Abstract
Polymorphisms in the coding region of the prion protein gene (PRNP) have been associated with the susceptibility and incubation period of prion diseases in humans and sheep. However, polymorphisms in this part of the bovine PRNP gene do not affect the classical bovine spongiform encephalopathy (BSE) susceptibility in cattle. Studies carried out in Germany have shown that insertion/deletion-type polymorphisms located in the promoter region of the bovine prion gene are possible genetic factors modulating BSE susceptibility by changing the level of PRNP expression. No such association was observed for atypical BSE cases; however, due to the rare nature of the disease, these results should be confirmed. Additionally, a single nonsynonymous mutation in PRNP codon 211 (E211K) was described in one H-type BSE case in the USA; however, it was not found in any other cases. Here, we performed genetic characterization of PRNP promoter indel variations and determined the polymorphism of open reading frames (ORFs) of PRNP and bovine prion-like Shadoo (SPRN) genes in six Polish atypical BSE cases and compared these results to the population of clinically healthy Polish Holstein cattle. No potentially pathogenic mutations were found in the PRNP ORF in atypical BSE-affected cattle, but our study showed a high frequency of deletions at the indel loci of PRNP promoter in these animals. Additionally, a rare sequence variation in the SPRN protein-coding sequence was found in one L-type atypical BSE-affected animal.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bovine spongiform encephalopathy (BSE) is a transmissible neurodegenerative disease of cattle, caused by the accumulation of partially protease resistant, pathogenic, misfolded cellular prion protein (PrPC) in the central nervous system (Prusiner 1998; Wilesmith et al. 1992). In the last few years, distinct from classical BSE, an atypical molecular and neuropathological variant of BSE was identified. The atypical BSE (BASE; bovine amyloidotic spongiform encephalopathy) is characterized by an unusual pattern of deposition and brain regional distribution of protease-resistant prion protein (PrPres) and by different PrPres banding patterns observed in Western blot analysis (Casalone et al. 2004; Biacabe et al. 2004). Later studies showed the existence of two atypical BSE variants, which, in comparison with classical BSE, showed molecular profiles of PrPres with a protease-resistant core of lower (L-type) or higher (H-type) molecular mass (Biacabe et al. 2004; Casalone et al. 2004; Polak et al. 2004; Buschmann et al. 2006). Most of the studies concerning atypical BSE suggest its sporadic or age-dependent etiology, except for a single American case of H-type BSE, which was shown to be connected with E211K mutation in the PrP sequence (Richt and Hall 2008).
In studies on classical BSE, the association with BSE susceptibility of two indel polymorphisms in the non-coding part of the prion protein gene (PRNP) was first described in a small case–control study (Sander et al. 2004). Deletion alleles of 23-bp indel polymorphism (in the region upstream of exon 1) and 12-bp indel (in intron 1) were significantly overrepresented in BSE-affected cattle compared to control animals. Because the deletion (del) of 23 bp removes the consensus binding site for RP-58 and the deletion of 12 bp removes the binding site for transcription factor Sp1, the authors assumed that different promoter haplotypes may cause differences in PRNP gene expression. Further in vitro studies showed that the promoter with the 23-bp del–12-bp del haplotype gives a higher level of reporter gene expression (Sander et al. 2005). In transgenic mice, higher expression of PRNP results in a shorter disease incubation period after BSE inoculation (Vilotte et al. 2001).
Studies on atypical BSE performed by Brunelle et al. (2007) showed no association between PRNP promoter genotypes and susceptibility of cattle to atypical BSE or other experimentally inoculated TSEs (transmissible spongiform encephalopathies). However, a low number of atypical BSE cases studied worldwide does not allow for reliable association analysis and more results are needed in order to better understand the etiology or putative genetic background of the disease.
