Abstract
Electrocardiogram analysis for the classification of several cardiac arrhythmias has gained a significant research importance in the medical field. Towards such objective, this paper proposed a novel approach based on the fusion of multiple features extracted from a signal through various methods and Convolution Neural Networks. The multiple features are precisely consisting of morphological features, temporal features, and statistical features. Every electrocardiogram signal is initially pre-processed to remove the base line and then processed for segmentation through a simple strategy. Further, for every heart beat segment, three different set of features are extracted. Among them, morphological features are obtained through Dual Tree Complex Wavelet Transform and remaining features are extracted through statistical measures. Further, Principal Component Analysis is applied over the morphological feature set to reduce the dimensionality. Finally, a composite and final feature vector is formulated and then fed to Convolutional Neural Networks classifier to predict the label for a given input heartbeat. Simulation experiments conducted through the MIT-BIH benchmark database exhibited that the proposed system achieves better classification accuracy and on an average, it of 98%. Compared with state-of-art methods, the improvement is approximately 5%.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The Electrocardiogram (ECG) is a mostly used signal for the analysis of heart related abnormalities. ECG signal captures the Electric activities of the heart by connecting some electric nodes at specific points on the chest. A medical expert or a cardiologist may get fail in analysing the condition of heart due to the non-stationary nature of ECG signal which consequences to various ‘Cardiovascular Diseases (CVDs)’. According to the World Health Organization (WHO), CVDs are the major reason for the worldwide deaths. An approximate value of 17.7 million people was died in 2015 with these CVDs and it is approximately 31% of global deaths. CVDs are increasing the worldwide mortality rate, particularly in the low and middle income countries (Mendis et al. 2011). Cardiac Arrhythmias are one of the CVDs which conquered major in these deaths. ‘Arrhythmia’ means a disturbance in the heart rate which occurs due to the improper impulse formation or electrical conduction in the heart. This improper functioning of heart can disrupt regular heart beat and also can affect the morphology of normal heartbeat. Bundle Branch Block Beats (BBBB) and Ectopic Beats (EB) are the two major consequences of an arrhythmia. BBBBs hinder the normal pathway of electrical impulses through the conduction system on the ventricles which causes the deterioration of heart function, asynchronous contraction in ventricles, which may consequences to the serious life-threatening issues (Rangayyan 2015). Further, the EBs are occurred due to the improper formation of an electrical impulse. Further classification of EBs results in premature atrial and ventricular contractions. These beats can disturb the normal functioning of regular rhythm of heart and can introduce some serious arrhythmias like atrial or ventricular fibrillation. Hence the researchers focused over the investigation and of both classification and detection (Ye et al. 2012) methods for cardiac arrhythmias.
An ECG is a most popular tool for the analysis of electrical activity of a heart. The depolarization and repolarization of heart muscles is measured with the help of ECG. Further, since ECG is an inexpensive technique, generally the cardiologists employ it in their daily clinical practices. ECGs can be used for the analysis of cardiac arrhythmias, because it can measure the electrical activity of a heart and the abnormalities in the heart rate can be detected through it. However, a manual detection of various abnormalities in the heart beats is a tedious task. Hence, by recognizing several types of heartbeats, the Computer Aided Diagnosis (CAD) plays a vital role in the detection and classification of cardiac arrhythmias (Meek and Morris 2002). This will help the cardiologists to observe the physical activity of heart at regular intervals. Several investigations are done earlier for the detection and classification of cardiac arrhythmias (Lenis et al. 2013; Tian et al. 2016). Since the ECG is a signal, the CAD system applies various methods to study and analyse the ECG in various domain viz., time-domain, frequency-domain, time–frequency domain and some non-linear methods (Martis et al. 2014).
This paper proposes a new cardiac arrhythmia detection and classification system based on the fusion of multiple features such as morphological, temporal and statistical features. Since the single model features are not effectively discriminate the sensitive variations in the ECG signals, we have focused on the hybrid features. For the extraction of morphological feature, this paper accomplished Dual Tree Complex Wavelet Transform (DTCWT) followed by principal component analysis. A composite feature vector is formulated by combining all the three features for every ECG signal and it is fed to the Convolution Neural Network (CNN) algorithm to find the arrhythmia hindered in that signal. Simulation experiments are conducted over a standard benchmark dataset, MIT-BIH to alleviate the performance effectiveness.
Remaining paper is organized as follows; Sect. 2 illustrates the details of related work. The details of MIT-BIH arrhythmia database, DWT, DTCWT and CNN algorithm are described in Sect. 3. Further Sect. 4 illustrates the details of proposed detection and classification system. Experimentation details are described in Sect. 5 and finally the concluding remarks are done in Sect. 6.
2 Related work
In recent years, several methods are developed to perform the detection and classification of heartbeats extracted from ECG signals. Generally, these methods consist of three stages such as, pre-processing, feature extraction and classification. In the pre-processing phase, the raw ECG signal is subjected to the removal of external noises and also to the correction of baseline wander. After baseline correction, the ECG signal is subjected to segmentation and then the obtained every heartbeat is processed for feature extraction. In the classification phase, the extracted features are fed to classifiers to find the class label of ECG beat under consideration. Here the literature survey is also done in the same manner. Initially the method focused over the pre-processing are focused, next the feature extraction methods and finally the classification methods.
