Abstract
Recent attempts to discover genetic factors affecting cattle resistance/susceptibility to bovine spongiform encephalopathy (BSE) have led to the identification of two insertion/deletion (indel) polymorphisms, located within the promoter and intron 1 of the prion protein gene PRNP, showing a significant association with the occurrence of classical form of the disease. Because the effect of the polymorphisms was studied only in few populations, in this study we investigated whether previously described association of PRNP indel polymorphisms with BSE susceptibility in cattle is also present in Polish cattle population. We found a significant relation between the investigated PRNP indel polymorphisms (23 and 12 bp indels), and susceptibility of Polish Holstein-Friesian cattle to classical BSE (P < 0.05). The deletion variants of both polymorphisms were related to increased susceptibility, whereas insertion variants were protective against BSE.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bovine spongiform encephalopathy (BSE) is a fatal, transmissible, neurodegenerative disease caused by the accumulation of partially protease-resistant, pathogenic prion protein (PrPSc) in the central nervous system. PrPSc is formed in the process of abnormal folding of cellular prion protein (PrPC) and the mechanism underlying this phenomenon has been proposed by Stanley B. Prusiner [1]. The accumulation of the misfolded protein in brains of infected cattle leads to the spongiosis, gliosis, and neuron death without inflammatory lesions [2].
The secondary and tertiary structure of proteins depends mainly on their amino acid sequences, which may be affected by mutations in the corresponding genes. PrPC is encoded by autosomal PRNP gene, which is expressed in most of the tissues [3]. A significant relationship between nucleic acid variations in PRNP protein-coding sequence and the occurrence of acquired or genetic prion diseases have been found in humans [4] and sheep [5]. The situation is different in cattle, in which polymorphisms identified within the open reading frame (ORF) do not affect the BSE susceptibility [6, 7]. Despite the large number of polymorphisms found in bovine PRNP locus [8], so far only two insertion/deletion (indel) variations in the promoter region (23 and 12 bp indel) have been associated with BSE susceptibility [9–12]. By the modification of consensus binding sites for transcription factors RP58 and Sp1, these polymorphisms affect expression level of prion protein gene, [13] and thus are considered as variations modulating the BSE incubation period.
Due to the restrictions in handling procedures for infectious material, studies showing association of PRNP indel polymorphism with BSE susceptibility in cattle are still relatively sparse. Furthermore, in some subject breeds/populations the effect of the indel polymorphisms on BSE susceptibility has not been observed [12, 14]. This encourages further studies on affected animals in order to extend the previously published results and to verify potential population-specific effect of the PRNP indel polymorphisms.
In this study, we investigated the frequency of PRNP indel polymorphisms in classical BSE-affected and in clinically healthy Polish Holstein-Friesian cattle. BSE cases were also characterized based on polymorphism of PRNP open reading frame. Moreover, PRNP indel polymorphisms were characterized in different Polish cattle breeds and the results were compared with worldwide studies in different breeds.
Materials and methods
Samples obtained from 28 BSE-affected Polish black-and-white Holstein-Friesian cattle (HO) were examined. The studied BSE cases were diagnosed between 2002 and 2010 and represented the classical form of the disease.
Initially, the brain stem tissue of prion-infected cattle was tested with rapid tests approved by European Commission for monitoring purposes. Then, every positive result was confirmed using either histopathology or immunohistochemistry and immunoblotting techniques.
Genomic DNA was isolated from brain stem of BSE-affected cattle and from the whole blood sample of randomly selected clinically healthy cattle of following breeds: HO (n = 651), Polish red-and-white Holstein-Friesian (RW; n = 76), Polish red (RP; n = 104), and Simmental (SM; n = 73). The isolation was performed with The Wizard® Genomic DNA purification kit (Promega) or MasterPure DNA purification kit (Epicentre Biotechnologies, USA). The genotypic data for part of the HO group (n = 450) were previously published by Czarnik et al. [15].
