Abstract
SPRN is a new prion-like gene coding for Sho, a protein with significant similarity to PrP. SPRN was initially described in zebrafish; however, the strong evolutionary conservation led to the hypothesis that SPRN might be the ancestral prion-like gene. We mapped SPRN in Bos taurus by comparative analysis of the locus and of the predicted flanking genes. BACs, spanning the whole SPRN genomic locus, were assigned to BTA26q23 by radiation hybrid mapping and fluorescent in situ hybridization (FISH). Sequencing of five genes flanking SPRN, namely, ECHS1, PAOX, MTG1, SPRN, and CYP2E1, high-resolution FISH on mechanically stretched chromosomes, and combed BAC DNA allowed us to establish their order and reciprocal orientation. The results confirmed that BTA26q23 corresponds to HSA10q24.3–26.3, which is the site where the human SPRN is located. The gene order in Bos taurus is the same as in man, cen-ECHS1-PAOX-MTG1-SPRN-CYP2E1-tel, but PAOX has a different orientation in the two species. SPRN has the typical two-exon PRNP arrangement, with the CDS fully contained within exon 2; furthermore, it codes for a 143-amino-acid protein with 74.8% identity and 84.7% similarity with the human PRNP. RT-PCR and Northern blot analysis showed that SPRN is expressed at high levels in brain and less in testis and lung.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the last decade, studies on prions have been extended from man and livestock species to a number of organisms as distant as mammals, birds, reptiles, amphibians, and fish. The results revealed a complex and somewhat unexpected variety of PRNP genes (Gabriel et al. 1992; Windl et al. 1995; Castiglioni et al. 1998; Lee et al. 1998; Wopfner et al. 1999; Simonic et al. 2000; Strumbo et al. 2001; Suzuki et al. 2002; Oidtman et al. 2003; Rivera-Milla et al. 2003). PRNP is the gene that causes prion diseases in man and TSE (transmissible spongiform encephalopathy) in animals. In eutherians it is adjacent to PRND, a prion-like gene presumably derived from PRNP via segmental duplication (Moore et al. 1999; Comincini et al. 2001; Uboldi et al. 2005). The human prion gene locus also contains PRNT, which has not been described in organisms other than man (Makrinou et al. 2002; Choi et al. 2006). A number of studies on prion genes have focused on the physiologic role of PRNP, but data are still fragmentary and rather undefined. Comparative studies on prions started by analyzing paralog genes across species (Lee et al. 1998) and represented a novel and powerful tool to shed light on the physiologic role of PRNP and possibly on the association of the coded PrP protein with prion diseases. Such studies are now greatly facilitated by the increasing number of available genome sequences and by the ease of access to integrated information in databases. In fact, comparative analysis of prion genes allowed the discovery of quite a number of PRNP-related genes that seem to be fish-specific and have the specific features of prion proteins (Suzuki et al. 2002; Rivera-Milla et al. 2003). Surprisingly, these fish-specific PRNP-related proteins are loosely conserved among vertebrates and also are moderately conserved within the main fish lineages. This unexpected result prompted a revisit of some of the accepted features of the consensus prion protein, suggesting a more accurate classification that embraces all known organisms (Rivera-Milla et al. 2006). In this complex scenario an interesting case is SPRN, or Shadoo, a PRNP paralog gene, originally found in zebrafish (Premzl et al. 2003) and later described in other fish, in man, in mouse, in rat (Premzl et al. 2004), and predicted in chimpanzee (GenBank accession No. XM508138). Conservation of SPRN across all tested organisms at a higher extent than PRNP itself has led to the hypothesis that SPRN might be the ancestor prion gene (Premzl et al. 2004). According to the model proposed, a duplication of the ancestor SPRN gene might have occurred in prevertebrates, thus generating SPRNA and SPRNB. SPRNA and SPRNB should have evolved independently under different evolutionary constraints, giving rise to the current organization of prion genes. Thus, SPRNA should have remained relatively unchanged throughout evolution because of a more stable genomic environment, and it is expected to be conserved in most vertebrates. Conversely, SPRNB should have been duplicated a second time before the separation of fish from tetrapods, yielding the genes SPRNB1 and SPRNB2. The higher rate of instability at the duplicated locus started two distinct groups of genes: the PRNP cluster in mammals (with PRNP, PRND, and PRNT), arising from SPRNB1, and the heterogeneous group of prion-like genes of fish, that originated independently from either SPRNB1 or SPRNB2. The model leaves unanswered questions concerning the existence and syntenic conservation of SPRNA in mammals other than man and the presence (or absence) in mammals of the SPRNB2 gene, which has been found only in fish so far. The model is predominantly based on bioinformatic data; therefore, it is important to accumulate experimental supporting evidence. Finally, the model would suggest that studies on the physiologic role of PRNP should benefit from the analysis of SPRN.