2.1 Pre-processing and segmentation
In this phase, the main focus is to enhance the quality of ECG signals and to achieve this objective; various techniques are proposed in earlier (Sameni et al. 2007; Tracey and Miller 2012; Singh and Tiwari 2006; Wang et al. 2015). Sameni et al. (2007), proposed a non-linear Bayesian filtering approach to filter the single channel ECG signals. A modified version of non-linear dynamic model is proposed by integrating several Bayesian filters, Unscented Kalman Filter, and Extended Kalman filter. Tracer and Miller (2012) proposed to apply nonlocal means (NLM) approach to the denoising of biomedical ECG signals. However, the NLM is not resilient to the baseline wander due to its non-stationary characteristics. Further, Singh and Tiwari (2006), considered the optimal selection of wavelet basis functions and the Wang et al. (2015), Parallel-type fractional zero-phase filtering for ECG signal denoising. These methods have provided an optimal performance between noise removal and the information retention. However, in these methods, the optimal selection of a wavelet basis function constitutes an extra computational burden over the system.
Some authors accomplished the wavelet transform (Wissam et al. 2016; Mounaim et al. 2017) and some used the thresholding techniques (Regis et al. 2018) for ECG signal denoising. These methods had accomplished the DWT decomposition of an ECG followed by thresholding. However, the signal reconstruction after denoising can not obtain an informative ECG. The reconstructed ECG has lost the information and this is due to the thresholding and quantization at inverse DWT. A low quality or less informative ECG can’t explore the type of arrhythmia present in it accurately. Moreover, the baseline wander noise is not effectively removed through a uniform thresholding.
Next, focusing over the baseline wander noise, Gustavo et al. (2017) conducted comparative analysis between various methods proposed for baseline removal and the preservation of ST information in ECG signal. In this paper, the comparison is done between Butterworth filter, Median, Spline, Wavelet Cancellation and Wavelet high pass filters and found that none of the methods was capable of reconstructing the original ECG without modifying the ST segment.
Next, Bono et al. (2014) proposed a new metric called Selvester Score which gives information about the depolarization changes in the ventricular regions, for the detection of QRS complex in ECG signals. However, these techniques constitutes more complexity and hence some simple QRS detection methods like Quadratic filter by Phukpattaranont (Phukpattaranont 2015), Improved Adaptive Threshold by Lu et al. (2018), and Crests and Troughs based fiducial points detection by Chen and Chuang (2017). The main motto of these methods is to detect the QRS segment and R peak by which the CA characteristics can be analyzed. But these are very simple and did not accomplish any preprocessing which has significant importance in the removal of noise.
2.2 Feature extraction
In the characterization of ECG signals, feature extraction is most important and most of the researches focused in this direction only. Basically the features are characterized as temporal, statistical (Grazia et al. 2015; Rashid Ghorbani et al. 2016; Kutlu and Kuntalp 2012) and morphological features (Rashid Ghorbani et al. 2016; Siva et al. 2018). Under the statistical features, Grazia et al. (2015) considered the mean, coefficient of variation, standard deviation, kurtosis, skewness and in phase-space diagrams for a sliding window of 10 beats. Further, in Rashid Ghorbani et al. (2016), evaluated the Kurtosis, Skewness and 5th moment as statistical parameters along with one temporal feature, RR-Interval. The Gaussian Mixture Model with Expectation Maximization is used for the fitting of probability density function of heartbeats. However, these approaches didn’t focused over the temporal features which have more importance in the detection of arrhythmias that can be analyzed through time based features such as QT-Interval, PR-Interval, and QRS interval. Moreover, the statistical features are most efficient in the discovery of any statistical deviations between normal ECG and arrhythmia existed ECG. Hence the statistical features have not much contribution towards the detection of CAs.
Next, in the case of morphological features, most of the authors used wavelet transform and based on this strategy, Siva et al. (2018) and Roghayyeh et al. (2017), developed new methods for the detection and classification of CAs in the ECG signal. Though the DWT is more effective in the detection of cardiac arrhythmias, the major drawback of DWT is lack of shift invariance and has a significant affect over the detection performance. To overcome this problem Manu et al. (2015) used DTCWT as a feature extraction technique. After extracting the sub bands through DTCWT, four statistical parameters such as AC power, Kurtosis, Skewness, Timing information are measured and then total set of features are processed for classifier. Further, Rajesh Kumar and Dandapat (2017) applied complex wavelet sub-band bi-spectrum (CWSB) as a feature extraction technique for the detection of heart ailments from a 12-lead ECG. This approach mainly focused over the detection of three CAs such as myocardial infarction (MI), heart muscle disease (HMD) and bundle branch block (BBB). Though the DTCWT has gained more effective results in the CAs detection, only morphological features won’t contribute to fruitful results. Moreover, for these studies we observed that to achieve an efficient accuracy in the detection of CAs, a deep analysis is required in which the characteristics of ECG is studied in all orientations.
Hence some authors focused on the fusion of multiple features and some approaches developed based on this strategy. Under this strategy, for a given ECG heartbeat, multiple features are extracted and they are fused and formulated into a single feature vector. In Ai et al. (2015), fused the multiple features and they are extracted based on Multi-linear sub-space learning. They used Wavelet Packet Decomposition (WPD) for feature extraction and Independent Component Analysis (ICA) for the feature fusion. Next, the method proposed by Das and Ari (2014), accomplishes two different systems, one system used S-Transform (ST) Features and Temporal features and the second system applies the mixture of temporal features along with ST features and Wavelet transform features. Compared to the single domain based features, these methods have gained an improved performance.
