Abstract
Interrupted blood flow to regions of the heart causes damage to heart muscles, resulting in myocardial infarction (MI). MI is a major source of death worldwide. Accurate and timely detection of MI facilitates initiation of emergency revascularization in acute MI and early secondary prevention therapy in established MI. In both acute and ambulatory settings, the electrocardiogram (ECG) is a standard data type for diagnosis. ECG abnormalities associated with MI can be subtle, and may escape detection upon clinical reading. Experience and training are required to visually extract salient information present in the ECG signals. This process of characterization is manually intensive, and prone to intra-and inter-observer-variability. The clinical problem can be posed as one of diagnostic classification of MI versus no MI on the ECG, which is amenable to computational solutions. Computer Aided Diagnosis (CAD) systems are designed to be automated, rapid, efficient, and ultimately cost-effective systems that can be employed to detect ECG abnormalities associated with MI. In this work, ECGs from 200 subjects were analyzed (52 normal and 148 MI). The proposed methodology involves pre-processing of signals and subsequent detection of R peaks using the Pan-Tompkins algorithm. Nonlinear features were extracted. The extracted features were ranked based on Student’s t-test and input to k-Nearest Neighbor (KNN), Support Vector Machine (SVM), Probabilistic Neural Network (PNN), and Decision Tree (DT) classifiers for distinguishing normal versus MI classes. This method yielded the highest accuracy 97.96%, sensitivity 98.89%, and specificity 93.80% using the SVM classifier.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The heart is an essential organ composed primarily of muscle tissue. The function of the coronary arteries is to supply oxygenated blood to the heart muscle. Thus, coronary artery disease obstructs the arteries, and reduces the blood supply to downstream muscle, potentially causing damage to the heart muscle and the possibility of myocardial infarction (MI) (Thomas 2015). If the extent of the MI is large, the affected poorly contracting wall segments can introduce increased mechanical stress to the heart, resulting in morphological and conformational alterations in response, i.e., left ventricular remodeling, which results in inefficient pump functioning and contributes to heart failure. The MI segments can undergo substrate alterations, e.g. fibrosis, that renders the tissue arrhythmogenic, which can be lethal. Symptoms of acute MI involve breaking into cold sweats, chest pain, shortness of breath, feeling faint, nausea, as well as discomfort that may radiate to the arm, shoulder, and neck (Setiawan et al. 2014). Major risk factors associated with MI are high cholesterol, diabetes, high blood pressure, physical inactivity, obesity, and unhealthy diets. These risk factors promote atherosclerotic plaque formation in the coronary artery wall that results in arterial narrowing. As fat content in the plaque increases, it becomes vulnerable and susceptible to surface rupture. The latter triggers a secondary pathophysiological phenomenon that activates blood clot formation within the coronary artery lumen to interrupt the distal blood supply suddenly and completely, resulting in MI (Roger 2007).
In 2017, 800,000 Americans died from MI (WHO fact 2012; Ley 2015). Of these, 280,000 had prior MI and the remainder were first-time presentations (Ley 2015) underscoring the need for accurate diagnostics. There are several diagnostic instruments utilized to characterize MI. The diagnostic tests include the exercise stress test (EST), cardiac catheterization, and electrocardiogram (ECG). Cardiac catheterization is an invasive procedure that requires expertise and training. In addition, patients are exposed to procedural risk, albeit small, as well as radiation and potentially nephrotoxic contrast. With EST, ECGs are recorded during treadmill exercise, which can be associated with a very small risk of cardiac arrest (Robert et al. 2013). Hence, not all MI patients can undergo this test due to the associated risk. Thus, the standard ECG remains the most common diagnostic tool for detection of MI, especially in the acute setting. As the ECG abnormalities associated with MI, both acute and chronic, can be subtle, experts’ readings are crucial to ensure the accuracy of interpretation. To overcome these drawbacks, computer aided diagnostic systems (CAD) should be designed to extract the pertinent parameters. These parameters can then be input to classifiers for the categorization of normal versus MI patients (Robert et al. 2013; Liu et al. 2014).
CAD tools are developed to detect a disorder and to minimize intra- and inter-observer variability (Jahmunah et al. 2019a, b). In general, CAD systems can be classified into online- and offline-based systems. Table 1 shows the summarized works for MI detection using a CAD system.
2 Literature review
Herein, a novel algorithm involving an offline processing system for diagnosing MI with the ECG is described and discussed. Acquired ECG signals were subjected to CAD after pre-processing to remove noise. A nonlinear feature extraction (Paul et al. 2019) operation was implemented to extract suitable features from the signals. These features were then subjected to a feature selection process, wherein the system selected the best features which would improve analysis speed. Student’s t-test was utilized to obtain significant features, which were ranked based on their t-values and then input to the classifier. The classifier was designed to distinguish between normal and MI signals.
3 Computer aided diagnostic systems (CAD)
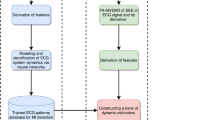
ACAD system comprises four main units: preprocessing of signals, nonlinear feature extraction, selection of the most significant features, and classification of these selected features into normal versus MI. Figure 1 illustrates a proposed CAD system.
3.1 Raw ECG signals
ECG signals were obtained from PhysioBank, the Physikalisch-Techische Bundesanstalt (PTB) Diagnostic ECG database (Goldberger et al. 2000). This database contains publicly available digitized ECG data from patients with different heart diseases, for training of models. The signals were recorded using a PTB sample recorder. The database contained 52 healthy and 148 subjects with acute and/or chronic MI and the entire dataset was used in this study. The normal and MI signals were sampled at a rate of 1000 Hz. In this work, only Lead II ECG signals were considered.
3.2 Pre-processing
At this phase, the discrete wavelet transform (DWT) was utilized for noise removal. DWT was employed to decompose the signals, using the Daubechies wavelet 6 (db6). The approximate and detail coefficients with high and low pass filters were obtained to eliminate noise including baseline wander (0–0.5 Hz) and power-line interference (50–150 Hz) (Acharya et al. 2016a, b).