The aim of this work was to search for SPRN in the bovine species. We exploited the public release of the whole bovine genome sequence (Build 2.1 at http://www.ncbi.nlm.nih.gov/mapview/map_search. cgi?taxid=9913), which was unavailable at the time of previous reports on SPRN (Premzl et al. 2004). Our results confirm the existence of an SPRN gene in the bovine species, with the expected features of SPRNA. The map position is exactly that predicted on a comparative basis; moreover, the bovine SPRN is flanked by the same genes that surround its human counterpart. However, we report a significantly low overall sequence homology at the locus and an inversion of one of the genes (PAOX) in cattle with respect to man. Finally, our data do not support the hypothesis that a second SPRNB, i.e., SPRNB2, exists in the bovine genome in addition to SPRNB1.
Materials and methods
EST selection and BACs isolation
Expressed sequence tags (ESTs) for the genes MTG1, PAOX, CYP2E1, and ECHS1 were selected using the ICCARE (Interspecific Comparative Clustering and Annotation foR Est) web interface at http://www.bioinfo.genopole-toulouse.prd.fr/iccare/. Among the bovine identified ESTs, four matching the human genes were selected (accession Nos. AV588698, AF226658, AJ001715, AY049961 for MTG1, PAOX, CYP2E1, and ECHS1, respectively). Primers were designed and used to screen two BAC libraries, the first described by Eggen et al. (2001) and the second created by Ross Miller (The Babraham Institute) and John Williams (Roslin Institute) and available at RPDZ (Berlin, Germany: library No. 750). Three clones were retrieved: bI0926F10, from the first and B0662Q2 and J03340Q2 from the second.
Genes annotation with STS, comparative PCR, and sequencing
Based on bovine ESTs retrieved from the ICCARE web site and GenBank, or using human genome sequence when bovine data were not available, oligonucleotides were designed and tested to amplify the predicted exons and introns of the MTG1, PAOX, CYP2E1, and ECHS1 genes on bovine BAC clones and on genomic DNA. The complete list of primers is given in the Supplementary Table. PCR products from BAC clones and genomic DNA were gel-purified (GeneElute, Sigma, St. Louis, MO) and sequenced with a Big Dye Terminator kit v1.1b (Applied Biosystems, Foster City, CA). Sequencing of SPRN was performed by walking 3′ of MTG1 on BACs B0662Q2 and bI0926F10, and the sequence obtained was later confirmed on genomic PCR products.
RT-PCR, cDNA synthesis, and Northern blot
RT-PCR was performed on total RNA extracted from spleen, heart, muscle, testis, brain, lung, kidney, and liver of a healthy friesian cow using Trizol (Invitrogen, Carlsbad, CA). RNA (3 μg) was reverse transcribed with Superscript II (Invitrogen) at 37°C for 2 h using an oligo(dT) primer, after which a PCR step (94°C for 20 sec, 60°C for 30 sec, 72°C for 90 sec for 32 cycles) was included with primers spanning the cDNA of the genes [namely, MTG1ex1-ex10, PAOXex1-ex7, CYP2E1ex1-ex9, ECHS1ex2-ex8, and SPRNex1-ex2 or SPRNex2 (see Supplementary Table for primer sequences)]. Genomic and cDNA sequences have been deposited in GenBank with accession Nos. DQ058602-10 and DQ591936.
For Northern blot analysis of SPRN, poly(A) mRNA was purified from the total RNAs used for RT-PCR, with the addition of bladder, using GeneElute mRNA Miniprep Kit (Sigma). Poly(A) mRNA (5 μg) of each sample was separated on a 1% agarose-formaldehyde gel and transferred to HyBond N+ (Amersham Pharmacia Biotech, Piscataway, NJ). The blot was hybridized with a SPRN cDNA, 710 bp (DQ531936), that was labeled with 32P-α-dCTP using HexaLabel Plus (MBI Fermentas, St. Leon-Rot, Germany).
DNA probes and probes labeling for FISH
Bio-16-dUTP and DIG-11-dUTP (Roche Molecular Biochemicals, Indianapolis, IN) were used to label BAC clones by nick-translation (Invitrogen). Gene-specific probes (Table 1) were labeled using a random priming system (Invitrogen). Fluorescent in situ hybridization (FISH) on metaphase chromosomes was performed as reported earlier (Iannuzzi et al. 2003).
Preparation of mechanically stretched chromosomes and of combed BAC DNA
Primary fibroblasts from bovine kidney were cultured according to standard protocols to perform mechanically stretched chromosome preparations (Haaf and Ward 1994). Mitotic cells were harvested, incubated in hypotonic buffer (10 mM HEPES, pH 7.3, 30 mM glycerol, 1 mM CaCl2, 0.8 mM MgCl2) and transferred onto slides by centrifugation at 200g using a Cytospin centrifuge (HERMLE).
DNA from BACs b10926F10 and B0662Q2 was purified with a MidiPrep kit (Qiagen, Valencia, CA), and used to prepare combed DNA as described by Henegariu et al. (2001). In brief, DNA was diluted to 5 ng/μl in 10 mM aminomethyl propenediol (Sigma) buffer, pH 8.2-8.5; 8 μl of each DNA solution were pipetted close to the free short edge of a previously sylanized microscope slide. A 24-mm × 50-mm coverslip was positioned at an angle with the slide so that it touched the DNA drop. The angle between the coverslip and the slide was gradually decreased and the drop spread laterally along the coverslip edge. The forced flow of liquid between the slide and the coverslip is the driving force that stretches the BAC DNA. Finally, the coverslip was removed, the slide air-dried, and fixed in ethanol.