2.3 Classification
In the classification phase, generally most of the researchers used machine learning algorithms like Support Vector Machines (SVMs) (Mustaqeem et al. 2018), Convolutional Neural Networks (CNNs) (Zhai and Tin 2018; Zubair et al. 2016; Kiranyaz et al. 2016) etc. Among the basic machine learning algorithms SVM has gained more efficiency. Considering the variants of SVM, a multiclass classification mechanism is proposed by Mustaqeem et al. (2018) for Cardiac Arrhythmia detection though improved feature selection. Though the SVM has obtained an optimal performance, the CNNs have gained an improved significance due to its deep analyzing capabilities in various applications. Considering this fact, An Automated ECG Classification is proposed by Zhai and Tin (2018) Using Dual Heartbeat Coupling and CNN. In this method, the ECG heartbeats are transformed into a dual beat coupling matrix as 2-D inputs to the CNN classifier, which are formulated by capturing both the morphology and beat-to-beat correlation in ECG. Next, Zubair et al. (2016) focused to accomplish the CNN for both feature extraction as well as for classification by which it negates the need of hand-crafted features. Further, Kiranyaz et al. (2016), proposed a patient-specific ECG classification system based on 1-D CNNs. Since this approach accomplished the CNN for both feature extraction and classification, it negated the need of feature extraction process.
Wu et al. (2020) developed a system called as Electrocardiographic Left Ventricular Hypertrophy Classifier (ELVHC) for the classification of Left Ventricular Hypertrophy. This system combines the profound neural networks with information from ECG signals. Six-layered deep neural network is employed for feature extraction after acquiring the ECG from IoT equipment followed by preprocessing. L2-regulariztion and drop out are used to avoid over fitting in each model iteration. Further, Huang et al. (2020) proposed an intelligent classifier using fast compression residual convolutional neural networks (FCResNet) for the classification of arrhythmias through ECG signal. As a feature extraction, this approach employed maximal overlap wavelet packet transform (MOWPT) which provides a comprehensive timescale paying pattern and poses the time-invariance property. This approach employed to classify totally five classes of arrhythmias; they are LBBB, RBBB, PVC, APC and Normal. MIT-BIH arrhythmia database is employed to test the performance of the proposed deep learning classifier. Kim et al. (2019) employed CNN for personal recognition based on 2-D coupling image of a ECG signal. Waveform of the 2-D coupling image which is the input data to the network cannot be visually confirmed and it has the advantage of being able to augment the QRS-complex which is a personal unique information.
2.4 Problem statement
Based on the study and analysis of earlier ECG classification methods, we have observed that a set of few features like either morphological or temporal or statistical features results in a higher false positive rate. The higher FPR is due to the more sensitive variations in ECG by which the CAs can be discriminated. Finding such sensitive variations through single set of features is very tough and hence this work focused over the multiple features. In the classification phase, the conventional machine learning algorithms have a higher computational complexity. In this regard even though CNN has more computational burden, it has gained an outstanding performance in the provision of sufficient discrimination between CAs. Moreover, due to the convolution of extracted multiple features with multiple convolutional filters at convolutional layers, even the sensitive differences by which the CAs are discriminated are also recognized effectively.
3 Materials and methods
3.1 MIT-BIT arrhythmia dataset
In this work, to evaluate the performance of proposed system, a standard and benchmark dataset, Massachusetts Institute of Technology-Beth Israel Hospital (MIT-BIH) is used. This dataset consists of 48 ECG records and each record is about 30 min. Each record is sampled at 360 Hz with 11-bit resolution from 48 different types of patients. This dataset consists of both normal beats and some life threatening arrhythmias. This dataset is created in 1980 to provide a standard reference for the detection of arrhythmias. In this dataset, each record is annotated by the cardiologic experts and the annotation file consist of much significant information like R-peak locations or class labels of the heartbeat signals. In the MIT-BIH dataset, totally there are 15 different types of heartbeats and they are summarized in the Table 1.
Further, the ECG records in this dataset are numbered with non-continuous integers from 100 to 234. All the 15 heart beat types are emerged from the 48 records only. According to the Association for the Advancement of Medical Instrumentation (AAMI) standard, the total 15 heart beat types are merged into five sub classes, namely, Non-ectopic or Normal (N), Supraventricular Ectopic Beat (SVEB), Ventricular Ectopic Beat (VEB), Fusion beat (F) and Unknown Beat (Q). Further, the AAMI recommends the non-utilization of paced beats and the respective records are numbered as 102, 104, 107, and 217. This dataset is highly imbalanced, almost 90% of beats are of normal class and the remaining beats are belongs to remaining classes such as SVEB, VEB, F and Q.
3.2 Wavelet transform
Wavelet Transform is a powerful transformation technique which provides a simultaneous representation of a signal in in both time and frequency domains. In Wavelet Transform, this simultaneous representation is obtained by decomposing the signal over a dilated (scale) and translator (time) version of a wavelet. Hence most of the applications such as signal, image and video processing prefer the wavelet transform for feature analysis. Based on the accomplishment of scaling and shifting through wavelet filters, wavelet family has so many varieties like Discrete Wavelet Transform (DWT), Dual Tree Complex Wavelet Transform (DTCWT), and Wavelet Packet Transform (WPT) etc. Among these varieties, DWT and DTCWT have gained more significance due to the effectiveness in analysing the hidden characteristics of a signal or image.
3.2.1 DWT
DWT is a most popular wavelet transform which represents the signal as a linear combination of its basis functions. In DWT, the signal is decomposed into several frequency bands called as Approximations (A) and Details (D). The main advantage of DWT is its ability of representation in both time and frequency domain. To obtain a time–frequency components of an ECG signal, a wavelet basis function, named as mother wavelet is formulated as
where, m and n are scaling and shifting parameters respectively. Further, the DWT decomposition of a signal \(Y\left( t \right)\) is formulated based on the mother wavelet and is as follows;
where, \(C_{m,n}\) represents the obtained DWT coefficients of a signal Y(t). In the case of signal representation from DWT coefficients, the inverse DWT is formulated as
Figure 1 shows the schematic of four level decomposition of a signal through DWT. Here the level of decomposition is completely depends on the applications. In the case of ECG signal, generally three or four level of decomposition is considered.