After DWT, empirical mode decomposition (EMD) (Pal and Mitra 2012) was then employed. Various intrinsic mode functions (IMFs) were obtained by decomposing the EMD signals, and low frequency components were removed using a notch filter. The denoised signals were reconstructed once the low frequency components were removed.
DWT is dependent on a prior choice of wavelet basis and taps on experts’ experience in determining the level of decomposition to extract the signals (Labate et al. 2013). EMD on the other hand, does not require the level of decomposition to be set beforehand and hence, overcomes the limitations of DWT (Labate et al. 2013). After the baseline wander and power-line interference were removed, the Pan-Tompkins algorithm was employed for R-peak detection (Pan and Tompkins 1985). Once the R point was detected, a two-second signal segmentation was done, considering 1749 samples to the left of R peak and 250 samples to the right of R peak at a 1 kilo Hertz sampling rate. Thus, a total of 2000 sample points were selected for each ECG beat analysis.
Two-second segments were extracted, comprising 16826 normal 3796 MI segments, respectively. Figure 2 shows the 2-s ECG signals in (a) normal and (b) MI.
3.3 Feature extraction
Naturally, ECG signals are disorderly and vary over time. These characteristics cause the extraction of nonlinear features to be used preferably in the development of CAD systems for MI diagnosis. Time, frequency, and time–frequency domain analyses are unable to detect the inherent variations in the ECG, but nonlinear techniques can be used to extract characteristic features. Many studies have reported using various nonlinear methods (Jahmunah et al. 2019a). The computational demands and time increase when nonlinear methods are utilized to obtain features from DWT coefficients, but it facilitates the extraction of large numbers of suitable features (Acharya et al. 2017a; b, c, d, e). In our study, we extracted the following nonlinear features: Bispectrum, Recurrence Qualitative Analysis (RQA), Approximate Entropy, Permutation Entropy, Detrended Fluctuation Analysis, Fractal Dimension, Largest Lyapunov Exponent, Sample Entropy, Rényi Entropy, Hurst Exponent, Tsallis Entropy, Kolmogorov Sinai Entropy, Fuzzy Entropy, Modified Multiscale Entropy (MMSE), Wavelet Entropy, and correlation Dimension.
3.3.1 Bispectrum
The Higher order spectrum (HOS) is a used to evaluate non-stationary and non-Gaussian signals (Acharya et al. 2015, 2017a; b, c, d, e; Pham et al. 2020a, b). It detects the diversion from phase correlations and Gaussian level among frequency components of the signal (Jahmunah et al. 2019a, b; Acharya et al. 2015). HOS retains phase information as it is more immune to noise.
3.3.2 Recurrence qualitative analysis (RQA)
The RQA computes the total number of recurrences in order to calculate ECG signal complexity (Webber and Zbilut 1994). The RQA includes parameters of transitivity, determinism, laminarity, mean diagonal line length, the entropy of diagonal length, trapping time, recurrence time, recurrence rate, and recurrence time entropy (Zbilut and Webber 1992; Zbilut et al. 2002).
3.3.3 Approximate entropy (ApEn)
This entropy helps to characterize the instability or irregularity (Pincus 1991) in the ECG signal. The approximate entropy is higher for more irregularity.
3.3.4 Fractal dimension (FD)
It is a powerful tool used to measure the complexity of fractals by changing the measurement scale. The fractal patterns are characterized by quantifying their complexity as the ratio of change in the detail to the change in scale (Acharya et al. 2011).
3.3.5 Permutation entropy (PEnt)
Permutation entropy is a measure of random time series data depending upon the examination of the permutation pattern. It measures the complexity in ECG signals by calculating the coupling between time series data (Bandt and Pompe 2002).
3.3.6 Detrended fluctuation analysis (DFA)
To obtain the self-similarity characteristics of ECG signals, DFA is used (Peng et al. 1996). The variants such as multi fractal DFA and root mean square (RMS) are implemented along with DFA features (Jahmunah et al. 2019a, b).
3.3.7 Hurst exponent (HE)
The Hurst Exponent (Hurst 1956) is an evaluation of self-similarity and predictability in the ECG. If the magnitude of the Hurst exponent is high, it indicates a smoother and simpler signal (Acharya et al. 2019).
3.3.8 Largest Lyapunov exponent (LLE)
The LLE is evaluated to assess the degree of chaos existing in the signals (Rosenstein et al. 1993). If the value of LLE is higher, then the signals are more complex.
3.3.9 Sample entropy (SampEn)
This technique calculates the complexity and regularity of a physiological time series signal, and is not constrained by the pattern length (Richman and Randall 2000; Song and Liò 2010). If the value of SampEn is high, then the time series signal is less predictable; if SampEn is of low value, then it implies that the signal is more predictable.
3.3.10 Tsallis entropy (TEnt)
It is used to calculate the differences and memory consequences in the signal (Acharya et al. 2018a, b). Tsallis coefficients help in characterizing ECG bursts, spikes, and continuous rhythms.
3.3.11 Fuzzy entropy (FEnt)
Fuzzy entropy is used to estimate ECG signal unpredictability (Acharya et al. 2018a, b). This entropy indicates the degree of randomness, and is calculated as the entropy of a fuzzy set whose elements have varying degrees of membership.
3.3.12 Kolmogorov-Sinai entropy (K-SEnt)
Kolmogorov Sinai entropy is used to calculate the uncertainty in the ECG over time (Acharya et al. 2019; Ziv and Lempel 1977).
3.3.13 Multivariate multi-scale entropy (MMSEn)
In MMSEn, nonstationary ECG signals are considered to calculate the intrinsic correlation and express the degree of correlation in the time series (Acharya et al. 2018b, a; Hu and Liang 2012).
3.3.14 Rényi entropy (RE)
Rényi entropy is commonly known as the generalized type of Shannon entropy. The sudden variations in the time series data can be elucidated through RE (Renyi 1961; Shannon 1948).
