Abstract
The prevalence and genetic diversity of human bocaviruses (HBoVs) in sewage water samples are largely unknown. In this study, 134 raw sewage samples from 25 wastewater treatment plants (WTPs) in Italy were analyzed by nested PCR and sequencing using species-specific primer pairs and broad-range primer pairs targeting the capsid proteins VP1/VP2. A large number of samples (106, 79.1 %) were positive for HBoV. Out of these, 49 were classified as HBoV species 2, and 27 as species 3. For the remaining 30 samples, sequencing results showed mixed electropherograms. By cloning PCR amplicons and sequencing, we confirmed the copresence of species 2 and 3 in 29 samples and species 2 and 4 in only one sample. A real-time PCR assay was also performed, using a newly designed TaqMan assay, for quantification of HBoVs in sewage water samples. Viral load quantification ranged from 5.51E+03 to 1.84E+05 GC/L (mean value 4.70E+04 GC/L) for bocavirus 2 and from 1.89E+03 to 1.02E+05 GC/L (mean value 2.27E+04 GC/L) for bocavirus 3. The wide distribution of HBoV in sewages suggests that this virus is common in the population, and the most prevalent are the species 2 and 3. HBoV-4 was also found, representing the first detection of this species in Italy. Although there is no indication of waterborne transmission for HBoV, the significant presence in sewage waters suggests that HBoV may spread to other water environments, and therefore, a potential role of water in the HBoV transmission should not be neglected.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human bocavirus (HBoV) is a single-stranded linear DNA virus belonging to the genus Bocavirus in the family Parvoviridae. It was first identified in 2005 in respiratory tract samples from children with clinical symptoms of respiratory infections in Sweden (Allander et al. 2005) and was later found, although less frequently, in stool samples collected from patients with gastroenteritis worldwide (Vicente et al. 2007; Arthur et al. 2009; Kapoor et al. 2009, 2010), suggesting that this virus is an enteric pathogen, besides being a respiratory pathogen. HBoV is responsible for a variety of symptoms and diseases, such as rhinitis, pharyngitis, cough, dyspnea, wheezing, pneumonia, acute otitis media, fever, nausea, vomiting, and diarrhea (Peltola et al. 2013). In immunocompromised patients, severe clinical cases have been described (Sadeghi et al. 2013; Akturk et al. 2015).
Four species of human bocaviruses (HBoV-1, -2, -3, and -4) are currently included in the Bocavirus genus (Kapoor et al. 2010), with HBoV-1 more frequently detected in clinical samples from patients with respiratory diseases, and HBoV-2, -3, and -4 more frequently associated with gastrointestinal infections. Several studies support an association of HBoV-1 with respiratory infections in children with pneumonia, acute wheezing, asthma, or bronchiolitis (Jartti et al. 2012), with higher viral loads more likely associated with respiratory disease.
However, the role of HBoV as enteropathogen remains still unclear (Schildgen 2013), due to the frequent coinfection of HBoV with other enteric viruses in patients with gastroenteritis, and the frequent detection of HBoV in asymptomatic individuals (Nawaz et al. 2012; Chhabra et al. 2013; Kim 2014; La Rosa et al. 2015). A recent review addressing whether HBoV present in stool could be considered as an independent risk factor for acute gastroenteritis concluded that most studies showed a comparable prevalence of HBoV in patients with gastroenteritis compared to asymptomatic patients. This observation, along with the high concomitant prevalence of other viruses in stool, suggests that HBoV in stool is more likely to be an innocent bystander rather than a true pathogen (Ong et al. 2016).
Besides fecal samples, HBoVs have also been detected in urine samples and in blood/serum, tonsils, and saliva of infected patients (Allander et al. 2005; Fry et al. 2007; Lau et al. 2007; Pozo et al. 2007; Vicente et al. 2007; Campe et al. 2008; Tozer et al. 2009; La Rosa et al. 2015). Through viral-shedding HBoV is discharged in untreated wastewaters. Only two studies addressed the presence of human bocavirus in sewages, demonstrating a wide circulation of the virus in the population served by the wastewater treatment plant (WTP) (Blinkova et al. 2009; Myrmel et al. 2015).
The aim of this work was to study the occurrence and genetic diversity of Human Bocaviruses (HBoVs) identified in sewage water samples from Italy.
Materials and Methods
Wastewater Samples
Grab wastewater samples were collected in the years 2014 (57 samples) and 2015 (77 samples) from 25 WTPs throughout Italy, in seven Italian regions. Figure 1 shows a geographic information system (GIS) map of the WTPs under study created using Quantum GIS (QGIS) version 2.0.1. Samples were handled and analyzed as previously described (La Rosa et al. 2014). In brief, 20 mL of untreated wastewater samples were seeded with a murine norovirus (MNV1), and treated with 2 mL of 2.5 M glycine pH 9.5. After incubation in ice for 30 min, 1/10 vol. chloroform was added and samples were centrifuged at 5000 rpm for 10 min. Viral nucleic acids were extracted from 10 mL of chloroform-treated samples, using the NucliSENS easyMAG (BioMerieux, Marcy l’Etoile, France) semi-automated extraction system with magnetic silica, according to the manufacturer’s instructions. Eluates (100 μL each) were divided into small aliquots and subsequently frozen at −70 °C until analyzed.