3.2.2 DTCWT
DTCWT is one of the most important and effective transform in the wavelet family. This was developed to overcome the major problem of DWT, i.e., lack of shift invariance. Though the traditional DWT has gained effective results in the signal decomposition, it suffers from several problems and the lack of shift invariance is the most distinct problem due to which the reconstructed signal will have distortions. The main reason behind this problem is the presence of a down sampling module at every stage of DWT implementation. Due to this reason, the shifts in the input signal can not be analysed through DWT coefficients. The best solution to overcome this problem an accomplishment of an un-decimated DWT. But, this solution consequences to a higher computational cost followed by very high redundancy in the obtained sub bands.
At this instant, DTCWT has come into picture which can overcome the DWT problem without any removal of down-sampler. By finding complex wavelet coefficients which are 900 out of phase with each other, this problem can be solved and here the DTCWT follows the same procedure which is inspired from the Fourier transform. Because, the Fourier coefficients obtained are of complex sinusoid form and they constitute a Hilbert transform Pair. Further the Fourier transform has no problem of shift invariance, the Fourier coefficients are perfectly shift variant. Considering these facts, the DTCWT employs a complex valued wavelet and scaling functions and they are defined as follows (Zubair et al. 2016);
where \(\varphi_{w} \left( t \right)\) is a wavelet coefficient obtained from the real wavelet coefficient \(\varphi_{rw} \left( t \right)\), and imaginary wavelet coefficient, \(\varphi_{iw} \left( t \right)\). Similarly, the term \(\varphi_{s} \left( t \right)\) is a scaled coefficient and it is obtained from the real scaled coefficient \(\varphi_{rs} \left( t \right)\), and imaginary scaled coefficient, \(\varphi_{is} \left( t \right)\). The DTCWT applies two wavelet filters, one extracts the real part and other extracts the imaginary part. These two filters combination is called as analytical filter. The structure of DTCWT is shown in Fig. 2, where the upper DWT is called as real part of Complex wavelet transform and the lower tree of DWT is called as imaginary part of complex wavelet transform. The wavelet filters applied at each tree are different. Let \(h_{0} \left( n \right)\) and \(g_{0} \left( n \right)\) be the low pass filters of Tree A and Tree B respectively, they should be designed in such a way that they are Hilbert Transform pair and this is obtained with the following constraint;
Here the DTCWT applies two different set of wavelet filters, one set is for first level and another set is for further levels. The set of filters used at higher levels are not strictly linear in phase and also having a group delay of 0.25, approximately. The general approximate group delay is 0.5 and this is achieved through the time inverse of filters accomplished at Tree B. Hence the DTCWT employs two DWTs and the overall complex valued coefficients are obtained after combining the real and imaginary coefficients.
3.3 Convolutional neural networks
Based on the recent studies, CNNs have gained an outstanding performance compared to the conventional classifiers in several applications related to visual and signal processing, speech recognition, machine vision, and several image processing tasks. Generally, the CNNs comprises of a hierarchical structure which is constructed by a stacked combination of Convolutional Layer, Rectified Linear Unit (ReLU), Pooling layer and a fully connected output layer. The output obtained at the first layer is given as an input to the next layer. The final output layer of CNN is a fully connected Multilayer Perceptron (MLP) that has the neurons which are same as the number of output class labels. Figure 3 shows the schematic of CNN architecture.
The details of individual layers of CNN are described below;
-
1.
Convolutional layer This layer of CNN consists of convolutional kernels of different sizes and they are used to extract the features of given input signal. For this layer, the input is in the form of (No. of signals) \(\times\) (No. of samples in the signal) \(\times\) (Every signal’s feature vector).
-
2.
ReLU layer These units are non-linear functions which performs threshold process for all the input values such that the any input value which was less than zero is set to be zero. Let’s consider x is the input to the ReLU, mathematically the output is formulated as;
$$ f\left( x \right) = max\left( {0,x} \right){ } $$(7) -
3.
Pooling layer This layer is generally used to reduce the dimensionality of feature set. The dimensionality reduction is accomplished by taking the maximum or average number that the filter convolves around every kernel. Based on this, the pooling layer is of maximum pooling and average pooling. The max pooling is based on the extraction of maximum value from each cluster of neurons. Further, the average pooling is based on the extraction of average value of each cluster of neurons. This work used average pooling to reduce the noise effects.
-
4.
Fully connected This is the final layer of CNN, which produces k output neurons, where k is the total number of class labels. This layer is in the principle of the conventional Multi-layer Perceptron (MLP) neural network.
4 Proposed system
The complete details of proposed arrhythmia detection system are illustrated in this section. The novelty of this system is that it is very simple and also effective. The novelty is composed in three phases. Initially at the pre-processing phase, a simple filter is accomplished to remove the baseline wander noise and the segmentation heart beat segmentation is done just based on the R-peak amplitude which is very simple. Next, in the feature extraction phase, this approach considered totally three different set of features by which the all varying characteristics of an ECG (with different CAs) is analysed. Moreover, the size of excess features obtained at features is reduced through PCA and finally CNN is accomplished for classification.
Totally, the proposed system is accomplished in two stages, namely, training and testing. In every stage, for a given input ECG signal, there are three phases and they are;
-
1.
Pre-processing and segmentation
-
2.
Feature extraction and
-
3.
Classification
In the pre-processing phase, the input ECG signal is processed for noise removal. Once the noise removed from the raw ECG signal, then it is subjected to segmentation to obtain heart beats and every heart beat have one R-peak. After segmenting, the obtained heart beats are processed for feature extraction and then subjected to classification. The overall block diagram of proposed system is shown in Fig. 4. The following subsections illustrate the details of all phases.