PRNP indel polymorphisms were analyzed using PCR method. Separate alleles were recognized based on the length of the amplified DNA fragments after electrophoresis in 2% agarose gel. PCR products from randomly selected samples from homozygous animals were sequenced to confirm accuracy of genotyping. The whole PRNP ORF was amplified using HotStarTaq polymerase (Qiagen). All used PCR primers, PCR mixtures, amplification thermal conditions as well as lengths of the amplicons are shown in Supplementary Tables 1, 2, and 3 (online resources).
ORFs of the PRNP gene as well as regions spanning indel sites of PRNP gene in chosen animals were sequenced bidirectionally using BigDye Terminator v3.1 cycle sequencing kit and 3130xl Genetic Analyzer (Applied Biosystems). Prior to sequencing, PCR products were purified with ExoSAP-IT enzymatic mixture (USB Corporation). Subsequently, the sequencing reaction products were cleaned up using BigDye XTerminator purification kit (Applied Biosystems).
Frequencies of haplotypes were estimated using EM (expectation–maximization) algorithm implemented in Arlequin v3.11 [17]. Only haplotypes occurring with frequencies higher than 1% were used for further analysis.
Differences in the distribution of separate markers between studied groups were tested using Fisher’s exact test in contingency tables and two-sided P-values were calculated.
Results and discussion
In previous studies, a significant association of two indel polymorphisms in promoter (23 bp indel) and intron 1 (12 bp indel) of PRNP gene with BSE susceptibility was found in German, British, and Swiss cattle of different breeds, and in general the deletion gene variants of both polymorphisms were overrepresented in affected animals as compared to controls [9–12]. The functional relevance of the polymorphisms was first described by Sander et al. [13]. The proposed model was based on the fact that deletions at both polymorphic sites remove/interfere with consensus binding sites for transcription factors RP58 and Sp1, and thus may affect the PRNP expression level. In studies carried out on cattle and cultured cells, promoter variants with deletions (especially at locus of 23 bp indel polymorphism) were related to higher expression levels of PRNP when compared with insertion gene variants [13, 18, 19]. Higher expression level of the prion gene was associated with reduced transmissible spongiform encephalopathy (TSE) incubation period in experimentally infected mice [20]. However, in a few studied Polish classical BSE cases PRNP expression level did not differ significantly from that observed in control cattle [21].
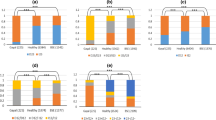
In this study we compared allele frequencies of PRNP indel polymorphisms in Polish BSE cases and the randomly selected population of clinically healthy cattle of the same breed. Results of this analysis showed a significantly higher frequency of deletion alleles of 23 and 12 bp indel polymorphisms in diseased than in control animals (P = 0.0242 and P = 0.0024, respectively). Genotype distribution of 23 bp indel polymorphism did not differ significantly between the groups; however, increased frequency of 23 bp del/del genotype was observed in BSE-affected cattle. Genotypic distribution of 12 bp indel polymorphism differed significantly between the diseased and healthy animals (P = 0.0066), with approximately two times higher frequency of 12 del/del genotype in BSE-affected animals (Table 1). A similar association of both polymorphisms with disease occurrence was observed in other studies [9–12].
Haplotypes spanning both polymorphic sites were established using EM algorithm, which allowed for the estimation of the combined effect of both polymorphisms. In the groups studied, four haplotypes occurring with frequency higher than 1% were identified (Table 2). The most common haplotype was spanning both deletions, haplotype 23del-12del. The rare haplotype 23ins-12del was observed only in a group of healthy animals (HO). Distribution of haplotypes differed significantly between diseased and healthy animals (P = 0.0111), and in general the deletion haplotype (23del-12del) was overrepresented in BSE-affected animals, while insertion haplotype (23ins-12ins) was more common in healthy animals. Similar results were reported by Juling et al. [11] and Haase et al. [12].
Our study identified a possible association of both studied PRNP promoter indel polymorphisms with susceptibility to BSE in Polish Holsteins. The low number of BSE cases studied here could lead to a false conclusion; however, the fact that our results are fully consistent with previously published data, makes these results sufficiently probable.