Nested PCR Assays
Viral DNA was used as a template for nested PCR using broad-range primers targeting the VP1/VP2 region of HBoV, as previously described (La Rosa et al. 2015).
PCR was performed using 2 µL of DNA and 1 µL (22 pmol) of each primer in a 25 µL reaction using the MyTaqTM Red Mix PCR kit (Bioline, London, UK). Cycling profile included an initial denaturing at 94 °C (10 min), followed by 35 cycles of denaturing at 94 °C (30 s), annealing at 51 °C (30 s), 72 °C (1 min), a final extension at 72 °C (5 min), and a hold at 4 °C. The amplification was carried out in a T100™ Thermal Cycler (Biorad). One microliter of the PCR product was used as a template in the nested PCR assay, lowering the annealing temperature at 50 °C.
Since the broad-range assay used in this study showed low sensitivity for HBoV-1 in a previous study (La Rosa et al. 2015), novel primers were designed, targeting the same region, specific for species 1 (see Table 1).
The sample process control, added in order to monitor nucleic acid extraction and the presence of potential inhibitors, was detected using previously published primers (Muscillo et al. 2013).
Standard precautions were followed to prevent PCR contamination: DNA extraction and PCR setup were performed in two different rooms, physically isolated from the PCR Thermal Cycler and the post-PCR processing area. Dedicated pipettes and reagents were used for each room. Negative controls were systematically used, and no indications of contamination were detected.
As positive PCR controls, HBoV DNAs were used and characterized in stool samples from patients with gastroenteritis in a previous study (La Rosa et al. 2015).
PCR products were purified using a Montage PCRm96 Micro-well Filter Plate (Millipore, Billerica, Mass), and purified PCR amplicons were subjected to direct automated sequencing with the same primers used in PCR (Bio-Fab Research, Rome, Italy). Inhibition in negative samples was ruled out using the sample process control (positive PCR signal for murine norovirus).
If the electropherograms suggested the presence of mixed species, cloning of PCR amplicons was performed on representative samples. PCR products were cloned in pGEM ®-T Vector Systems (Promega), and half ligation was used to transform NEB® Turbo Competent E. coli (BioLabs inc). Forty white colonies for each ligation were grown in 3 mL 2xTY medium. The DNA plasmids were extracted by the one-step FastPlasmid mini purification kit (5Prime.com) and quantified by ethidium bromide-stained agarose gel. Recombinant plasmids were then subjected to nucleotide sequencing using universal SP6 and T7 primers.
The raw forward and reverse ABI files obtained by both PCR and plasmid sequencing were aligned and assembled into a consensus sequence using MEGA 6.06 software, and sequences were submitted to BLAST analysis for genotyping at http://blast.ncbi.nlm.nih.gov/Blast.cgi. The phylogenetic comparison was performed by a distance matrix/UPGMA analysis using the Kimura 2-parameter via MEGA 6.06 software package program.
Real-Time PCR Assay
Standard quantification plasmids were obtained for Bocavirus species 2, 3, and 4 by cloning the Vp1/Vp2 fragment of each species. Plasmid DNA concentrations were determined by measuring the optical density at 260 nm. The DNA content in micrograms was converted to genomic copies using Avogadro’s number and the number of nucleotide pairs in the plasmid. We then performed an absolute quantification of the HBoV copies contained in the samples, using an external calibration curve generated through tenfold serial dilutions of the standard, corresponding to 5.5E+04 to 5.5E+00 GC per reaction.
Primers and probe targeting the Vp1/Vp2 gene (see Table 1) were designed using the Genescript software (https://www.genscript.com/ssl-bin/app/primer). Reactions were performed in triplicate in a 25-μL mixture containing 12.5 μL of 2× SensiMixT II Probe No-ROX Kit (Bioline), 400 nM (each) of forward and reverse primers, 100 nM of fluorescence-labeled probe, and 5 μL of DNA template. Cycling conditions were 95 °C for 10 min, 45 cycles at 95 °C for 5 s, and 60 °C for 1 min. Real-time PCRs were carried out in a MiniOpticon Real-Time PCR System (Bio Rad) with CFX Manager software control. Run acceptability was defined as a correlation coefficient (R2) >0.98 and a slope between −3.6 and −3.1, corresponding to reaction efficiencies between 90 and 110 %, according to the equation:
Results
From the total of 134 sewage samples analyzed, HBoV was positive in 106 samples (79.1 %) and was detected throughout the year with no seasonal prevalence. Positive and negative controls gave expected results. Genomic sequencing and BLAST searches showed that 49 samples were classified as HBoV species 2, and 27 samples as species 3. In the remaining 30 samples, PCR-direct sequencing results showed mixed electropherograms containing two fluorescent signals in different positions. The alignment of the study sequences with prototype sequences from GeneBank showed that both species 2 and 3 were present in 29 samples, since two fluorescent signals were detected simultaneously in different positions, corresponding to the alternative nucleotide present in HBoV-2 or HBoV-3. In one sample (ID 2225), a mixture of human bocavirus 2 and 4 was suggested, in accordance with the degenerate positions (data not shown).