4.1 Pre-processing and segmentation
In this phase, the raw ECG signal is fed to the pre-processing module that involves the removal of baseline, and removal of noise etc. To find the baseline, here two consecutive median filters 200–600 ms was applied and then the obtained baseline is subtracted from the original signal, obtaining a baseline corrected ECG signal. Once the baseline free ECG signal is obtained then it is subjected to segmentation. In the segmentation process, first, the R-peaks are detected and then a segment of 280 samples are extracted, 140 samples are from right side of R-peak and the remaining 140 samples are extracted from the left-side of R-peak. At the starting of ECG signal, if the total number of samples left to the first R-peak is less than 140, then that heart beat is neglected. Similarly, at the ending of ECG signal, if the total number of samples right to the last R-peak is less than 139, then that heart beat is also neglected. Further, the obtained every heartbeat is normalized through Z-score evaluation such that the scale of obtained normalized heart beat segments is within the range of zero mean and unity standard deviation. The Z-score normalization is accomplished through the following formulation;
where Y is the heart beat segment, \(\mu\) is the mean of segment and \(\sigma\) is the standard deviation of that segment.
4.2 Feature extraction
In the detection of cardiac arrhythmias through ECG signal analysis, feature extraction is very important and it is a key step in classification system. Generally, the features of an ECG signal were extracted through various methods like temporal feature extraction methods, frequency feature extraction methods and spectral feature extraction methods etc. The features must be extracted in such way that they have to represent the entire signal information in a compact format. Moreover, the performance of a classification system also depends on the set of features considered as whole or only a partial subset. Every feature extraction method has its own advantages and disadvantages. Hence considering the robustness, noise resilience, computational efficiency, and discriminative capability, this work employed various well-known feature extraction methods. Totally, this work employs three techniques such as Morphological features, temporal features and statistical features.
4.2.1 Morphological features
Morphological features are also termed as shape based features and they are extracted through the accomplishment of DTCWT over the ECG heart beat signal. The obtained detailed and approximation coefficients of the heartbeat help in the computation of morphological features. In this work, Dabuchies (db1) wavelet function is used and the beat is decomposed up to five levels. The feature extraction process through the DTCWT is summarized as follows;
-
1.
Perform 1D-DTCWT over the heart beat segment up to five scales.
-
2.
Choose the fourth and fifth scale detailed coefficients and approximation coefficients as features. The approximation and detailed coefficients of upper Tree are real part and the same ones of lower Tree are imaginary part. The absolute values are computed from these real and imaginary coefficients.
-
3.
Append the absolute values of approximation and detailed coefficients (both fourth and fifth scales) to form a complete morphological feature set.
-
4.
Apply Principal Component Analysis (PCA) to reduce the dimensionality of morphological feature set to reduce the computational efficiency.
4.2.2 Temporal and statistical features
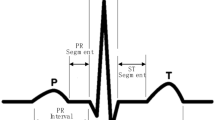
Under this phase of feature extraction, the proposed system focused to compute the temporal and statistical features for every heart beat segment. According to some earlier studies, the basic characteristics of the ECG signal such as QRS complex, duration, ST-Interval, PR-Interval, TT-Interval, PP-Interval, QT-Interval, the consecutive RR-Interval and the average of last ten betas RR-Intervals are considered as the temporal features.
Under the statistical features, the features like Sum, Variance, Mean, Mean Absolute Deviation, Route Mean Square, Skewness and Kurtosis are computed. Along with these features, two more features namely, AC power and Timing Information are also measured for every heartbeat segment. Once the features are extracted, then they are subjected to Z-score normalization. Let \(x\left[ n \right]\) be the heart beat segment, the statistical features are measured as follows;
-
1.
Skewness: it measures the distribution symmetry of a signal
$$ Skew\left( X \right) = { }\frac{{E\left[ {\left( {X - \mu } \right)^{3} } \right]}}{{\sigma^{3} }}{ } $$(9) -
2.
Kurtosis: it measures the peakedness of given heartbeat segment
$$ Kurt\left( X \right) = { }\frac{{E\left[ {\left( {X - \mu } \right)^{4} } \right]}}{{\sigma^{4} }}{ } $$(10) -
3.
AC power: it measures the total power of the given input signal. This power explores the difference between features between two different ECGs with two different CAs.
$$ { }P = { }E\left[ {X\left( n \right)^{2} } \right]{ } $$(11) -
4.
Timing information: it is computed based on the ratio of RR-Interval and if there is any deviation in the rate of heartbeat, this metric can identifies it.
$$ IR_{i} = \frac{{T_{i} - T_{i - 1} }}{{T_{i + 1} - T_{i} }}{ } $$(12)
where \(T_{i}\) represents the occurrence of time instance at which the R-peak occurs in the given heart beat signal.
At the ending of feature extraction process, the obtained morphological features, temporal features and statistical features are fused into a single feature vector. The fusion process here is accomplished through the concatenation. Finally, the feature vector is processed for classification through CNN classifier.
