M.W. Wynes, Y. Kato, C. Tran, B.R. Asuncion, J.M. Zhao, M. Gustavson, J. Ranger-Moore, F. Gaire, J. Matsubayashi, T. Nagao, K. Yoshida, T. Ohira, N. Ikeda, F.R. Hirsch, EGFR protein expression in non-small cell lung cancer predicts response to an EGFR tyrosine kinase inhibitor–a novel antibody for immunohistochemistry or AQUA technology. Clin Cancer Res 17, 7796–7807 (2011)
W.D. Travis, E. Brambilla, H.K. Muller-Hermerlink, C.C. Harris, World Health Organization Classification of Tumors. Pathology and Genetics. Tumors of the Lung, Pleura, Thymus and Heart (IARC Press, Lyon, 2004)
L. Sobin, M. Gospodarowicz and C. Wittekind, TNM classification of malignant tumors, 7th ed, Wiley-Blackwell, (2009)
E.A. Eisenhauer, P. Therasse, J. Bogaerts, L.H. Schwartz, D. Sargent, R. Ford, J. Dancey, S. Arbuck, S. Gwyther, M. Mooney, L. Rubinstein, L. Shankar, L. Dodd, R. Kaplan, D. Lacombe, J. Verweij, New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45, 228–247 (2009)
D.A. Eberhard, B.E. Johnson, L.C. Amler, A.D. Goddard, S.L. Heldens, R.S. Herbst, W.L. Ince, P.A. Janne, T. Januario, D.H. Johnson, P. Klein, V.A. Miller, M.A. Ostland, D.A. Ramies, D. Sebisanovic, J.A. Stinson, Y.R. Zhang, S. Seshagiri, K.J. Hillan, Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 23, 5900–5909 (2005)
L. Cao, Y. Yu, I. Darko, D. Currier, L.H. Mayeenuddin, X. Wan, C. Khanna, L.J. Helman, Addiction to elevated insulin-like growth factor I receptor and initial modulation of the AKT pathway define the responsiveness of rhabdomyosarcoma to the targeting antibody. Cancer Res 68, 8039–8048 (2008)
M.G. Myers Jr., J.M. Backer, X.J. Sun, S. Shoelson, P. Hu, J. Schlessinger, M. Yoakim, B. Schaffhausen, M.F. White, IRS-1 activates phosphatidylinositol 3′-kinase by associating with src homology 2 domains of p85. Proc Natl Acad Sci USA 89, 10350–10354 (1992)
F. Cappuzzo, L. Toschi, G. Tallini, G.L. Ceresoli, I. Domenichini, S. Bartolini, G. Finocchiaro, E. Magrini, G. Metro, A. Cancellieri, R. Trisolini, L. Crino, P.A. Bunn Jr., A. Santoro, W.A. Franklin, M. Varella-Garcia, F.R. Hirsch, Insulin-like growth factor receptor 1 (IGFR-1) is significantly associated with longer survival in non-small-cell lung cancer patients treated with gefitinib. Ann Oncol 17, 1120–1127 (2006)
M.J. Fidler, S. Basu, L. Buckingham, K. Walters, S. McCormack, M. Batus, J. IV Coon, P. Bonomi, Utility of insulin-like growth factor receptor-1 expression in gefitinib-treated patients with non-small cell lung cancer. Anticancer Res 32, 1705–1710 (2012)
Acknowledgements
This research was funded by the NCI/SPORE in Lung Cancer CA580187 (F.H.), the International Association for the Study of Lung Cancer (N.P.) and the Fulbright Foundation (N.P.).
Disclosure
F.H: Consultant/Advisory Boards: Astra Zeneca, Roche, Lilly, Pfizer, Boehringer-Ingelheim, Merck Serono, Ventana-Roche, Glasxo Smith Kline, BMS/Imclone; Research Funding: OSI, Genentech, AstraZeneca, Merck (USA), Syndax, Ventana-Roche; Patent: EGFR FISH as a predictive marker for EGFR Inhibitors.
N.P., M.W., N.I., T.O., K.Y., M.I., J.Q., R.B., Y.K., C.M.: nothing to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Peled, N., Wynes, M.W., Ikeda, N. et al. Insulin-like growth factor-1 receptor (IGF-1R) as a biomarker for resistance to the tyrosine kinase inhibitor gefitinib in non-small cell lung cancer. Cell Oncol. 36, 277–288 (2013). https://doi.org/10.1007/s13402-013-0133-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-013-0133-9