To further confirm the obtained results we performed a meta-data analysis with previously published study of Juling et al. [11] performed on German Holstein cattle (Table 3). We found that alleles and genotypes distribution did not differ significantly between German and Polish healthy cattle neither between German and Polish BSE-affected cattle. Analysis performed with combined groups of Polish and German cattle (BSE combined vs. healthy combined) showed that the distribution of alleles, but not genotypes at locus of 23 bp indel polymorphism, differed significantly between diseased and healthy animals (P < 0.01). At the locus of 12 bp indel polymorphism, both allelic and genotypic distribution differed highly significantly between healthy and diseased animals (P < 0.01) (Table 3).
In the previous studies, no clear association between PRNP ORF polymorphism and BSE susceptibility in cattle has been found [6, 7]. Only one H-type atypical BSE case in the USA was found to be caused by nonsynonymous mutation E211 K [22]. In Polish classical BSE cases in PRNP ORF only three previously described polymorphisms were found: two silent SNPs in codons 78 (234 G → A) and 192 (576 C → T) and one 24 bp indel (one octapeptide repeat indel) (Table 4) [6–8]. Frequencies of alleles of both observed SNPs were similar to the previously published data for healthy Holstein cattle [23, 24]. The 24 bp indel was found in one BSE case in heterozygous state (5/6 repeats) (Table 4). Since no clear association between 24 bp indel polymorphism and BSE susceptibility in cattle has been found in larger studies [6, 7, 10], we did not undertake further investigation, even though we found, that in a previous study on healthy Polish black-and-white cattle the frequency of the 5/6 genotype was clearly higher (0.180) [25] than in BSE-affected animals studied (0.037).
Frequencies of PRNP promoter indel polymorphisms (23 and 12 bp indel) were also compared between the subject cattle groups of different breeds (HO, RW, RP, and SIM). To avoid excessive analysis, only frequencies of haplotypes are shown in Table 5. Of all the breeds analyzed, the highest frequency of the haplotype 23del-12del was found in the Simmental breed (0.575), and the lowest in the Polish red breed (0.346). The haplotype distribution differed significantly only between the Polish red breed and the remaining breeds (P < 0.05). These differences resulted mainly from lower frequency of 23del-12del haplotype in the Polish red breed and simultaneously higher frequency of potentially BSE-protective haplotype 23ins-12ins.
Establishing the allele frequency of the BSE-associated PRNP polymorphisms in different breeds of cattle was the subject of many studies. Such analysis allows comparison of individual breeds and identification of breeds which are genetically susceptible/resistant to BSE. In Figs. 1 and 2 we combine the results of most studies to date showing frequencies of deletion alleles and deletion genotypes of both BSE-associated PRNP indel polymorphism in different cattle breeds and populations. Breeds were ranked based on growing deletion allele frequency; however, del/del genotype frequency has been shown to partially visualize genotype distribution within the breeds. The results allowed for general evaluation of which populations may potentially be most susceptible to BSE and showed that the Polish cattle breeds are characterized by relatively low or medium frequency of BSE-associated deletion alleles. Nevertheless, a comparatively high frequency of the predisposing alleles was found in the Simmental breed which is of importance as the breed is abundant in Poland. This comparison also showed that the deletion allele frequency seems to be lower in native or primitive breeds than in modern, highly selected cattle breeds.
Deletion allele and deletion genotype frequency of PRNP 23 bp indel polymorphisms in different cattle breeds and populations. References are given in brackets. For the Braunvieh breed the authors did not provide genotype frequency. *Group spans cattle of Swiss Schwarzfleck and Swiss Simmental × Red Holstein breeds, [W] - this study
Deletion allele and deletion genotype frequency of PRNP 12 bp indel polymorphisms in different cattle breeds and populations. References are given in brackets. For the Braunvieh breed the authors did not provide genotype frequency. *Group spans cattle of Swiss Schwarzfleck and Swiss Simmental × Red Holstein breeds, [W] - this study
In summary, the association analysis performed in this study indicated a significant relationship between the investigated PRNP indel polymorphisms and susceptibility of Polish Holstein-Friesian cattle to classical BSE (P < 0.05). In the population studied the deletion variants of both polymorphisms were related to increased BSE susceptibility, whereas insertion variants were recognized as protective. This suggests that both polymorphisms can be considered as potential susceptibility markers in the Polish HO breed.