3.3.15 Wavelet entropy (WE)
Wavelet entropy is used to measure the level of disorder as well as to compute the energy levels in the various frequency bands of the ECG signals (Rosso et al. 2001).
3.3.16 Correlation dimension (D2)
Correlation dimension calculates self-similarity in the ECG signals (Renyi 1961). The correlation integral C(r) is calculated first, and then the gap between N pairs of data points are measured (Jahmunah et al. 2019a, b).
3.4 Feature selection
Feature selection determines the best features and inputs them to the classification algorithm, to increase the speed of the CAD system. In the current work, the t test is used to select highly significant features. The t-distribution is defined as a continuous probability distribution for calculating population mean when sample size is small and the variance of the population is unknown. The t distribution is similar to the normal distribution. It is bell-shaped and symmetrical about the mean (Student Biometrika 1908). Features with a p-value lower than threshold are considered to be significant (Acharya et al. 2013). Here, the significant level corresponds to the probability of rejecting the null hypothesis, given that the condition is true. Figure 3 presents the 45 nonlinear parameters that were extracted from the ECG signals of MI patients and normal subjects using lead II.
3.5 Classification
Classification of data is broadly grouped into two types: parametric and nonparametric classifiers. The parametric type classifies the data based upon the statistical distribution of the class, whereas nonparametric classifiers rely upon conditions of the unknown density function. In this work, K-Nearest Neighbor and Decision Tree are implemented, and they are categorized as nonparametric classifiers.
3.5.1 K-Nearest Neighbor (KNN)
KNN is a nonparametric classification method. KNN classifies the test sample by estimating the distance between the training and testing set. The class of the test sample is decided based on the nearest samples present in the training set. The distance is estimated using Euclidean distance (Acharya et al. 2013).
3.5.2 Decision Tree (DT)
DT is a graph in which a branching technique is used to decide possible outcomes. The decision tree splits the complex decisions into simpler decisions in order to extract the rules for the recognition of the class of data (Peng et al. 1996). The output of the decision tree is a binary structure. Here the input and outputs are deliberated as the root and terminal/leaf nodes respectively.
3.5.3 Support vector machine (SVM)
This is a supervised learning tool which simultaneously minimizes the empirical classification error and maximizes the geometric margin. SVM classifiers have two types of data: separable and non-separable data. For non-separable data, kernels such as radial basis aid in the classification process (Liu et al. 2013).
3.5.4 Probabilistic Neural Network (PNN)
PNN is a supervised learning algorithm. It comprises an input layer, representing the feature vector. This input layer is totally interconnected to the concealed layer, wherein these layers connect to the output representing the choices in which the input data can be classified (Acharya et al. 2019).
The three classifiers are trained and tested using ten-fold cross-validation strategy.
4 Results
20,622 beats were segmented in total, from 52 normal and 148 MI subjects. The two second segmentation was done in order to find the R-peak. Performance of the classifier was estimated based on the ranked features and was assessed by calculating the average accuracy, Positive Predictive Value (PPV), sensitivity, and specificity (Acharya et al. 2011). Table 2 provides a summary of the diagnostic performance for detection of MI versus normal using recurrence time, HOS, entropy, bispectrum, DFA, LLE, Hurst exponent, fractal dimension, permutation entropy, MMSE, Tsallis entropy, wavelet entropy, sample entropy, and approximate entropy features extracted from ECG signals with a duration of two seconds. A feature is statistically significant if p < 0.05; the t value indicates ranking of features (Hagiwara et al. 2018; Bishop 2006) If the t-value is higher, it represents features that are more significant. Hence, there is an inverse relationship between t-value and p-value.
Table 3 shows the best performance of KNN, DT, SVM, and PNN classifiers used in our study. The highest accuracy, sensitivity, and specificity values of 97.96%, 98.89%, 93.81% were achieved respectively, with the SVM classifier.
5 Discussion
In this work, a nonlinear feature extraction method was investigated for the detection of normal versus MI from lead II ECG signals. This technique was used because it detects intrinsic complexity existing in the time series (Hurst 1956). Table 1 is a comparison of the results obtained from different CAD systems using a similar database.
From Table 1, it is apparent that various methods were proposed to develop CAD systems. The nonlinear analysis and time–frequency domain approaches were commonly employed (Hariharan et al. 2012). Hence, CAD systems along with a feature extraction technique yielded enhanced diagnostic accuracy as compared to single-type feature extraction techniques (Hagiwara et al. 2018). Bhaskar (2015), Dohare et al. (2018), Kora (2017), Padhy and Dandapat (2017), Sadhukhan et al. (2018), Bharadwaj et al. (2018), Han and Shi (2019), Seenivasagam and Chitra (2016) achieved lower classification accuracies as compared to our study. Although Acharya et al. (2016a, b), Kumar et al. (2017), Sharma et al. (2018), Lin et al. (2020) achieved higher classification accuracies, the methods they had proposed are computationally intensive as compared to the Pan-Thompkins algorithm, which is computationally more efficient. Although Liu et al. (2018a, b), Zhang et al. (2019), Tripathy et al. (2019), Alghamdi et al. (2020), Han and Shi (2020), Fu et al. (2020), Ramesh et al. (2020), Huang et al. (2020) achieved relatively higher classification accuracies than our study, these authors explored deep learning techniques, which require a lot of time to train the model as compared to our technique. Also, these authors have used small data sizes to validate their models; hence robustness is not guaranteed. Deep learning models should be validated with a larger data. Ribeiro et al. (2020) had employed a deep neural network with larger data, but still obtained a lower accuracy than our study. Acharya et al. (2017a, b, c, d, e) also achieved higher classification accuracies, but the authors investigated a different classification from our study. They did a 3-class classification and 4-class classification study, respectively. Venu et al. (2019), Haddadi et al. (2019) had employed deep learning methods but still achieved lower accuracies than our study. Hence, our method is technically sound as we have achieved a relatively high classification accuracy with the usage of a less computationally intensive algorithm. Therefore, a faster diagnosis of MI is possible.