Some studies involving association analysis or QTL mapping suggest that the genetic resistance/susceptibility of cattle to BSE may be modulated by genome regions other than the PRNP locus (Hernández-Sánchez et al. 2002; Zhang et al. 2004; Murdoch et al. 2010). Also, the involvement of protein molecules other than PrPC in prion deposit formation was presumed (Kaneko et al. 1997). One of the proposed candidate genes/proteins is SPRN gene encoding Shadoo protein (shadow of prion protein, Sho). SPRN is a prion gene paralog (prion-like gene) localized on bovine chromosome 26. The protein product of this gene (Sho) shares several structural and biochemical features with PrP, and it was predicted to be an extracellular, N-glycosylated, and glycosylphosphatidylinositol-anchored molecule (Mo et al. 2001; Premzl et al. 2003). Sho shows prion-like neuroprotective activity and participates in nervous system development, suggesting that its physiological function may be closely related with that of PrP (Watts et al. 2007). The most homologous region of Sho and PrP is a hydrophobic alanine-rich sequence. This sequence in the prion protein was proved to be an indispensable factor for PrPC–PrPSc interactions (Norstrom and Mastrianni 2005). In the brains of mammals, the expression of SPRN showed overlapping with the PRNP profile and some studies suggested the co-regulation of activity of both genes (Premzl et al. 2003; Uboldi et al. 2006; Lampo et al. 2007; Premzl and Gamulin 2007). The potential role of Sho in TSE pathogenesis was first suggested by Watts et al. (2007), who showed a dramatic reduction of endogenous protein in the brains of mice inoculated with mice-adapted scrapie strain. The subsequent results showed that the variations in the SPRN open reading frame (ORF) may be a potential risk factor for vCJD (variant Creutzfeldt–Jakob disease) and sporadic CJD in humans (Beck et al. 2008) and for classical scrapie in sheep (Lampo et al. 2010). All these data suggest the potential role of Sho in TSE development or in prion biology and encourage further studies.
In this study, we investigated the protein-coding sequences of PRNP and SPRN genes, as well as PRNP promoter and intron 1 indel polymorphisms in six atypical BSE cases (five L-type and one H-type) and in a control group composed of clinically healthy cattle of the Polish Holstein-Friesian breed. According to our knowledge, the SPRN sequence of atypical BSE cases has not been analyzed to date, so this is probably the first study describing this issue.
Materials and methods
Genomic DNA was isolated from the brain stems of six atypical BSE-affected cattle (five L-type and one H-type) of the Polish Holstein-Friesian breed, using the Wizard® Genomic DNA Purification Kit (Promega). Brain stem samples were initially tested with an approved rapid test and confirmed as BSE-positive using Western blot and immunohistochemistry. The studied atypical BSE cases accounted for 50 % of all atypical BSE cases identified in Poland and for 10 % of all atypical BSE cases identified worldwide (April, 2012).
Genomic DNA from the whole blood of 105 clinically healthy animals of the Polish Holstein-Friesian breed was isolated using the Wizard® Genomic DNA Purification Kit (Promega). Analysis of DNA fragments spanning PRNP indel polymorphisms and whole PRNP ORFs was performed as described earlier by Gurgul et al. (2012). The amplification of SPRN gene fragments containing the ORF and 12-bp indel was performed with the primers shown in Table 1. The polymerase chain reaction (PCR) cycling conditions and reaction mixtures used are shown in Tables 2 and 3.
The ORFs of the studied genes were sequenced from both complementary strands using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Prior to sequencing, PCR products were purified with ExoSAP-IT enzyme mixture (USB Corporation). Sequencing products were purified using the BigDye XTerminator Purification Kit (Applied Biosystems) and sequence analysis was made using a 3130xl Genetic Analyzer (Applied Biosystems).
Differences in the distribution of separate markers between groups of diseased and control cattle were studied using Fisher’s exact test in contingency tables. This test allows the analysis of a small number of samples with no limitations regarding the effective sample size.
Results and discussion
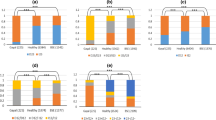
PRNP promoter (23-bp) and intron 1 (12-bp) indel variations have been found to be associated with classical BSE susceptibility in cattle and the deletion variants of both polymorphisms were related to increased susceptibility, whereas insertion variants were recognized as being protective (Sander et al. 2004; Haase et al. 2007; Gurgul et al. 2012). These polymorphisms were also connected with different levels of PRNP expression using various experimental approaches (Sander et al. 2005; Kashkevich et al. 2007; Msalya et al. 2010). No such proof was found for a few (seven H-type and two L-type) atypical BSE-affected French and US cattle, in which deletion allele frequencies were similar to those described for healthy animals (Brunelle et al. 2007). However, in four studied French and Polish H-type atypical BSE cases, the expression level of PRNP in brain stem samples was significantly elevated (p = 0.001) compared to control cattle (Larska et al. 2010). In this study, we investigated six atypical BSE cases (five L-type and one H-type) diagnosed in Polish cattle. All these animals were homozygous for deletion allele at the locus of the 23-bp indel polymorphism. A high frequency of deletion was also observed at the locus of the 12-bp indel polymorphism and only one L-type case was homozygous for the insertion allele. The frequencies of alleles and genotypes at both loci differed significantly (p > 0.05) between animals with atypical BSE and healthy cattle (Tables 4 and 5) and, generally, deletion gene variants were overrepresented in diseased animals. Similar findings were reported earlier for Polish cattle with classical BSE (Gurgul et al. 2012). The results obtained for atypical BSE cases were statistically significant, even after Bonferroni correction for multiple testing (except for the allele frequency of the 12-bp polymorphism), which suggests a potential role of both polymorphic sites in atypical BSE susceptibility. The low number of studied atypical BSE cases does not allow for reliable association analysis; however, our results show that previously published data suggesting no association of the PRNP indel polymorphism and atypical BSE susceptibility should be confirmed with more atypical BSE cases and in animals of different breeds.