FISH on stretched chromosomes and on combed BAC DNA; probes detection
The FISH protocol in this case was basically as that in Raimondi et al. (1991). Briefly, slides were denatured in 70% formamide/2×SSC. Labeled probes were coprecipitated with a tenfold excess of salmon sperm DNA, resuspended in hybridization buffer (50% formamide, 10% dextran sulfate, 1× Denhart’s solution, 0.1% SDS, 40 mM Na2HPO4, pH 6.8, 2×SSC) at a final concentration of 5 mg/ml, and denatured 10 min at 80°C. Each hybridization was performed combining two or three differently labeled probes for 24 h at 37°C. Stringent washings were done with 50% formamide/2×SSC at 42°C.
For triple- or double-signal detection, slides were incubated first with FITC-conjugated avidin DCS and rhodamine-conjugated sheep anti-digoxigenin antibody (Roche), then with biotin-conjugated anti-avidin D antibody and rhodamine-conjugated rabbit anti-sheep antibody (Chemicon, Temecula, CA), and finally with FITC-conjugated avidin. Avidin and antibodies were used at a final concentration of 5 mg/ml. Eventually, slides were counterstained with DAPI (0.01 mg/ml) (Sigma) and mounted in 2% 1 M Tris-HCl (pH 7.5) 90% glycerol containing 0.2 M DABCO [1,4 diazabicyclo-(2.2.2) octane] antifade (Sigma).
Analysis was performed on a Zeiss Axioplan fluorescent photomicroscope equipped with a cooled CCD camera (Photometrics, Tucson, AZ). Images were captured with IPlab spectrum P software (Scanalytics, Billerica, MA).
RH mapping
A 3000-rad bovine-hamster radiation hybrid (RH) panel (Williams et al. 2002) was amplified with tags for MTG1 (487 bp, exon 1 to intron 1), PAOX (287 bp, exon 2), CYP2E1 (190 bp, exon 1), and ECHS1 (160 bp, exon 9) (Supplementary Table). Cycling conditions were 32 cycles of 94°C for 20 sec, 60°C for 30 sec, and 72°C for 45 sec. A preliminary assignment of the markers to BTA26 was obtained from the analysis of LOD score values for all the pairwise combinations of markers in a public data set (Williams et al. 2002). Then, vector data for the markers were used to produce a more detatiled BTA26 map using CarthaGene (de Givry et al. 2005).
Results
Isolation of BACs and chromosomal localization of the gene cluster by FISH
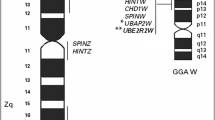
Three BAC clones, matching the human genomic region predicted to span the SPRN gene, i.e., ECHS1, PAOX, MTG1, and CYP2E1, were isolated using four ESTs selected at the ICCARE web site. FISH was performed on bovine metaphase chromosomes. The BACs bI0926F10 and B0662Q2, spanning the entire gene cluster, were used as probes; chromosomes were identified by RBPI banding. Strong hybridization signals were present on BTA26q23 only, thus allowing the unequivocal assignment of the gene cluster (Fig. 1); BTA26q23 corresponds to HSA10q26.3, where the human SPRN gene is known to be located.
Mapping of the bovine Shadoo gene SPRN by FISH. BACs bI0926F10 and B0662Q2 containing ECHS1, PAOX, MTG1, SPRN, and CYP2E1 were hybridized to RBPI-banded chromosomes by FISH. The top panel shows a single-color FISH with a mix of the two BACs and signals on BTA26q23. In the bottom panel, differentially labeled BACs bI0926F10 (in red) and B0662Q2 (in green) were used as probes; the partially overlapping signals derive from ECHS1-PAOX-MTG1-SPRN which is some 100-120 kb from CYP2E1 (see further details on the gene cluster arrangement in Fig. 2).
Molecular cytogenetic analysis of SPRN locus by means of FISH on elongated chromosomes and on combed DNA of BAC clones. (A) The table summarizes the gene content of the BACs used: B0662Q2 contains only a partial MTG1 gene (from exon 5 to exon 10). (B) BAC bI0926F10 (in red) and B0662Q2 (in green) were hybridized to elongated chromosomes. The position of the signals with respect to the telomere and centromere of BTA26 clearly suggested that B0662Q2 is distal, whereas bI0926F10 is proximal. Thus, CYP2E1 is the most telomeric gene in the cluster. (C) The order and reciprocal arrangement of individual genes were deduced with a set of three-color FISH on combed DNA of BACs. Probes were the exonic portions that are shown to the left with colors matching the corresponding hybridization signal (see also Table 1). The yellow color derives from mixed dual-labeling of the relevant probe. (D) The use of multiple exonic portions of each gene allowed the establishment of the different arrangement and order of genes at the cluster. Arrowed bars mark the orientation of genes, and the size is proportional to the genomic size of individual genes; however, the entire cluster is not drawn to scale.