It should be noted that recurrence time yielded more statistically significant features. The means for normal class features are greater than for the MI class. Figures 4, 5, 6 indicate recurrence, bispectrum and cumulant plots, respectively. These plots are different for normal versus MI classes. Figure 4 shows recurrence plots for normal and MI. In this, the recurrence parameter indicates the degree of feature repetition in the ECG signals. In the plot, blue dots indicate recurrence at a specific time interval. Figure 5 represents bispectrum and contour plots. The cumulant and contour plots of normal and MI signals are presented in Fig. 6. It is evident that the recurrence plots are distinct in the normal and MI classes. Also, the patterns in the bispectrum and cumulant plots are unique and show good separation between the two classes. Hence, the non-linear features used in our study are highly discriminatory. There are some benefits and limitations of our study, following.
5.1 Benefits
-
1.
The recommended system enables rapid diagnosis of MI.
-
2.
The system has been validated by ten-fold cross-validation; hence it is reliable.
-
3.
The system could be used to find anomalies associated with MI in real-time analysis.
-
4.
The top few features exhibit a high degree of separation and hence, can be used to develop the index (Pham et al. 2020a, b)
5.2 Limitations
-
1.
Only a small data size can be used for this technique.
-
2.
The manual extraction and selection of features can be a laborious task.
A CAD system is advantageous due to its versatility, rapidness, reliability, and the fact that it can be used to find anomalies associated with MI in real-time analysis.
5.3 Challenges in myocardial infarction prediction
The features of ECG, including its peak deflections, time domain amplitude, and duration, provide information regarding myocardial characteristics. The time domain features are not able to provide exact discrimination between normal and MI beats (Martis et al. 2014). The wavelet domain can be used to distinguish two dissimilar ECG signals with the same magnitude due to an increase in time resolution and a compromise in the frequency resolution. The challenge lies in selecting an optimal wavelet basis function.
Assuming that noise and ECG signals are seen in separate frequency components, linear models can be used. However when ECG signals and noise overlap the same frequency spectrum, random processes cannot be described using linear models (Chua et al. 2010). Nonlinear methods have some limitations including: (i) these techniques do not follow both principles of superposition and homogeneity, (ii) they are computationally rigorous, (iii) linear shift invariance is not valid for a nonlinear system, and (iv) reflection and symmetry properties are not followed.
6 Future work
The amplitude peaks provide information concerning cardiac characteristics. However, minute alterations in peak deflection morphology and position cannot be identified clearly by visual inspection. In signal processing terms, the time domain features are not able to provide discriminative information for normal versus abnormal beats (Lih et al. 2020; Clifford et al. 2006). Herein, we have listed different non-linear features used by different authors for testing (Kannathal et al. 2006). The linear method provides satisfactory classification accuracy. Hence, for future work, testing should be done on noise-free data using nonlinear methods, as they can be implemented even under noisy conditions (Ansari et al. 2017; Acharya et al. 2017a, b, c, d, e). Additionally, more data should be added to test our recommended model to improve performance.
6.1 ECG in mobile healthcare and telecare
Apart from modifications done for ECG algorithmic analysis, there is additional scope for mobile healthcare technology, wherein ECG signals are recorded with mobile devices (Constantinescu and Jinman 2012). Patients are connected to Holter-like devices for ECG signal recording. Any emergency in patient health can be readily reported to the critical care unit. Adeli and Sankari (2011) invented a cost-effective mobile medical device for the purpose of real-time monitoring of cardiac health. A future challenge is to develop an efficient remote monitoring device to rapidly provide information about patient health and suitable treatments, using a decision support system (DSS) (Tamura 2012). There is a requirement for DSS in cardiac telecare, and a necessity for an ECG monitoring unit via wearable electrodes. This can help to capture data via a web-based unit, and can be employed for instantaneous screening of ECG signals by doctors. The system should be cost-effective, low in power consumption, and should be affordable to the patient (Venkatesan et al. 2018; Pandey et al. 2012) Fig. 7 presents the ECG recognition system in a clinical setting.
In this system, mobile devices and sensors are connected wirelessly through cloud computing devices. Without processing ECG data, signals are sent to mobile devices from different sensing devices. As the data is not processed and is generated from different devices with continuous monitoring, it will require more space. To store large volumes of data, cloud storage space is utilized. The unknown data is sent to the developed model at the cloud for diagnosis. The diagnostic results can be sent to healthcare centers and physicians (Acharya et al. 2018a, b).
The advent of cloud computing has enabled host software packs to analyze ECG data, including Software-as-a-Service (SaaS), Platform-as-a-Service (PaaS), and Infrastructure-as-a-Service (IaaS). The SaaS layer is used for organizing custom-designed analysis of current and historic ECG data. The PaaS layers supervise the execution of software into three major units: (i) Workflow Engine, (ii) Container scaling manager, and (iii) Aneka. The IaaS layers are self-service models for monitoring, accessing, and managing remote datacenter infrastructures, such as compute, storage, and networking services (LeCun et al. 2015).
Deep learning is the part of machine learning methods in which hidden layers of neurons are used to construct inherent features. For big datasets, deep learning methods can be used as it performs better than machine classification and classic analysis methods. The word “deep” is obtained from various hidden layers in an artificial neural network (ANN) (Fukushima and Miyake 1982). The ANN structure comprises the input, hidden, and output layers. Connection link is used to connect every nerve cell of one layer to every nerve cell of another layer. These computational nerve cells are composed of dendrites (input), axon (output), nucleus (activation function), node (soma), and synapses (weights) (Goodfellow et al. 2016). Here, the input signals are dendrites, weight models are synapses, and the activation function is the nucleus in the biological neuron. The ANN structure adversely affects the performance of classification due to susceptibility to translation and shift deviation (LeCun and Bengio 1995). To overcome this drawback, a Convolutional Neural Network (CNN) can be used. The CNN ensures shift invariance and translation. CNN is a feedforward network which consists of pooling, convolution, and fully connected layers (Hinton and Salakhutdinov 2006; Lih et al. 2020; Murat et al. 2020).