4.3 Classification
In the classification phase, CNN is accomplished to classify the heartbeat. For a CNN classifier, the input is a final feature vector which has all the features in a concatenated form. For example, Let’s consider an ECG signal X and the total number of beats segmented are 100, 280 be the total number of samples present in each heartbeat. After subjecting every beat to the feature extraction, we will get a final feature vector. Totally three convolutional layers and two pooling layers are considered here. In the proposed model, the first convolutional layer (\(Conv_{1}\)) used 8 convolutional filters and the size of each filter is \(1 \times 7\). The next two convolutional layers such as (\(Conv_{2}\)), and (\(Conv_{3}\)), used 16, and 32, convolutional filters and the size of each convolutional filter is \(1 \times 5\). A Rectifier Linear Unit (ReLu) follows every convolutional layer and it is an activation function to increase the non-linearity. Next, the proposed model uses two max-pooling layers. At every max-pooling layer, the stride is considered as 1. Finally, a Fully Connected Layer (FCL) with the size equal to the total number of CAs is considered and it is the final stage of feature extraction. The following Table 2 shows the details of parameters and their values considered at convolutional layers and pooling layers.
Initially, several ECG signals acquired from MIT-BIH dataset are trained through the CNN algorithm and then testing is accomplished to test the remaining signals acquired from the same dataset. Here the signal used for training and testing won’t be same. The classification procedure follows the AAMI standard, i.e., total classes considered here for classification are five and they are N, S, V, F and Q. For every test heartbeat, the CNN classifier produces an output label to which it belongs.
5 Simulation experiments
To alleviate the performance of proposed approach, many experiments are conducted by considering different ECG signals with different arrhythmias. The complete simulation is done using MATLAB software. The complete details of dataset considered, performance metrics and the comparative analysis are described in the following sections.
5.1 Dataset
In our simulation experiments, the required EC records are acquired from the MIT-BIH dataset. According the AAMI standards, totally five classes are considered for evaluation. The entire 48 ECG records are considered and they are divided into two sets namely, Training Set and Testing Set. Training Set considered the ECG records numbered with 101, 106, 108, 109, 112, 114, 115, 116, 118, 119, 122, 124, 201, 203, 205, 207, 208, 209, 215, 220, 223, and 230; and the test set considered the ECG records numbered with 100, 103, 105, 111, 113, 117, 121, 123, 200, 202, 210, 212, 213, 214, 219, 221, 222, 228, 231, 232, 233, and 234. Totally 50,604 heart beats are extracted from both training and testing records. Among these, 44,982 are Normal, 1473 are SVEB, 3729 are VEB, 410 are F and the remaining 10 are of Q. For every class, 70% heart betas (35,423) are considered for training and remaining 30% (15,181) are considered for testing. The summary of both training and testing heartbeats considered for simulation are illustrated in the following Table 3.
5.2 Performance metrics
To analyse the proposed approach through subjective assessment, some performance metrics are measured for the obtained results. Firstly, some secondary metrics such as True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN) are calculated. The secondary metrics are measured through the confusion matrix, as shown in Table 4. The model of sample confusion matrix is shown in Table 2, for a test case of Normal class only. In the case of signals testing are related to normal class, the heartbeats labelled as Normal are counted as TP and remaining are counted as FN. Similarly, this is applied for remaining classes also. The confusion matrices obtained after testing all the heartbeats are represented in Table 5.
Table 5 shows the confusion matrix constructed after the simulation of proposed system only through the morphological features. Under this simulation, for each heart beat signal in the training and testing sets, only the morphological feature is extracted through DTCWT. According to the morphological feature extraction process described in the Sect. 4.2.1, the ECG heart beat is decomposed through DTWCT into five levels and the obtained details coefficients at four and fifth levels and approximation coefficients are considered as features. These features are fed to CNN classifier to predict the class label for an input heartbeat.
Next, Table 6 shows the confusion matrix constructed after the simulation of proposed system only through the morphological features. Under this simulation, for each heart beat signal, only the Temporal and Statistical features are extracted according to the process described in Sect. 4.2.2. After extraction these feature, they are subjected to z-score normalization and then concatenated and then fed to CNN classifier to predict the class label of input hear beat signal.
Finally, Table 7 shows the confusion matrix constructed after the simulation of proposed system through the fusion of features. Under this case, for each heart beat signal totally three set of features are extracted and they are fused into a single feature vector. This feature vector is fed to CNN classifier to predict the class label for an input heartbeat. From the above tables, it can be observed that the obtained TPs are more in Table 6 compared to the values in Tables 4 and 5. For example, for a Normal class, the proposed fused vector system has detected totally 13,292 heart beats as normal for a total 13,494 normal test signals, whereas the morphological feature assisted system has obtained only 13,089 TPs and the temporal and statistical assisted system has obtained only 13,011 TPs. Similarly, the TPs of SVEB class is 375, 353, and 362 for Fused Feature vector, morphological and temporal system respectively. The further TPs, TNs, FPS and FNs can also be observed from these tables.
Based on these secondary metrics, the performance metrics such as Sensitivity (True Positive Rate), Specificity (True Negative Rate), Positive Predictive Value (PPV) or Precision, False Negative Rate (FNR) or Miss Rate, False Discovery Rate (FDR) and Accuracy metrics are measured according to (13), (14), (15), (16) and (17) respectively.
After testing the all heart beats specified under testing Column in Table 2, the performance metrics are measured according to formula described above and they are represented in the following tables.
Table 8 shows the details of performance metrics measured after the accomplishment of proposed detection through different feature sets. As it can be observed from the values represented in Table 7, almost all the performance metrics has obtained an optimal value for the proposed fused feature vector assisted detection system. Since this system can get more knowledge about the characteristics of an ECG heartbeat through the analysis multiple features, it can classify more effectively which results in a more sensitivity and Precision and less FNR and FDR. In the case of Morphological features assisted detection system, the system acquires only the knowledge about the shape of heartbeat signal but not about the basis characteristics like time intervals and also the relation with other beats. Similarly, if we consider only the temporal and statistical features, the basic characteristics can be studied followed by the relation with other beats, For example, consider a feature called mean of two different heart beats. It reveals a cumulated behaviour of heart beat and the mean will be different for two different heart beats. To get a perfect discrimination between two beats in a simple manner, the statistical features are effective paradigm which can be extracted in a very simple manner and also effective in the provision of a proper discrimination knowledge. But, the alone temporal and statistical features won’t analyse the morphology of a heartbeat. Hence the sensitivity and precision are high and FNR and FPR are low for the system accomplished through the fused feature vector.