RH mapping
Four ESTs—AV588698, AF226658, AJ001715, and AY049961—corresponding to the genes MTG1, PAOX, CYP2E1, and ECHS1, respectively, were amplified in the 3000-rad panel described by Williams et al. (2002). Two-point analysis of LOD scores identified the closest markers to the mapped gene tags, namely, MAF36, BM804, BM7237, and MAF92 (LOD scores > 4). RH vector data were processed with the software CarthaGene, thus obtaining a map of BTA26 with the ECHS1, PAOX, MTG1, and CYP2E1 genes clustered in a telomeric/subtelomeric position, compatible with FISH data (not shown).
Gene order and arrangement at the SPRN bovine locus
The BACs containing the ECHS1-PAOX-MTG1-CYP2E1 cluster were further analyzed by PCR using a set of oligonucleotides (Supplementary Table) to assign single genes and gene portions to each BAC. STS-content mapping showed that BAC bI0926F10 harbored three complete genes, ECHS1, PAOX, and MTG1, that BAC B0662Q2 contained a portion of the MTG1 gene (exons 5-10) plus the complete CYP2E1 gene, and that BAC J0334Q2 contained the CYP2E1 gene only. The results allowed us to establish that MTG1 is in a central position, flanked by ECHS1 and PAOX on one side and by CYP2E1 on the other side. This approach was unsuccessful in assigning SPRN, because none of the STS selected from the human gene worked comparatively in the bovine. The human SPRN gene is in an opposite direction with respect to MTG1 and overlaps its 3′ untranslated region (UTR); therefore, sequencing was performed on BAC bI0926F10 and B0662Q2 starting from the mapped 3′ end of MTG1. The CDS of the bovine SPRN was found 2693 bp from the stop codon of MTG1 in an opposite orientation with respect to MTG1, exactly as in man.
To define the exact order and reciprocal arrangement of the genes, a long-range analysis, based on a molecular-cytogenetic approach, was used. High-resolution FISH was performed on mechanically stretched chromosomes and on combed BAC DNA, using as probes labeled BACs and selected portions of the genes, respectively (Fig. 2A–C).
BACs bl0926F10 and B0662Q2 were oriented with respect to the centromere of BTA26 by performing a two-color high-resolution FISH on mechanically stretched chromosomes. Twenty metaphase spreads from normal bovine fibroblast with properly stretched chromosomes were scored. Both BAC clones hybridized to the telomeric portion of the long arm of BTA26, the red signal (bl0926F10) being centromeric with respect to the green signal (B0662Q2) (Fig. 2B). Because STS-content PCR mapping previously showed that BAC B0662Q2 harbors only CYP2E1, the FISH data demonstrated that the order of the genes in the cluster is cen-ECHS1/PAOX-MTG1/SPRN-CYP2E1-tel, with the relative positions of ECHS1/PAOX and MTG1/SPRN still undefined.
To establish the arrangement and reciprocal orientation of all the genes in the cluster, six independent high-resolution three-color FISH experiments were performed on combed DNA purified from BACs bl0926F10 and B0662Q2. The target region to be analyzed was dissected into nine exon-intron segments that were used as FISH probes, as summarized in Figure 2C. For each experiment 15 combed BAC DNA fibers were scored. The comparison and alignment of the FISH images suggested the following order and orientation of the genes: cen - 3′ ECHS1 5′ - 3′ PAOX 5′ - 5′ MTG1 3′ - 3′ SPRN 5′ - 5′ CYP2E1 3′ - tel (Fig. 2D). These results confirmed that similarity between BTA26q23 and HSA10q24.3-q26.3 is maintained at the gene order level and showed an inversion of PAOX because it has an opposite orientation in Bos taurus with respect to man. (http://www.ncbi.nlm.nih.gov/mapview/maps. cgi?taxid=9606&chr=10&MAPS=genec,ugHs, genes-r&cmd=focus&fill=40&query=uid(14239983)& QSTR=PAOX).
Definition of the individual gene structure and expression pattern analysis
The exon-intron structure of each gene was determined by sequencing PCR products obtained from BACs and from genomic DNA after amplification with exon-specific primers for the coding region, and with interexonic primers for the intronic regions (see Materials and methods). Table 2 summarizes information obtained for each gene.
ECHS1
The ECHS1 gene (enoyl coenzyme A hydratase, short chain, 1, mitochondrial) is 8171 bp long and is divided into eight exons as in man. The gene is transcribed into a 1326-nt mRNA coding for a 290-amino-acid protein and is highly homologous to its human counterpart (85% sequence similarity in the CDS and 83% conservation at the protein level). The expression profile of ECHS1 was analyzed by means of RT-PCR in brain, lung, liver, kidney, and spleen (data not shown). The gene is expressed at high levels in all tissues and no evidence was found of differential tissue-specific expression.
PAOX
The PAOX gene (peroxisomal N1-acetyl-spermine/spermidine oxidase) is composed of seven coding exons like the human, but it is shorter: 8272 bp compared with 12,458 bp. We detected a 1810-nt mRNA that corresponds to the longest isoform of the human protein, isoform 1, and codes for a 511-amino-acid polypeptide. The gene is expressed in all the tissues analyzed by RT-PCR; the highest expression level was found in brain, kidney, and spleen, an intermediate expression level was found in liver, and the lowest expression level was in lung (data not shown). We did not find transcripts corresponding to the human isoforms 2, 3, and 4. Conservation with respect to man is higher at the DNA level (83% similarity in the CDS) than at the protein level (65%).