Other deep learning techniques include the autoencoder, Recurrent Neural Network (RNN), and deep generative models, which are used to evaluate physiological signals. For the autoencoder, the input dimension is the same as the output, and it is an unsupervised neural network. The autoencoder has three layers: input, output, and hidden. For the autoencoder, encoding and decoding are two important steps (Hinton et al. 2006; Hopfield 1987).
The Deep Generative model comprises the Restricted Boltzmann Machine (RBM) and Deep Belief Network (DBN). The RBM consists of two layers: (i) visible and (ii) hidden layers. The DBN is the probabilistic model with various hidden layers. In RBM, the first layer is trained to reconstruct the input (Hinton et al. 2006).
The RNN is a recurrent network which follows a recursive approach. The Long Short-Term Memory (LSTM) network, which has the capability to learn long term dependencies, is a common type of RNN. The LSTM consists of three gates to incorporate memory block; they are: input, output, and forget gate. These gates assist in adding and removal of information from the network, based on cell state (Hopfield 1987).
For future work, we intend to employ a deep learning model and use a larger dataset to train our model. We hope to integrate this developed model to the cloud system such that information about patient health from the acquired ECG signals could be sent to the clinicians to aid them in their diagnostic and treatment decisions.
7 Conclusion
MI is an irreversible damage to the myocardium due to coronary artery blockage. The expansion of MI can be rapid if it is not treated expediently. If left untreated, further damage can occur to myocardial function and the structure of left ventricle. The recommended approach to distinguish between normal and MI signals involves a non-invasive tool. In this work, nonlinear parameters were extracted from lead II ECG signals. The performance of each classifier was measured using accuracy, sensitivity, and specificity. The highest accuracy of 97.96% was achieved with the SVM classifier using nonlinear features with a ten-fold cross-validation strategy. We have also proposed unique recurrence and HOS plots for normal and MI ECG classes to identify the two classes qualitatively. The proposed system outperforms other recently developed techniques, and has the potential to be readily deployed in hospitals and clinics, even in remote areas. CAD systems provide rapid and accurate results, and can be implemented for real-time diagnosis of MI.
Availability of data and material
The data used in this study is available in the publicly available PTB ECG database.
References
Acharya UR, Chua ECP, Faust O, Lim TC, Lim LFB (2011) Automated detection of sleep apnea from electrocardiogram signals using nonlinear parameters. Physiol Meas. https://doi.org/10.1088/0967-3334/32/3/002
Acharya UR, Faust O, Sree SV, Ghista DN, Dua S, Joseph P et al (2013) An integrated diabetic index using heart rate variability signal features for diagnosis of diabetes. Comput Methods Biomech Biomed Eng 16:222–234. https://doi.org/10.1080/10255842.2011.616945
Acharya UR, Sudarshan VK, Adeli H, Santhosh J, Koh JEW, Adeli A (2015) Computer-aided diagnosis of depression using EEG signals. Eur Neurol. https://doi.org/10.1159/000381950
Acharya UR et al (2016a) Automated detection and localization of myocardial infarction using electrocardiogram: a comparative study of different leads. Knowledge Based Syst 99(2016):146–156. https://doi.org/10.1016/j.procs.2015.01.043
Acharya UR, Fujita H, Sudarshan VK, Lih OS, Adam M (2016b) Automated detection and localization of myocardial infarction using electrocardiogram: a comparative study of different leads. Knowl Based Syst 9(8):1–39. https://doi.org/10.1016/j.knosys.2016.01.040
Acharya UR et al (2017a) Application of higher-order spectra for the characterization of Coronary artery disease using electrocardiogram signals. Biomed Signal Process Control. https://doi.org/10.1016/j.artmed.2019.07.006
Acharya UR, Fujita H, Oh SL, Hagiwara Y, Tan JH, Adam M (2017b) Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inf Sci 415–416:190–198. https://doi.org/10.1016/j.ins.2017.06.027
Acharya UR, Fujita H, Adam M, Lih OS, Sudarshan VK et al (2017c) Automated characterization and classification of coronary artery disease and myocardial infarction by decomposition of ECG signals: a comparative study. Inf Sci 377:17–29. https://doi.org/10.1016/j.ins.2016.10.013
Acharya UR et al (2017d) Automated characterization and classification of coronary artery disease and myocardial infarction by decomposition of ECG signals: a comparative study. Inf Sci 377:17–29. https://doi.org/10.1016/j.ins.2016.10.013
Acharya UR et al (2017e) Automated characterization of coronary artery disease, myocardial infarction, and congestive heart failure using contourlet and shearlet transforms of electrocardiogram signal. Knowl Based Syst 132:156–166. https://doi.org/10.1016/j.knosys.2017.06.026
Acharya UR et al (2018a) Entropies for automated detection of coronary artery disease using ECG signals: a review. Biocybern Biomed Eng. https://doi.org/10.1016/j.bbe.2018.03.001
Acharya UR et al (2018b) Automated identification of shockable and non-shockable life-threatening ventricular arrhythmias using convolutional neural network. Futur Gener Comput Syst. https://doi.org/10.1016/j.future.2017.08.039