Next, the simulation is accomplished in the presence of noise. Under this simulation, the ECG signal is contaminated with noise and then subjected to classification system. After the simulation the performance metrics are measured and they are represented in the following figures.
Figure 5 shows the details of obtained performance metrics under the simulation of ECG signal with noise. In the case of noise presence, the signal amplitudes will change which results in an improper discrimination between different classes. Compared to the noise free signals, the noisy signals are less qualitative and hence the system may misclassify the signals which results in more FNR and FDR. But it is not much more less because of the proposed fusion of features. Due to this fusion strategy, the system can acquire the perfect discrimination even in the presence of noise and classifies effectively.
5.3 Comparative analysis
To show the effectiveness of proposed approach, a simple comparative analysis with state-of-art methods is done here. Table 9 shows the comparative analysis between the proposed and conventional approaches. Since the wavelet transform is most prominent transform through which the morphological characteristics can be studies, most of the researchers accomplished DWT and its derivatives for feature extraction. For instance, the methods proposed by Siva et al. 2018 and Roghayyeh et al. (2017), applied DWT as feature extractors. In these methods, the input ECG signal is decomposed through DWT into sub bands and then the features are processed for classification through ANN and LS-SVM respectively and the average accuracy obtained through these methods is observed as 94.1400% and 95.7500% respectively. However, the DWT is lack of shift invariant and hence all the CAs is not recognized accurately. Next R. Rajesh Kumar and Dandapat (2017), proposed an ECG classification method in which the features are extracted through an extended version of DWT, called as CWSB and the classification is done through two classification algorithms such as Extreme learning machine and SVM. On an average, this method has gained an accuracy of 96.4000% and an improvement is of 0.75% form DWT assisted methods. Further, to improve the accuracy, Zhai and Tin (2018), considered two set of features such as Morphological features and correlation features and CNN is accomplished for classification. This approach has gained a great improvement in accuracy and it is approximately 97.3000%. From this observation, we noticed that compared to the single set of features, multiple features have gained more accuracy. Next, some methods like Kiranyaz et al. (2016) and Zubair et al. (2016), utilized the advantages of CNN and applied directly on ECG to extracts patient specific features and gained an accuracy of 96.4000% and 92.7000% respectively. There is a need of careful design of CNN, i.e., total number of filters, filter size and pooling stride to achieve an improved accuracy through CNN. Finally, the proposed approach gained an accuracy of 98.2500% and this is due to the integration of multiple set of features and CNN. Since we have considered totally three different set of features through which the complete study of an ECG is possible, we have gained a more accuracy compared to the conventional approaches.
6 Conclusion
Cardiac Arrhythmia detection and classification is one of the most significant research fields in medical related computer-aided diagnosis. A study of multiple features set based deep learning mechanism is proposed in this paper. Under the multiple feature set, this paper extracted morphological features, statistical features and temporal features for every ECG heart beat signal. Further, for classification purpose, this work accomplished CNN algorithm which has gained a significant results in various applications, Simulation experiments are conducted over a standard MIT-BIH cardiac arrhythmia database and the performance is measured under both noise free and noisy conditions. Totally four classes such as Normal, SVEB, VEB, and F are considered for from this database and the performance is measured through metrics like sensitivity, Precision, False Negative Rate, False Discovery Rate and Accuracy. Based on the obtained results, we can conclude that the performance of proposed mechanism is more distinguished than that of single feature based detection systems. On an average, the proposed approach has gained an accuracy of 98.2500% and the average accuracy of conventional approaches is 94.5700%. This shows an improvement of 3.6785% from the conventional approaches.
In this work, we have focused on the fusion of multiple features by which a super vector is constructed. This super vector is much effective and also discriminative which helps the classifier in the perfect distinguish of CAs. However, the super vector is of larger size by which the classifier suffers from additional computational burden during the classification. Though this method applied PCA for dimensionality reduction, during the simulation we have noticed that the classification time is very much high and also have more information loss. Hence, as a next contribution of future scope, this work is decided to focus on the dimensionality reduction with less information loss.