The whole sequence of the PRNP ORF in atypical BSE-affected animals was concordant with those previously published for healthy animals. The only polymorphism observed in this region in the diseased animals was the 24-bp insertion/deletion occurring in the sequence encoding octapeptide repeats in the N-terminal part of PrP. One L-type atypical BSE case was homozygous for the 5-repeat (5/5) allele and the other animals were homozygous for the 6-repeat allele. Although the 5/5 genotype is rare and was not observed in some populations studied (Premzl et al. 2000; Leone et al. 2002), it was found with relatively high frequency in healthy Polish Holstein cattle (Walawski and Czarnik 2003). In previous studies, no association of octapeptide repeat polymorphism and classical BSE incidence was found (Hunter et al. 1994); however, an allele containing five octapeptide repeats was associated with increased stability of prion protein in transgenic mice expressing bovine PrP (Brun et al. 2007). In accordance with this, no potentially pathogenic effect of the 5/5 genotype can be assumed. Additionally, the mutation E211K described by Richt and Hall (2008) was not detected in any Polish animals with H- or L-type atypical BSE.
In the SPRN protein-coding sequence in atypical BSE-affected and in healthy animals, five single-nucleotide polymorphisms (SNPs) and one indel were identified (Table 6). Of the SNPs identified, three (110G>C, 125C>T, and 128G>A) were nonsynonymous nucleotide substitutions and only two silent SNPs (288A>G and 360G>A) were previously described by Stewart et al. (2009). Of the newly observed SNPs in bovine (Bos taurus) SPRN gene, two (125C>T and 128G>A) were previously described in homologous sequences obtained from the B. grunniens species (yak) (Stewart et al. 2009). The allele and genotype frequencies of the identified SNPs did not differ significantly between atypical BSE and control animals (p > 0.05) and only common polymorphisms (SNP 110G>C, 288A>G, and 360G>A) were observed in both diseased and control animals (Table 6).
The deletion of 12 bp in the coding region of SPRN (nucleotides 201 to 212) observed by Stewart et al. (2009) in domestic cattle was identified in one L-type atypical BSE-affected animal in a heterozygous state. This mutation causes the indel of one (aa 67–70) of five tetra-amino-acid repeats in the repetitive alanine-rich sequence of bovine Sho. The repetitive region spans amino acids from 59 to 78 in bovine Sho and forms the hydrophobic region of the protein. A homologous, hydrophobic sequence was also found in the N-terminal part of the prion protein, and it was shown to be crucial for PrPC–PrPSc interaction (Norstrom and Mastrianni 2005). A special role is played in this sequence by palindrome sequence AGAAAAGA, which alone shows strong neurotoxic activity (Tagliavini et al. 2001). In bovine Sho, a similar peptide with the AGAAAGA motif was identified. The deletion of 12 bp, observed here, occurs in the region encoding this peptide, but due to the repetitive nature of the region, the AGAAAGA sequence is not being deleted from the Sho sequence (Fig. 1). The deletion of 12 bp was identified only in one atypical BSE-affected animal and none of 100 healthy animals studied carried the deletion gene variant. To test whether this mutation is a natural rare sequence variation in the Polish cattle population, 112 other randomly selected animals were tested (with the use of the PCR test). In general, 212 samples from healthy animals (424 genes) have been tested and no deletion allele of a SPRN 12-bp indel has been found. This mutation could be considered as potentially pathogenic; however, previous studies showing similar gene variants in some healthy animals of B. taurus (cattle), B. gaurus (gaur) (EU605794.1), and Bison bison (American bison) (HM179105.1, HM179104.1) do not allow for such a conclusion.
Conclusions
Here, we found a significant association of two PRNP promoter and intron 1 indel variations and atypical BSE susceptibility in Polish Holstein-Friesian cattle. These results are inconsistent with previously published studies of atypical BSE from France and the USA (Richt and Hall 2008) and encourage further studies comprising a greater number of diseased animals. Also, a rare variant of the SPRN ORF sequence was identified in one of the studied atypical BSE-affected animals. This gene variant (12-bp deletion) was not observed in 212 clinically healthy animals of the control group. The same variant was previously described in other animals of the Bovidae family but, however, not in association with the possible susceptibility to atypical BSE. This may suggest its natural occurrence in some populations, rather than its pathogenic effect.