Acharya UR et al (2019) Application of nonlinear methods to discriminate fractionated electrograms in paroxysmal versus persistent atrial fibrillation. Comput Methods Programs Biomed. https://doi.org/10.1016/j.cmpb.2019.04.018
Alghamdi A et al (2020) Detection of myocardial infarction based on novel deep transfer learning methods for urban healthcare in smart cities. Multimed Tools Appl. https://doi.org/10.1007/s11042-020-08769-x
Ansari S, Farzaneh N, Duda M, Horan K, Andersson HB, Goldberger ZD, Nallamothu BK, Najarian K (2017) A review of automated methods for detection of myocardial ischemia and infarction using electrocardiogram and electronic health records. IEEE Rev Biomed Eng 10:264–298. https://doi.org/10.1109/RBME.2017.2757953
Baloglu UB, Talo M, Yildirim O, Tan RS, Acharya UR (2019) Classification of myocardial infarction with multi-lead ECG signals and deep CNN. Pattern Recognit Lett 122:23–30. https://doi.org/10.1016/j.patrec.2019.02.016
Bandt C, Pompe B (2002) Permutation entropy: a natural complexity measure for time series. J Physiol 88(16):3–17. https://doi.org/10.1103/PhysRevLett.88.174102
Bharadwaj AV, Upadhyaya SM, Sharath L, Srinivasan R (2018) Early diagnosis and automated analysis of myocardial infarction (STEMI) by detection of ST segment elevation using wavelet transform and feature extraction. Int Conf Des Innov 3Cs Comput Commun Control ICDI3C, pp 24–28. https://doi.org/10.1109/ICDI3C.2018.00014
Bhaskar NA (2015) Performance analysis of support vector machine and neural networks in detection of myocardial infarction. Procedia Comput Sci 46:20–30
Bishop CM (2006) Pattern recognition and machine learning. Springer-Verlag, Berlin, Heidelberg
Chua KC, Chandran V, Acharya UR, Lim CM (2010) Application of higher order statistics/spectra in biomedical signals—a review. Med Eng Phys 32:679–689. https://doi.org/10.1016/j.medengphy.2010.04.009
Clifford GD, Azuaje F, McSharry P (2006) Advanced methods and tools for ECG data analysis. Artech House, Norwood
Constantinescu L, Jinman K (2012) Feng DD (2012) Spark Medical: a framework for dynamic integration of multimedia medical data in to distributed m-health systems. IEEE Transl Inf Technol Biomed 16:40–52. https://doi.org/10.1109/TITB.2011.2174064
Dohare AK, Kumar V, Kumar R (2018) Detection of myocardial infarction in 12 lead ECG using support vector machine. Appl Soft Comput 64:138–147. https://doi.org/10.1016/j.asoc.2017.12.001
Feng K, Pi X, Liu H, Sun K (2019) Myocardial infarction classification based on convolutional neural network and recurrent neural network. Appl Sci 9(9):1–12. https://doi.org/10.3390/app9091879
Fu L, Lu B, Nie B, Peng Z, Liu H, Xi P (2020) Hybrid network with attention mechanism for detection and location of myocardial infarction based on 12-lead electrocardiogram signals. Sensors (Basel). https://doi.org/10.3390/s20041020
Fukushima K, Miyake S (1982) Neocognitron: a self-organizing neural network model for a mechanism of visual pattern recognition, In: Competition and cooperation in neural nets. Springer, pp 267–285. https://doi.org/10.1007/BF00344251
Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC (2000) PhysioBank, Physiotool Kit, and Physionet components of a new research resource for complex physiologic signals. Circulation 101(4):215–220
Goodfellow I, Bengio Y, Courville A (2016) Deep learning. MIT Press, Cambridge
Haddadi R, Abdelmounim E, El Hanine M, Belaguid A (2019) A wavelet-based ECG delineation and automated diagnosis of myocardial infarction in PTB database, 2284216
Hagiwara Y, Fujita H, Oh SL, Acharya UR et al (2018) Computer-aided diagnosis of atrial fibrillation based on ECG signals: a review. Inf Sci 32(12):2–38. https://doi.org/10.1016/j.ins.2018.07.063
Han C, Shi L (2019) Automated interpretable detection of myocardial infarction fusing energy entropy and morphological features. Comput Methods Programs Biomed 175:9–23. https://doi.org/10.1016/j.cmpb.2019.03.012
Han C, Shi L (2020) ML–ResNet: a novel network to detect and locate myocardial infarction using 12 leads ECG. Comput Methods Programs Biomed. https://doi.org/10.1016/j.cmpb.2019.105138
Hariharan M, Vijean V, Yaacob (2012) Objective analysis of vision impairments using single trial VEPs. Int Conf Biomed Eng (ICoBE). https://doi.org/10.1109/CCDC.2013.6561736
Hinton GE, Salakhutdinov RR (2006) Reducing the dimensionality of data with neural networks. Science 313(5786):504–507. https://doi.org/10.1126/science.1127647
Hinton GE, Osindero S, The YW (2006) A fast learning algorithm for deep belief nets. Neural Comput 18(7):1527–1554. https://doi.org/10.1162/neco.2006.18.7.1527
Hopfield JJ (1987) Neural networks and physical systems with emergent collective computational abilities, in: spin glass theory and beyond: an introduction to the replica method and its applications. World Sci. https://doi.org/10.1073/pnas.79.8.2554
Hu M, Liang H (2012) Adaptive multiscale entropy analysis of multivariate neural data. IEEE Trans Biomed Eng 59:12–15. https://doi.org/10.1109/TBME.2011.2162511
Huang JS, Chen BQ, Zeng NY, Cao XC, Li Y (2020) Accurate classification of ECG arrhythmia using MOWPT enhanced fast compression deep learning networks. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02110-y
Hurst HE (1956) Methods of using long-term storage in reservoirs. Proc Inst Civ Eng 5(5):519–543. https://doi.org/10.1680/iicep.1956.11503
Jahmunah V et al (2019a) Automated detection of schizophrenia using nonlinear signal processing method. Artif Intell Med. https://doi.org/10.1016/j.artmed.2019.07.006