MTG1
In a previous build of the NCBI human genome database (build 35), MTG1 (mitochondrial GTPase 1 homolog) was misannotated as SPRN. Now in build 36.1, MTG1 (accession No. BN000518) is correctly identified as the gene adjacent to SPRN. Indeed, the bovine homolog of the human MTG1 gene is a GTPase-like protein, identified as LOC509768 (accession No. DQ058604). The gene is composed of ten exons like in the human gene, but it is shorter, spanning 12,048 bp compared with 27,124 bp; this difference is mainly a result of the length of intron 8, which is 2853 bp in cattle and 16,706 bp in man. In addition, the human CDS starts in exon 1 and stops within exon 7, whereas in the bovine it spans exons 1-10. Thus, the bovine 1356-nt mRNA codes for a 332-amino-acid polypeptide, whereas the human polypeptide is only 267 amino acids long. The two aligned CDS show only 63% similarity. The reason for this low score is a 563-nt insertion within the coding region of exon 7 in the human gene. This insertion provides an in-frame stop codon that terminates the polypeptide at amino acid 267. Notably, the 563-bp insert is flanked by 5-bp repeats (CAGGT) and its removal restores the reading frame of the human gene, extending it through all the exons down to exon 10, exactly as it is in cattle. In this case, the sequence of the genes and the coded proteins in the two species align to a much higher degree.
The analysis of RT-PCR profiles for the bovine MTG1 gene revealed a ubiquitous expression pattern; however, expression levels were lower when compared with those observed for ECHS1, PAOX, and CYP2E1.
CYP2E1
The CYP2E1 gene (cytochrome P450 subfamily IIE polypeptide 1) is composed of nine exons, as in the human gene, and spans a genomic region of comparable size, 10,396 bp in cattle vs. 11,756 bp in man. All the exons are coding and the 1921-nt mRNA translates into a 495-amino-acid polypeptide. The homology with the human gene is 80% at the nucleotide (CDS) level and 78% at the protein level. Expression of the bovine CYP2E1 was found restricted to liver, as was expected given the known function of the protein and according to literature data (MIM 124040).
SPRN
Because the comparative PCR with STS tags did not work, to find out the bovine homolog of SPRN we walked some 5.4 kb from exon 10 of MTG1 in the 3′ direction on BACs B0662Q2 and bI0926F10. The bovine gene has only two exons like the SPRN gene described in other species (Premzl et al. 2004) and as expected for a PRNP-like gene. The entire CDS (432 bp) is within exon 2 and starts 2693 bp 3′ of the TGA stop codon of MTG1 exon 10. A 726-bp intron, showing the standard GT-AG splicing consensus signals, separates exon 2 from exon 1, which is 111 bp long.
It is noticeable that the 5466-bp sequence that we obtained at the 3′ side of the stop codon of the adjacent MTG1 gene spanning SPRN is very GC rich (average 68%, with regions above 85%). As shown in Figure 3A, the CpG plot tool on the EMBL-EBI server (http://www.ebi.ac.uk/emboss/cpgplot/) identified two CpG islands: the first, 333 bp, completely covers the SPRN exon 1, the second, 232 bp, is totally embedded in the CDS of exon 2. The high GC content made the analysis of SPRN expression difficult. Specifically, the presence of a stretch of 138 bp with a GC content around 86% within the CDS (91-228 bp in exon 2) prevented recovery of a RT-PCR product representative of the entire reading frame. We presume that this was a result of polymerase slippage during the reaction; indeed, the RT-PCR products that were sequenced invariably lacked the GC-rich region (data not shown). The use of DMSO in the RT reaction was of no help. To circumvent the problem, RT-PCR products were obtained with two alternative approaches: (1) by using primers designed inside the GC-rich stretch of the SPRN CDS, pointing in opposite directions, combined with primers at the 5′ and 3′ and (2) by excluding the GC-rich region in the design of RT-PCR products (Fig. 3A). This allowed us to obtain a 710-bp partial cDNA comprising exon 1 (111 bp) and a portion of exon 2 (599 bp). Evidence in support of the existence of the partial transcript defined here has come from the recent publication of an additional incomplete cDNA (DV828917, 449 bp) in the NCBI database: DV828917 perfectly aligns with our cDNA, including 111 bp of exon 1 and 338 bp of exon 2.