In PRNP ORF in Polish classical BSE cases no novel, potentially pathogenic mutations have been found and the observed variations have occurred with low frequency, which is similar to the findings of previous studies performed with healthy Holstein cattle [23, 24].
References
Prusiner SB (1998) Prions. Proc Natl Acad Sci USA 95:13363–13383
van Keulen LJM, Schreuder BEC, Meloen RH, Poelen-van den Berg M, Mooji-Harkes G, Vromans M, Langeveld JPM (1995) Immunohistochemical detection and localization of prion protein in brain tissue of sheep with natural scrapie. Vet Pathol 32:299–308
Tichopad A, Pfaffl MW, Didier A (2003) Tissue-specific expression pattern of bovine prion gene: quantification using real-time RT-PCR. Mol Cell Probes 17:5–10
Hilton DA, Ghani AC, Conyers L, Edwards P, Mccardle L, Ritchie D, Penney M, Hegazy D, Ironside JW (2004) Prevalence of lymphoreticular prion protein accumulation in UK tissue samples. J Pathol 203:733–739
Bossers A, Schreuder BE, Muileman IH, Belt PB, Smits MA (1996) PrP genotype contributes to determining survival times of sheep with natural scrapie. J Gen Virol 77:2669–2673
Hunter N, Goldmann W, Smith G, Hope J (1994) Frequencies of PrP gene variants in healthy cattle and cattle with BSE in Scotland. Vet Rec 135:400–403
Neibergs HL, Ryan AM, Womack JE, Spooner RL, Williams JL (1994) Polymorphism analysis of the prion gene in BSE-affected and unaffected cattle. Anim Genet 25:313–317
Clawson ML, Heaton MP, Keele JW, Smith TP, Harhay GP, Laegreid WW (2006) Prion gene haplotypes of U.S. cattle. BMC Genet 7:51
Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, Groschup MH, Ziegler U, Distl O, Leeb T (2004) Analysis of sequence variability of the bovine prion protein gene (PRNP) in German cattle breeds. Neurogenetics 5:19–25
Geldermann H, He H, Bobal P, Bartenschlager H, Preuss S (2006) Comparison of DNA variants in the PRNP and NF1 regions between bovine spongiform encephalopathy and control cattle. Anim Genet 37:469–474
Juling K, Schwarzenbacher H, Williams JL, Fries R (2006) A major genetic component of BSE susceptibility. BMC Biol 4:33
Haase B, Doherr MG, Seuberlich T, Drögemüller C, Dolf G, Nicken P, Schiebel K, Ziegler U, Groschup MH, Zurbriggen A, Leeb T (2007) PRNP promoter polymorphisms are associated with BSE susceptibility in Swiss and German cattle. BMC Genet 8:15
Sander P, Hamann H, Drogemuller C, Kashkevich K, Schiebel K, Leeb T (2005) Bovine prion protein gene (PRNP) promoter polymorphisms modulate PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem 280:37408–37414
Kashkevich K, Humeny A, Ziegler U, Groschup MH, Nicken P, Leeb T, Fischer C, Becker CM, Schiebel K (2007) Functional relevance of DNA polymorphisms within the promoter region of the prion protein gene and their association to BSE infection. The FASEB J 21:1547–1555
Czarnik U, Strychalski J, Zabolewicz T, Pareek CS (2011) Populationwide investigation of two indel polymorphisms at the prion protein gene in Polish Holstein-Friesian cattle. Biochem Genet 49:303–312
Hills D, Comincini S, Schlaepfer J, Dolf G, Ferretti L, Williams JL (2001) Complete genomic sequence of the bovine prion gene (PRNP) and polymorphism in its promoter region. Anim Genet 32:231–232
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinf Online 1:47–50