Jahmunah V et al (2019b) Computer-aided diagnosis of congestive heart failure using ECG signals—a review. Phys Med. https://doi.org/10.1016/j.ejmp.2019.05.004
Kannathal N, Lim CM, Rajendra Acharya U, Sadasivan PK (2006) Cardiac state diagnosis using adaptive neuro-fuzzy technique. Med Eng Phys. https://doi.org/10.1109/IEMBS.2005.1615304
Kora P (2017) ECG based myocardial infarction detection using hybrid firefly algorithm. Comput Methods Programs Biomed 152:141–148
Kumar N, Pachori RB, Acharya UR (2017) Automated diagnosis of myocardial infarction ECG signals using sample entropy in flexible analytic wavelet transform framework. Entropy. https://doi.org/10.3390/e19090488
Labate D, Foresta FL, Occhiuto G, Morabito FC, Lay-Ekuakille A, Vergallo P (2013) Empirical mode decomposition vs. wavelet decomposition for the extraction of respiratory signal from single-channel ECG: a comparison. IEEE Sens J 13(7):2666–2674. https://doi.org/10.1109/JSEN.2013.2257742
Lecun Y, Bengio Y (1995) Convolutional networks for images, speech, and time series. In: Arbib MA (ed) The hand book on brain theory neural networks. MIT Press
Lecun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
Ley K (2015) Atherosclerosis, thrombosis and vascular biology. J Am Heart Assoc 15(9):436–448. https://doi.org/10.1161/ATVBAHA.117.309813
Lih OS, Jahmunah V, San TR, Ciaccio EJ, Yamakawa T, Tanabe M, Kobayashi M, Faust O (2020) Comprehensive electrocardiographic diagnosis based on deep learning. Artif Intell Med 103:101789
Lin Z, Gao Y, Chen Y, Ge Q, Mahara G, Zhang J (2020) Automated detection of myocardial infarction using robust features extracted from 12-lead ECG. Signal Image Video Process. https://doi.org/10.1007/s11760-019-01617-y
Liu C, Wang H, Lu Z (2013) EEG classification for multiclass motor imagery BCI. Control and decision conference (CCDC) 25th Chinese, pp 4450–4453. https://doi.org/10.1109/CCDC.2013.6561736
Liu B, Liu J, Wang G, Huang K, Li F et al (2014) A novel electrocardiogram parameterization algorithm and its application in myocardial infarction detection. Jpn J Clin Med 27(12):2892–2900. https://doi.org/10.1016/j.compbiomed.2014.08.010
Liu W, Huang Q, Chang S, Wang H, He J (2018a) Multiple-feature-branch convolutional neural network for myocardial infarction diagnosis using electrocardiogram. Biomed Signal Process Control 45:22–32. https://doi.org/10.1016/j.bspc.2018.05.013
Liu W et al (2018b) Real-time multilead convolutional neural network for myocardial infarction detection. IEEE J Biomed Heal Inf 22(5):1434–1444. https://doi.org/10.1109/JBHI.2017.2771768
Liu W, Wang F, Huang Q, Chang S, Wang H, He J (2020) MFB-CBRNN: a hybrid network for MI detection using 12-Lead ECGs. IEEE J Biomed Health Inf 24(2):503–514. https://doi.org/10.1109/JBHI.2019.2910082
Lodhi AM et al (2018) A novel approach using voting from ECG leads to detect myocardial infarction. IntelliSys. https://doi.org/10.1007/978-3-030-01057-7_27
Lui HW, Chow KL (2018) Multiclass classification of myocardial infarction with convolutional and recurrent neural networks for portable ECG devices. Inf Med Unlocked 13:26–33. https://doi.org/10.1016/j.imu.2018.08.002
Martis JR, Acharya UR, Adeli H (2014) Current methods in electrocardiogram characterization. Comput Biol Med 48:133–149. https://doi.org/10.1016/j.compbiomed.2014.02.012
Murat F, Yildirim O, Talo M, Baloglu UB, Demir Y, Acharya UR (2020) Application of deep learning techniques for heartbeats detection using ECG signals-analysis and review. Comput Biol Med 120:103726
Padhy S, Dandapat S (2017) Third-order tensor based analysis of multilead ECG for classification of myocardial infarction. Biomed Signal Process Control 31:71–78. https://doi.org/10.1016/j.bspc.2016.07.007
Pal S, Mitra M (2012) Empirical mode decomposition based ECG enhancement and QRS detection. Comput Biol Med 42(1):83–92. https://doi.org/10.1016/j.compbiomed.2011.10.012
Pan J, Tompkins WJ (1985) A real time QRS detection algorithm. IEEE Trans Biomed Eng 32(3):230–236. https://doi.org/10.1109/TBME.1985.325532
Pandey S, Voorsluys W, Niu S, Khandoker A, Buyya R (2012) An autonomic cloud environment for hosting ECG data analysis services. Future Gener Comput Syst 28:147–154. https://doi.org/10.1016/j.future.2011.04.022
Paul JK, Iype T, Hagiwara DRY, Koh JEW, Acharya UR (2019) Characterization of fibromyalgia using sleep EEG signals with nonlinear dynamical features. Comput Biol Med. https://doi.org/10.1016/j.compbiomed.2019.103331
Peng CK, Havlin S, Hausdorf JM, Mietus JE et al (1996) Fractal mechanisms and heart rate dynamics. J Electro Cardiol 8(15):59–64. https://doi.org/10.1016/S0022-0736(95)80017-4
Pham TH, Vicnesh J, Wei JKE, Oh SL, Arunkumar N, Abdulhay EW (2020a) Autism spectrum disorder diagnostic system using HOS bispectrum with EEG signals. Int J Environ Res Public Health 17(3):971
Pham TH, Raghavendra U, Koh JEW, Gudigar A, Chan WY, Hamid MTR, Rahmat K, Fadzil F, Ng KH, Ooi CP, Ciaccio EJ, Fujita H, Acharya UR (2020b) Development of breast papillary index for differentiation of benign and malignant lesions using ultrasound images. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02310-6
Pincus SM (1991) Approximate entropy as a measure of system complexity. Proc Natl Acad Sci 88(12):2297–2301. https://doi.org/10.1073/pnas.88.6.2297
Ramesh G, Satyanarayana D, Sailaja M (2020) Composite feature vector based cardiac arrhythmia classification using convolutional neural networks. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02259-6