Genomic structure and analysis of expression of the bovine SPRN gene. (A) A 5466-bp DNA region distal to MTG1 (of which exon 10 is visible) and spanning SPRN was analyzed with a CpG plot tool from the EBI server. The top profile shows the GC% and the arrowed empty boxes above the gene structure mark CpG islands. In the genomic view of SPRN, exon 2 is shown with a solid line to represent the cDNA product obtained by RT-PCR; otherwise, it is drawn with a dashed line down to a predicted potential polyadenylation site (solid circle) that is flanked by a GU-rich stretch cleavage signal (open circle). The mRNA shown is deduced from the primary transcript found by Northern blot analysis (panel C) in the hypothesis that transcription starts at exon 1. DQ531936 and DV828917 are the SPRN cDNA produced by RT-PCR in this work and a partial cDNA homologous to SPRN retrieved from GenBank, respectively. The black box inside the CDS is a 138-bp sequence with >86% GC content. (B) RT-PCR of SPRN in spleen (Sp), heart (He), muscle (Mu), testis (Te), kidney (Ki), liver (Li), lung (Lu), brain (Br). Primer pairs 1-2 and 3-4 (oligonucleotides SPRN ex1U-SPRNex2L and SPRN GC-richU-SPRN es2L_A, respectively; Supplementary Table) yielded fragments of 469 bp and 166 bp. Asterisks suggest that on genomic and BAC templates the PCR product includes the SPRN intron (726 bp); thus, the fragment size is 892 bp. Controls were genomic (GE), BAC (BA), and no DNA (-) samples. Size markers (MA) were a mix of the 50- and 100-bp ladders (MBI Fermentas). (C) Northern blot analysis of SPRN: poly(A) mRNA from testis (Te), liver (Li), spleeen (Sp), brain (Br), lung (Lu), kidney (Ki), bladder (Bl), heart (He), and muscle (Mu) were blotted and hybridized to a labeled cDNA (DQ531936) probe of SPRN. The SPRN transcript migrates exactly as the 3-kb band of an RNA ladder standard (New England Biolabs, data not shown).
To evaluate SPRN expression in tissues, RT-PCR was performed with two sets of primers that amplified DNA proximal and distal to the GC-rich stretch in the CDS (Fig. 3A, B). The first primer pair was designed to amplify 166 bp, comprising exon 1 and the proximal 55 bp of exon 2. The second primer pair amplified 459 bp, starting from nt 141 of exon 2 toward the 3′ UTR. The results (Fig. 3B) suggested that bovine SPRN expression was not as ubiquitous as might be expected for a gene with CpG islands and a TATA-less promoter region. In fact, expression was found predominantly in brain but also in testis and lung, whereas no expression was detected in muscle, heart, kidney, and liver. RT-PCR data were confirmed with the analysis of SPRN mRNA by Northern blot analysis (Fig. 3C). A probe corresponding to the SPRN CDS (Table 1) revealed a signal in brain, testis, and lung. The apparent size of the mRNA, about 3 kb, is similar to that of the human transcript of 3226 nt (NM001012508), which starts exactly with the first nucleotide of human SPRN exon 1. In the case of the bovine transcript, a potential polyadenylation signal (AAUAAA) is placed 1846 bp distal of the SPRN CDS (Fig. 3) and is followed by a GU-rich stretch that might represent the transcript cleavage signal. However, the tool used to predict the polyadenylation signal (Polyadq, http://www.rulai.cshl.org/tools/polyadq/polyadq_form.html) assigned a low score to the sequence (data not shown). Furthermore, if the bovine transcript would start with the first nucleotide of exon 1 as in the human mRNA (NM001012508), the size of the mRNA that is polyadenylated at the predicted signal would be 2554 nt, much shorter than the transcript observed in the Northern blot analysis (3 kb). Two explanations can be proposed: either there is another polyadenylation signal distal to the predicted one, or the bovine SPRN mRNA has a 5′ extension proximal to exon 1. Because we sequenced the entire sequence 3′ of SPRN and could not find an additional polyadenylation site using specific software tools, we are inclined to believe that the extension of the SPRN mRNA should be at the 5′ of the gene.
Discussion
In this work experimental evidence is presented to support the existence of the prion-like gene SPRN in Bos taurus. Interest in SPRN comes from the hypothesis that the gene might represent the prion gene-family ancestor (Premzl et al. 2004). Our results have demonstrated that SPRN exists in cattle and that it presents the typical features of a PRNP-like two-exon gene, with the entire CDS in exon 2. The deduced protein is 143 amino acids long (151 and 147 amino acids for the human and murine polypeptides, respectively) and aligns nicely with the human, mouse, and rat Sho (with an average similarity of 84% and identity of 74%) and, to a lesser extent, with Sho from Danio rerio (34% similarity and identity).
Like PRNP, SPRN has the appearance of a housekeeping gene in that it contains a CpG island spanning the entire exon 1 and a second one overlapping exon 2. Our data have shown that no TATA-like promoter is present, while we could find a number of Sp1 and housekeeping transcription factor binding sites (data not shown). Despite these features, the gene does not seem to be ubiquitously expressed. As documented by RT-PCR and by Northern blot analysis, the gene is predominantly expressed in brain and to a lesser extent in testis and lung. It should be noted that the whole SPRN genomic locus is particularly GC rich (68% average, with highs at over 85%); thus, the CpG islands found might simply reflect the great divergence in base pair composition of this particular sequence with respect to the whole bovine genome. However, in agreement with our findings, the genomic context in which the human, mouse, and rat SPRN genes are embedded is also GC rich. In these species, SPRN expression is similarly confined to brain (Premzl et al. 2003). In addition, the PRNP genes of hamster (Li and Bolton, 1997), man, sheep, mouse (Lee et al. 1998), and cattle (Hills et al. 2001), have a GC-rich exon 1 and TATA-less promoters.