References
Beck JA, Campbell TA, Adamson G, Poulter M, Uphill JB, Molou E, Collinge J, Mead S (2008) Association of a null allele of SPRN with variant Creutzfeldt–Jakob disease. J Med Genet 45:813–817
Biacabe AG, Laplanche JL, Ryder S, Baron T (2004) Distinct molecular phenotypes in bovine prion diseases. EMBO Rep 5:110–115
Brun A, Gutiérrez-Adán A, Castilla J, Pintado B, Díaz-San Segundo F, Cano MJ, Alamillo E, Espinosa JC, Torres JM (2007) Reduced susceptibility to bovine spongiform encephalopathy prions in transgenic mice expressing a bovine PrP with five octapeptide repeats. J Gen Virol 88:1842–1849
Brunelle BW, Hamir AN, Baron T, Biacabe AG, Richt JA, Kunkle RA, Cutlip RC, Miller JM, Nicholson EM (2007) Polymorphisms of the prion gene promoter region that influence classical BSE susceptibility are not applicable to other transmissible spongiform encephalopathies in cattle. J Anim Sci 85:3142–3147
Buschmann A, Gretzschel A, Biacabe AG, Schiebel K, Corona C, Hoffmann C, Eiden M, Baron T, Casalone C, Groschup MH (2006) Atypical BSE in Germany—proof of transmissibility and biochemical characterization. Vet Microbiol 117:103–116
Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, Monaco S, Caramelli M (2004) Identification of a second bovine amyloidotic spongiform encephalopathy: molecular similarities with sporadic Creutzfeldt–Jakob disease. Proc Natl Acad Sci USA 101:3065–3070
Gurgul A, Czarnik U, Larska M, Polak MP, Strychalski J, Słota E (2012) Polymorphism of the prion protein gene (PRNP) in Polish cattle affected by classical bovine spongiform encephalopathy. Mol Biol Rep 39:5211–5217. doi:10.1007/s11033-011-1318-9
Haase B, Doherr MG, Seuberlich T, Drögemüller C, Dolf G, Nicken P, Schiebel K, Ziegler U, Groschup MH, Zurbriggen A, Leeb T (2007) PRNP promoter polymorphisms are associated with BSE susceptibility in Swiss and German cattle. BMC Genet 8:15
Hernández-Sánchez J, Waddington D, Wiener P, Haley CS, Williams JL (2002) Genome-wide search for markers associated with bovine spongiform encephalopathy. Mamm Genome 13:164–168
Hunter N, Goldmann W, Smith G, Hope J (1994) Frequencies of PrP gene variants in healthy cattle and cattle with BSE in Scotland. Vet Rec 135:400–403
Kaneko K, Zulianello L, Scott M, Cooper CM, Wallace AC, James TL, Cohen FE, Prusiner SB (1997) Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc Natl Acad Sci USA 94:10069–10074
Kashkevich K, Humeny A, Ziegler U, Groschup MH, Nicken P, Leeb T, Fischer C, Becker CM, Schiebel K (2007) Functional relevance of DNA polymorphisms within the promoter region of the prion protein gene and their association to BSE infection. FASEB J 21:1547–1555
Lampo E, Van Poucke M, Hugot K, Hayes H, Van Zeveren A, Peelman LJ (2007) Characterization of the genomic region containing the Shadow of Prion Protein (SPRN) gene in sheep. BMC Genomics 8:138
Lampo E, Duchateau L, Schepens B, Van Poucke M, Saelens X, Erkens T, Van Zeveren A, Peelman LJ (2010) Identification of polymorphisms in the ovine Shadow of prion protein (SPRN) gene and assessment of their effect on promoter activity and susceptibility for classical scrapie. Anim Genet 41:169–178
Larska M, Polak MP, Zmudzinski JF, Torres JM (2010) Comparison of mRNA expression levels of selected genes in the brain stem of cattle naturally infected with classical and atypical BSE. Brain Res 1351:13–22
Leone P, Castiglioni B, Sechi T, Cassini P, Stella A (2002) Prion gene octarepeat variability in the Italian cattle breeds. In: Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, August 19–21 2002, communication no. 13-40
Mo H, Moore RC, Cohen FE, Westaway D, Prusiner SB, Wright PE, Dyson HJ (2001) Two different neurodegenerative diseases caused by proteins with similar structures. Proc Natl Acad Sci USA 98:2352–2357