Reasat T, Shahnaz C (2018) Detection of inferior myocardial infarction using shallow convolutional neural networks. In: 5th IEEE Reg 10 Humanit Technol Conf 2017, R10-HTC 2017, pp 718–721. https://doi.org/10.1109/R10-HTC.2017.8289058
Renyi A (1961) On measures of entropy and information. Math Stat Probab 1(4):547–561
Ribeiro AH, Ribeiro MH, Paixão GMM, Oliveira DM, Gomes PR, Canazart JA (2020) Automatic diagnosis of the 12-lead ECG using a deep neural network. Nature Commun 11(1):1–9
Richman JS, Randall MJ (2000) Physiological time-series analysis using approximate entropy and sample entropy. Am J Physiol Heart Circ Physiol 278:H2039–H2049
Robert H, Mangner N, Schuler G, Erbs S (2013) Physical exercise training and coronary artery disease. Rev Health Care 4(3):1–17
Roger VL (2007) Epidemiology of myocardial infarction. Med Clin North Am 91(7):537–552. https://doi.org/10.1016/j.mcna.2007.03.007
Rosenstein M, Colins JJ, Luca CJ (1993) A practical method for calculating largest Lyapunov exponents from small data sets. Physica D 65:117–134. https://doi.org/10.1016/0167-2789(93)90009-P
Rosso OA, Blanco S, Yordanova J, Kolev V et al (2001) Wavelet entropy: a new tool for analysis of short duration electrical signals. J Neurosci Methods 105:65–75. https://doi.org/10.1016/S0165-0270(00)00356-3
Sadhukhan D, Pal S, Mitra M (2018) Automated identification of myocardial infarction using harmonic phase distribution pattern of ECG Data. IEEE Trans Instrum Meas 67(10):2303–2313. https://doi.org/10.1109/TIM.2018.2816458
Sankari Z, Adeli H (2011) Heart Saver: a mobile cardiac monitoring system for auto-detection of arial fibrillation, myocardial infarction, and atrio-ventricular block. Comput Biol Med 41:211–220. https://doi.org/10.1016/j.compbiomed.2011.02.002
Seenivasagam V, Chitra R (2016) Myocardial infarction detection using intelligent algorithms. Neural Netw World 26(1):91–110
Setiawan NA, Prabowo DW, Nugroho HA (2014) Benchmarking of feature selection techniques for coronary artery disease diagnosis. In: 6th International Conference on Information Technology and Electrical Engineering, Yogyakarta, Indonesia, vol 3(1), pp 123–131. https://doi.org/10.1109/ICITEED.2014.7007898
Shannon C (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x
Sharma LN, Tripathy RK, Dandapat S (2015) Multiscale energy and eigenspace approach to detection and localization of myocardial infarction. IEEE Trans Biomed Eng 62(7):1827–1837
Sharma M, Tan RS, Acharya UR (2018) A novel automated diagnostic system for classification of myocardial infarction ECG signals using an optimal biorthogonal filter bank. Comput Biol Med 102:341–356. https://doi.org/10.1016/j.compbiomed.2018.07.005
Song Y, Liò P (2010) A new approach for epileptic seizure detection: sample entropy based feature extraction and extreme learning machine. J Biomed Sci Eng 3(6):556–567. https://doi.org/10.1152/ajpheart.2000.278.6.H2039
Strodthoff N, Strodthoff C (2019) Detecting and interpreting myocardial infarction using fully convolutional neural networks. Physiol Meas 40(1):1–11. https://doi.org/10.1088/1361-6579/aaf34d
Student Biometrika (1908) The probable error of a mean. Biometrika 6(1):1–25. https://www.jstor.org/stable/2331554
Tamura T (2012) Home geriatric physiological measurements. Physiol Meas 33(10):R47. https://doi.org/10.1088/0967-3334/33/10/R47
Thomas FL (2015) Myocardial infarction: mechanisms, diagnosis, and complications. Eur Heart J 36(16):947–949. https://doi.org/10.1093/eurheartj/ehv071
Tripathy RK, Bhattacharyya A, Pachori RB (2019) A novel approach for detection of myocardial infarction from ECG signals of multiple electrodes. IEEE Sens J 19(12):4509–4517
Venkatesan C, Karthigaikumar P, Satheeskumaran S (2018) Mobile cloud computing for ECG telemonitoring and real-time coronary heart disease risk detection. Biomed Signal Process Control 44:138–145. https://doi.org/10.1016/j.bspc.2018.04.013
Venu K, Natesan P, Krishnakumar B, Sasipriya N (2019) Classification of myocardial infarction using convolution neural network. Int J Recent Technol Eng 8(4):12763–12768
Webber CL, Zbilut JP (1994) Dynamical assessment of physiological systems and states using recurrence plot strategies. J Appl Physiol 76(2):965–973. https://doi.org/10.1152/jappl.1994.76.2.965
WHO fact sheet (2012) https://www.who.int/mediacentre/factsheets/fs310/en/index.html. Accessed 19 Jan 2016
Zbilut JP, Webber CL (1992) Embeddings and delays as derived from quantification of recurrence plots. Phys Lett 171(4):199–203. https://doi.org/10.1016/0375-9601(92)90426-M
Zbilut JP, Thomasson N, Webber CL (2002) Recurrence quantification analysis as a tool for nonlinear exploration of nonstationary cardiac signals. Med Eng Phys 24(4):53–60. https://doi.org/10.1016/S1350-4533(01)00112-6
Zhang J et al (2019) Automated detection and localization of myocardial infarction with staked sparse autoencoder and treebagger. IEEE Access 7:70634–70642. https://doi.org/10.1109/ACCESS.2019.2919068
Ziv J, Lempel A (1977) A universal algorithm for sequential data compression. IEEE Trans Inf Theory 23:337–343. https://doi.org/10.1109/TIT.1977.1055714
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sridhar, C., Lih, O.S., Jahmunah, V. et al. Accurate detection of myocardial infarction using non linear features with ECG signals. J Ambient Intell Human Comput 12, 3227–3244 (2021). https://doi.org/10.1007/s12652-020-02536-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-020-02536-4