According to the hypothesis of Premzl et al. (2004), the bovine SPRN described here should be the ortholog of the human and mouse SPRN genes already reported. As such, it should represent SPRNA, which is one of two duplicates of the ancestor SPRN gene. Our mapping data, analysis of expression profiles, and database searches did not support the existence of a second SPRN gene, SPRNB; the same conclusion was reached recently by Choi et al. (2006). A possible explanation is that in mammals SPRNB might have diverged so much from SPRNA that it is undetectable when using homology as search criterium.
To study the stability at the SPRN locus in terms of gene content and gene orientation, we have produced a physical map of the genomic region. The results obtained confirm that the genes mapping at this locus form a highly conserved syntenic block, thus supporting the hypothesis that SPRN played a key role throughout evolution, from fish to mammals. It seems worth discussing some points about gene order and the current annotation of the SPRN locus in the NCBI bovine genome database. Specifically, the bovine region does contain the genes described in man and in other mammalian species at the corresponding locus; however, according to our results, an inversion changed the orientation of the bovine PAOX gene. Similar to what has been reported for man, the maximum distance between SPRN and CYP2E1 in cattle should be within 100-120 kb because the two genes are on the same BAC (B0662Q2).
We could highlight some inconsistencies in the NCBI bovine genome database (build Btau 2.1) via direct analysis of the organization of the locus by high-resolution FISH on “combed” BAC DNA filaments. A large sequence contig that contains the genes that we mapped is available (NW_930534, 916997 bp), but it suggests a different gene order and orientation: ECHS1, MTG1, PAOX, and SPRN instead of our ECHS1, PAOX, MTG1, and SPRN. A rearrangement in the BAC clones could explain the discrepancy, but we are inclined to exclude this. In fact, we sequenced the MTG1-SPRN region directly on BACs, but also amplified and sequenced the corresponding fragment on genomic DNA using oligonucleotides derived from the BACs sequencing, and obtained exactly the same result. In addition, we performed sequencing on three independent BACs recovered from two different libraries, again finding the same sequence.
Another inconsistency is that despite the bulky size (916,997 bp), contig NW_930534 does not contain CYP2E1, which is represented in the bovine genome database within an unassigned 126-kb-long contig (NW_993476). Our FISH data on combed BAC DNA and RH data have clearly shown that CYP2E1 is tightly associated with the ECHS1-PAOX-MTG1-SPRN cluster at an estimated distance of 100-120 kb. We also showed by FISH that the BAC containing CYP2E1 maps to BTA26q23, as already reported by Gautier et al. (2001, 2003).
In conclusion, we have demonstrated the existence of the prion-like SPRN gene in the bovine species and have annotated a 200-kb BAC contig containing SPRN itself and the flanking genes ECHS1, PAOX, MTG1, and CYP2E1. We have shown that the gene content at the genomic locus is that expected on a comparative basis and predicted by the authors who described the SPRN gene in other mammals (Premzl et al. 2004). However, our results have highlighted substantial inconsistencies in the annotation of the SPRN region in the bovine genome database. It is likely that these inconsistencies will be corrected with time once the many unfinished sequence bits in the relevant contigs are filled and a newer build replaces the current one. In any case, attention should be paid when extracting sequence information from databases, especially for comparative mapping purposes.
References
Castiglioni B, Comincini S, Drisaldi B, Motta T, Ferretti L (1998) Comparative mapping of the prion gene (PRNP) locus in cattle, sheep and human with PCR-generated probes. Mamm Genome 9, 853–855
Choi SH, Kim IC, Kim DS, Kim DW, Chae SH, et al. (2006) Comparative genomic organization of the human and bovine PRNP locus. Genomics 87, 598–607
Comincini S, Foti MG, Tranulis MA, Hills D, Di Guardo G, et al. (2001) Genomic organization, comparative analysis, and genetic polymorphisms of the bovine and ovine prion Doppel genes (PRND). Mamm Genome 12, 729–733
de Givry S, Bouchez M, Chabrier P, Milan D, Schiex T (2005) CARHTA GENE: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21, 1703–1704
Eggen A, Gautier M, Billaut A, Petit E, Hayes H, et al. (2001) Construction and characterization of a bovine BAC library with four genome-equivalent coverage. Genet Sel Evol 33, 543–548
Gabriel JM, Oesch B, Kretzschmar H, Scott M, Prusiner SB (1992) Molecular cloning of a candidate chicken prion protein. Proc Natl Acad Sci U S A 89, 9097–9101