Xue G, Sakudo A, Kim CK, Onodera T (2008) Coordinate regulation of bovine prion protein gene promoter activity by two Sp1 binding site polymorphisms. Biochem Biophys Res Commun 372:530–535
Msalya G, Shimogiri T, Nishitani K, Okamoto S, Kawabe K, Minesawa M, Maeda Y (2009) Indels within promoter and intron 1 of bovine prion protein gene modulate the gene expression levels in the medulla oblongata of two Japanese cattle breeds. Anim Genet 41:218–221
Vilotte JL, Soulier S, Essalmani R, Stinnakre MG, Vaiman D, Lepourry L, Da Silva JC, Besnard N, Dawson M, Buschmann A, Groschup M, Petit S, Madelaine MF, Rakatobe S, Le Dur A, Vilette D, Laude H (2001) Markedly increased susceptibility to natural sheep scrapie of transgenic mice expressing ovine PrP. J Virol 75:5977–5984
Larska M, Polak MP, Zmudzinski JF, Torres JM (2010) Comparison of mRNA expression levels of selected genes in the brain stem of cattle naturally infected with classical and atypical BSE. Brain Res 1351:13–22
Richt J, Hall SM (2008) BSE case associated with prion protein gene mutation. PLoS Pathogenesis 4:9
Jeong BH, Sohn HJ, Lee JO, Kim NH, Kim JI, Lee SY, Cho IS, Joo YS, Carp RI, Kim YS (2005) Polymorphisms of the prion protein gene (PRNP) in Hanwoo (Bos taurus coreanae) and Holstein cattle. Genes Genet Syst 80:303–308
Nakamitsu S, Miyazawa T, Horiuchi M, Onoe S, Ohoba Y, Kitagawa H, Ishiguro N (2006) Sequence variation of bovine prion protein gene in Japanese cattle (Holstein and Japanese black). J Vet Med Sci 68:27–33
Walawski K, Czarnik U (2003) Prion octapeptide-repeat polymorphism in Polish black-and-white cattle. J Appl Genet 44:191–195
Ün C, Oztabak K, Ozdemir N, Tesfaye D, Mengi A, Schellander K (2008) Detection of bovine spongiform encephalopathy-related prion protein gene promoter polymorphisms in local Turkish cattle. Biochem Genet 46:820–827
Czarnik U, Grzybowski G, Zabolewicz T, Strychalski J, Kaminski S (2009) Deletion/insertion polymorphism of the prion protein gene (PRNP) in Polish red cattle, Polish white-backed cattle and European bison (Bison bonasus L., 1758). Russ J Genet 45:453–459
Kim Y, Kim JB, Sohn H, Lee C (2009) A national survey on the allelic, genotypic, and haplotypic distribution of PRNP insertion and deletion polymorphisms in Korean cattle. J Genet 88:99–103
Brunelle BW, Kehrli ME Jr, Stabel JR, Spurlock DM, Hansen LB, Nicholson EM (2008) Allele, genotype, and haplotype data for bovine spongiform encephalopathy-resistance polymorphisms from healthy US Holstein cattle. J Dairy Sci 91:338–342
Czarnik U, Zabolewicz T, Strychalski J, Grzybowski G, Bogusz M, Walawski K (2007) Deletion/insertion polymorphism of the prion protein gene (PRNP) in Polish Holstein-Friesian cattle. J Appl Genet 48:69–71
Hreško S, Mojžiš M, Tkáčiková L (2009) Prion protein gene polymorphism in healthy and BSE-affected Slovak cattle. J Appl Genet 50:371–374
Kerber AR, Hepp D, Passos DT, Weimer TA (2008) Polymorphisms of two indels at the PRNP gene in three beef cattle herds. Biochem Genet 46:1–7
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s11033-012-1609-9.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gurgul, A., Urszula, C., Larska, M. et al. Polymorphism of the prion protein gene (PRNP) in Polish cattle affected by classical bovine spongiform encephalopathy. Mol Biol Rep 39, 5211–5217 (2012). https://doi.org/10.1007/s11033-011-1318-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1318-9