Change history
04 July 2022
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s12652-022-04298-7
References
Ai D, Yang J, Wang Z et al (2015) Fast multi-scale feature fusion for ECG heartbeat classification. EURASIP J Adv Signal Process. https://doi.org/10.1186/s13634-015-0231-0
Bono V, Mazomenos EB, Chen T, Rosengarten AJA, Maharatna A et al (2014) Development of an automated updated Selvester QRS scoring system using SWT-based QRS fractionation detection and classification. IEEE J Biomed Health Inform 8:193–204
Chieh C, Te Chuang C (2017) A QRS detection and R point recognition method for wearable single-lead ECG devices. Sensors 17:1–19
Das MK, Ari S (2014) ECG beats classification using mixture of features. Int Sch Res Not 2014:178436–1–178436–12
Grazia C, Das S, Mazomenos EB, Maharatna K, Koulaouzidis G, Morganand J, Pudd PE (2015) A statistical index for early diagnosis of ventricular arrhythmia from the trend analysis of ECG phase-portraits. Physiol Meas 36:107–131
Gustavo L, Pilia N, Loewe A, Schulze WHW, Dössel O (2017) Comparison of baseline wander removal techniques considering the preservation of ST changes in the ischemic ECG: a simulation study. Comput Math Methods Med 2017:9295029–1–9295029–13
Huang J-S, Chen B-Q, Zeng N-Y, Cao X-C, Li Y (2020) Accurate classification of ECG arrhythmia using MOWPT enhanced fast compression deep learning networks. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02110-y
Kim JS, Kim SH, Pan SB (2019) Personal recognition using convolutional neural networks with ECG coupling image. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-019-01401-3
Kiranyaz S, Ince T, Gabbouj M (2016) Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans Biomed Eng 63:664–675
Kutlu Y, Kuntalp D (2012) Feature extraction for ECG heartbeats using higher order statistics of WPD coefficients. Comput Methods Progr Biomed 105:257–267
Lenis G, Baas T, Dossel O (2013) Ectopic beats and their influence on the morphology of subsequent waves in the electrocardiogram. Biomed Eng 58:109–119
Lenis G, Pilia N, Oesterlein T, Luik A, Schmitt C, Dossel O (2016) P wave detection and delineation in the ECG based on the phase free stationary wavelet transform and using intracardiac atrial electrograms as reference. Biomed Eng 61:37–56
Lu X, Pan M, Yu Y (2018) QRS detection based on improved adaptive threshold. J Healthc Eng 2018:5694595–1–5694595–8
Manu T, Das M, Ari S (2015) Automatic ECG arrhythmia classification using dual tree complex wavelet based features. Int J Electron Commun 69:715–721
Martis RJ, Acharya UR, Adeli H (2014) Current methods in electrocardiogram characterization. Comput Biol Med 48:133–149
Meek S, Morris F (2002) Introduction. I – Leads, rate, rhythm, and cardiac axis. BMJ 324:415–418
Mendis S, Puska P, Norrving B et al (2011) Global atlas on cardiovascular disease prevention and control. World Health Organization, Geneva
Mounaim A, Jbari A, Bourouhou A (2017) ECG signal denoising by discrete wavelet transform. IJOE 137:51–68
Mustaqeem A, Muhammad Anwar S, Majid M (2018) Multiclass classification of cardiac arrhythmia using improved feature selection and SVM invariants. Comput Math Methods Med. 2018:7310496–1–7310496–10
Phukpattaranont P (2015) QRS detection algorithm based on the quadratic filter. Expert Syst Appl 42:4867–4877
Rajesh Kumar T, Dandapat S (2017) Automated detection of heart ailments from 12-lead ECG using complex wavelet sub-band bi-spectrum features. Healthc Technol Lett 4:57–63
Rangayyan RM (2015) Biomedical signal analysis, vol 33. Wiley, Hoboken
Rashid Ghorbani A, Azarnia G, Ali Tinati M (2016) Cardiac arrhythmia classification using statistical and mixture modeling features of ECG signals. Pattern Recogn Lett 70:45–51
Regis N, Cláudio A, Veiga P (2018) Electrocardiogram signal denoising by clustering and thresholding. IET Signal Proc 12:1165–1171
Roghayyeh A, Sabalan D, Hadi S, Oshvarpour A (2017) Classification of cardiac arrhythmias using arterial blood pressure based on discrete wavelet transform. Biomed Eng Appl Basis Commun 29(5):1750034. https://doi.org/10.4015/S101623721750034X
Sameni R, Shamsollahi MB, Jutten C, Clifford GD (2007) A non-linear Bayesian filtering framework for ECG denoising. IEEE Trans Bio-med Eng 54:2172–2185
Singh BN, Tiwari AK (2006) Optimal selection of wavelet basis function applied to ECG signal denoising. Digit Signal Process 16:275–287
Siva A, Hari Sundar M, Siddharth S, Nithin M, Rajesh CB (2018) Classification of arrhythmia using wavelet transform and neural network model. J Bioeng Biomed Sci 8:1
Tian Y-M, Zhang C, Wang H-W (2016) Review of ECG signal identification research, 2016 joint international conference on artificial intelligence and computer engineering (AICE 2016) and international conference on network and communication security (NCS 2016). ISBN: 978-1-60595-362-5
Tracey BH, Miller EL (2012) Non local means denoising of ECG signals. IEEE Trans Biomed Eng 59:2383–2386
Wang J, Ye Y, Pan X, Gao X (2015) Parallel-type fractional zero-phase filtering for ECG signals denoising. Biomed Signal Process Control 18:36–41
Wissam J, Latif R, Toumanari A, Dliou A, Bcharri O, Maoulainine MR (2016) An efficient algorithm of ECG signal denoising using the adaptive dual threshold filter and the discrete wavelet transform. Bio-Cybern Bio-med Eng 36:499–508
Wu JM-T, Tsai M-H, Xiao S-H, Liaw Y-P (2020) A deep neural network electrocardiogram analysis framework for left ventricular hypertrophy prediction. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-020-01826-1
Ye C, Kumar BV, Coimbra MT (2012) Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans Biomed Eng 59:2930–2941
Zhai X, Tin C (2018) Automated ECG classification using dual heartbeat coupling based on convolutional neural network. IEEE Access 6:27465–27472
Zubair M, Kim J, Yoon C (2016). An automated ECG beat classification system using convolutional neural networks. In: 6th International conference on IT convergence and security (ICITCS), pp 1–5
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1007/s12652-022-04298-7
About this article
Cite this article
Ramesh, G., Satyanarayana, D. & Sailaja, M. RETRACTED ARTICLE: Composite feature vector based cardiac arrhythmia classification using convolutional neural networks. J Ambient Intell Human Comput 12, 6465–6478 (2021). https://doi.org/10.1007/s12652-020-02259-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-020-02259-6