Gautier M, Hayes H, Taourit S, Laurent P, Eggen A (2001) Syntenic assignment of sixteen genes from the long arm of human chromosome 10 to bovine chromosomes 26 and 28: refinement of the comparative map. Chromosome Res 9, 617–621
Gautier M, Hayes H, Eggen A (2003) A comprehensive radiation hybrid map of bovine Chromosome 26 (BTA26): comparative chromosomal organization between HSA10q and BTA26 and BTA28. Mamm Genome 14, 711–721
Haaf T, Ward DC (1994) High resolution ordering of YAC contigs using extended chromatin and chromosomes. Hum Mol Genet 3, 629–633
Henegariu O, Grober L, Haskins W, Bowers PN, State MW, et al. (2001) Rapid DNA fiber technique for size measurements of linear and circular DNA probes. Biotechniques 31, 246–250
Hills D, Comincini S, Schlaepfer J, Dolf G, Ferretti L, et al. (2001) Complete genomic sequence of the bovine prion gene (PRNP) and polymorphism in its promoter region. Anim Genet 32, 231–232
Iannuzzi L, Perucatti A, Di Meo GP, Schibler L, Incarnato D, et al. (2003) Chromosomal localization of sixty autosomal loci in sheep (OVIS ARIES, 2n = 54) by fluorescence in situ hybridization and R-banding. Cytogenet Genome Res 103, 135–138
Lee IY, Westaway D, Smit AF, Wang K, Seto J, et al. (1998) Complete genomic sequence and analysis of the prion protein gene region from three mammalian species. Genome Res 8, 1022–1037
Li G, Bolton DC (1997) A novel hamster prion protein mRNA contains an extra exon: increased expression in scrapie. Brain Res 751, 265–274
Makrinou E, Collinge J, Antoniou M (2002) Genomic characterization of the human prion protein (PrP) gene locus. Mamm Genome 13, 696–703
Moore RC, Lee IY, Silverman GL, Harrison PM, Strome R, et al. (1999) Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein doppel. J Mol Biol 292, 797–817
Oidtmann B, Simon D, Holtkamp N, Hoffmann R, Baier M (2003) Identification of cDNAs from Japanese pufferfish (Fugu rubripes) and Atlantic salmon (Salmo salar) coding for homologues to tetrapod prion proteins. FEBS Lett 538, 96–100
Premzl M, Sangiorgio L, Strumbo B, Marshall Graves JA, et al. (2003) Shadoo, a new protein highly conserved from fish to mammals and with similarity to prion protein. Gene 314, 89–102
Premzl M, Gready JE, Jermiin LS, Simonic T, Marshall Graves JA (2004) Evolution of vertebrate genes related to Prion and Shadoo proteins – clues from comparative genomic analysis. Mol Biol Evol 21, 2210–2231
Raimondi E, Ferretti L, Young BD, Sgaramella V, De Carli L (1991) The origin of a morphologically unidentifiable human supernumerary minichromosome traced via sorting molecular cloning and in situ hybridization. J Med Genet 28, 92–96
Rivera–Milla E, Stuermer CA, Malaga–Trillo E (2003) An evolutionary basis for scrapie disease: identification of a fish prion mRNA. Trends Genet 19, 72–75
Rivera–Milla E, Oidtmann B, Panagiotidis CH, Baier M, Sklaviadis T, et al. (2006) Disparate evolution of prion protein domains and the distinct origin of Doppel- and prion-related loci revealed by fish-to-mammal comparisons. FASEB J 20, 317–319
Simonic T, Duga S, Strumbo B, Asselta R, Ceciliani F, et al. (2000) cDNA cloning of turtle prion protein. FEBS Lett 469, 33–38
Strumbo B, Ronchi S, Bolis LC, Simonic T (2001) Molecular cloning of the cDNA coding for Xenopus laevis prion protein. FEBS Lett 508, 170–174
Suzuki T, Kurokawa T, Hashimoto H, Sugiyama M (2002) cDNA sequence and tissue expression of Fugu rubripes prion protein-like: a candidate for the teleost orthologue of tetrapod PrPs. Biochem Biophys Res Commun 294, 912–917
Uboldi C, Del Vecchio I, Foti MG, Azzalin A, Paulis M, et al. (2005) Prion-like Doppel gene (PRND) in the goat: genomic structure, cDNA, and polymorphisms. Mamm Genome 16, 963–971
Williams JL, Eggen A, Ferretti L, Farr CJ, Gautier M, et al. (2002) A bovine whole-genome radiation hybrid panel and outline map. Mamm Genome 13, 469–474
Windl O, Dempster M, Estibeiro P, Lathe R (1995) A candidate marsupial PrP gene reveals two domains conserved in mammalian PrP proteins. Gene 159, 181–186
Wopfner F, Weidenhofer G, Schneider R, von Brunn A, Gilch S, et al. (1999) Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J Mol Biol 289, 1163–1178
Acknowledgments
This work was supported by grants from the Italian MIUR, Progetti di Ricerca di Rilevante Interesse Nazionale (2005), and from the Istituto Superiore di Sanità (ISS) “UO9 – Fattori genetici, patogenetici e biochimici responsabili della sensibilità/resistenza alle EST.”
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Uboldi, C., Paulis, M., Guidi, E. et al. Cloning of the bovine prion-like Shadoo (SPRN) gene by comparative analysis of the predicted genomic locus. Mamm Genome 17, 1130–1139 (2006). https://doi.org/10.1007/s00335-006-0078-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-006-0078-7