Msalya G, Shimogiri T, Nishitani K, Okamoto S, Kawabe K, Minesawa M, Maeda Y (2010) Indels within promoter and intron 1 of bovine prion protein gene modulate the gene expression levels in the medulla oblongata of two Japanese cattle breeds. Anim Genet 41:218–221
Murdoch BM, Clawson ML, Laegreid WW, Stothard P, Settles M, McKay S, Prasad A, Wang Z, Moore SS, Williams JL (2010) A 2cM genome-wide scan of European Holstein cattle affected by classical BSE. BMC Genet 11:20
Norstrom EM, Mastrianni JA (2005) The AGAAAAGA palindrome in PrP is required to generate a productive PrPSc–PrPC complex that leads to prion propagation. J Biol Chem 280:27236–27243
Polak MP, Rożek W, Rola J, Żmudziński JF (2004) Prion protein glycoforms from BSE cases in Poland. Bull Vet Inst Pulawy 48:201–205
Premzl M, Gamulin V (2007) Comparative genomic analysis of prion genes. BMC Genomics 8:1
Premzl M, Bozic P, Gamulin V (2000) PRNP octarepeat allele genotype frequencies among the modern and rare cattle breeds in Croatia. Anim Genet 31:408–409
Premzl M, Sangiorgio L, Strumbo B, Marshall Graves JA, Simonic T, Gready JE (2003) Shadoo, a new protein highly conserved from fish to mammals and with similarity to prion protein. Gene 314:89–102
Prusiner SB (1998) Prions. Proc Natl Acad Sci USA 95:13363–13383
Richt JA, Hall SM (2008) BSE case associated with prion protein gene mutation. PLoS Pathog 4:e1000156
Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, Groschup MH, Ziegler U, Distl O, Leeb T (2004) Analysis of sequence variability of the bovine prion protein gene (PRNP) in German cattle breeds. Neurogenetics 5:19–25
Sander P, Hamann H, Drögemüller C, Kashkevich K, Schiebel K, Leeb T (2005) Bovine prion protein gene (PRNP) promoter polymorphisms modulate PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem 280:37408–37414
Stewart P, Shen C, Zhao D, Goldmann W (2009) Genetic analysis of the SPRN gene in ruminants reveals polymorphisms in the alanine-rich segment of shadoo protein. J Gen Virol 90:2575–2580
Tagliavini F, Forloni G, D’Ursi P, Bugiani O, Salmona M (2001) Studies on peptide fragments of prion proteins. Adv Protein Chem 57:171–201
Uboldi C, Paulis M, Guidi E, Bertoni A, Meo GP, Perucatti A, Iannuzzi L, Raimondi E, Brunner RM, Eggen A, Ferretti L (2006) Cloning of the bovine prion-like Shadoo (SPRN) gene by comparative analysis of the predicted genomic locus. Mamm Genome 17:1130–1139
Vilotte JL, Soulier S, Essalmani R, Stinnakre MG, Vaiman D, Lepourry L, Da Silva JC, Besnard N, Dawson M, Buschmann A, Groschup M, Petit S, Madelaine MF, Rakatobe S, Le Dur A, Vilette D, Laude H (2001) Markedly increased susceptibility to natural sheep scrapie of transgenic mice expressing ovine PrP. J Virol 75:5977–5984
Walawski K, Czarnik U (2003) Prion octapeptide-repeat polymorphism in Polish Black-and-White cattle. J Appl Genet 44:191–195
Watts JC, Drisaldi B, Ng V, Yang J, Strome B, Horne P, Sy MS, Yoong L, Young R, Mastrangelo P, Bergeron C, Fraser PE, Carlson GA, Mount HT, Schmitt-Ulms G, Westaway D (2007) The CNS glycoprotein Shadoo has PrPC-like protective properties and displays reduced levels in prion infections. EMBO J 26:4038–4050
Wilesmith JW, Ryan JB, Hueston WD (1992) Bovine spongiform encephalopathy: case–control studies of calf feeding practices and meat and bonemeal inclusion in proprietary concentrates. Res Vet Sci 52:325–331
Zhang C, De Koning DJ, Hernández-Sánchez J, Haley CS, Williams JL, Wiener P (2004) Mapping of multiple quantitative trait loci affecting bovine spongiform encephalopathy. Genetics 167:1863–1872
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gurgul, A., Polak, M.P., Larska, M. et al. PRNP and SPRN genes polymorphism in atypical bovine spongiform encephalopathy cases diagnosed in Polish cattle. J Appl Genetics 53, 337–342 (2012). https://doi.org/10.1007/s13353-012-0102-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-012-0102-4