PCR amplicons of samples ID 2225 (HBoV-2 + HBoV-4 suggested in mixed electropherograms) and ID 2189 (HBoV-2 + HBoV-3) were cloned and randomly selected clones (20 clones for each sample) were sequenced. Results confirmed the presence of mixed Bocavirus 2 and 3 in sample 2189 (representative of 29 samples with similar mixed electropherograms), and mixed bocavirus 2 and 4 in sample ID 2225.
HBoV-1 was not detected with the broad-range assay. Due to the low sensitivity of this assay for species 1, all negative samples (25 samples) were tested again with the newly designed assay specific for HBoV-1. They were all confirmed as negative for HBoV-1. Positive controls for HBoV-1, selected in the previous study on gastroenteritis patients (La Rosa et al. 2015), were tested positive with the PCR specific for species 1, thus indicating that the reagents, primers, and procedure were functional.
The result of the phylogenetic tree constructed with HBoV detected in this study is presented in Fig. 2. The study sequences were compared with 18 genomic sequences available from Genbank representing species 1 (a.n. DQ778300, KJ684074, FJ375128, GQ455988, KP710213, and DQ000495), species 2 (EU082214, GU048663, FJ170278, FJ948860, GU048662, and KM624025), species 3 (GQ867667, GQ867666, FJ948861, and JN086998), and species 4 (FJ973561, KC461233, and KM624027). The TN93 + G model was selected as the best fitted model using the MEGA 6.06 program. Bootstrap values represented on the tree are greater than 60 %. The sequences characterized as species 1 detected from clinical samples and used as positive controls were also included (samples IDs 44D, 49D, 50D, 64D, 67D 138Dbis). Sequences from sewage samples were classified within two separate groups corresponding to species 2 and 3, in accordance with blast analysis.
Phylogenetic analysis of human bocavirus sequences. The tree is based on a partial sequence (298 nt) of the VP1/2 region of the HBoV genome. The strains identified in this study are indicated with ID number. Other strains in the tree are shown by their GenBank accession number and strain name. These include species 1 strain (DQ778300, KJ684074, FJ375128, GQ455988, KP710213, and DQ000495), 2 strains (EU082214, GU048663, FJ170278, FJ948860, GU048662, and KM624025), 3 (GQ867667, GQ867666, FJ948861, and JN086998), and 4 strains (FJ973561). The evolutionary history was inferred using the UPGMA method. The evolutionary distances were computed using the Kimura 2-parameter method. Evolutionary analyses were conducted in MEGA6. The robustness of branching patterns was tested by 1000 bootstrap pseudo-replications; bootstrap values >80 are indicated
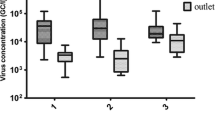
A portion of the samples (15 bocavirus 2 and 15 bocavirus 3) were randomly selected and further analyzed in order to obtain quantitative data on viral concentrations in wastewater samples. Viral concentrations ranged from 5.51E+03 to 1.84E+05 GC/L (mean value 4.70E+04 GC/L) for bocavirus 2 and from 1.89E+03 to 1.02E+05 GC/L (mean value 2.27E+04 GC/L) for bocavirus 3.
Discussion
Untreated sewage samples from 25 WTPs in Italy were screened for the presence of HBoV. The screening of sewage water for human viruses began with the detection and monitoring of polioviruses (Sinclair et al. 2008; La Rosa and Muscillo 2013), and since then it has been successfully extended to study the circulation of different enteric and nonenteric viruses in the human population (La Rosa et al. 2010, 2013, 2014; Cantalupo et al. 2011; Di Bonito et al. 2014; Smith et al. 2015; Victoria et al. 2016).
Only few studies have addressed the presence of human bocavirus in sewages. Blinkova and coworkers analyzed untreated sewage samples in the United States and detected HBoV in 17 of 21 samples (81 %). HBoV was the most frequently detected virus compared to cardioviruses, cosaviruses, and circoviruses (Blinkova et al. 2009). Sequences were characterized as HBoV-2 (67 %), HBoV-3 (26 %), and HBoV-1 (6 %). Bibby and Peccia conducted a metagenomic analysis on sewage sludge samples collected from wastewater treatment facilities located throughout the United States (Bibby and Peccia 2013) and detected HBoV among the most abundant (found in more than 90 % of the samples) and ubiquitous viruses identified (along with Adenovirus, Herpesvirus, and Papillomavirus). Information on the species of HBoV identified was not available. More recently, Myrmel and coworkers detected HBoV in two WTPs in Norway, in 54 and 64 % of the samples, with subtype 3 and 2 prevalent compared to subtype 1, while subtype 4 was not detected (Myrmel et al. 2015).
Our results corroborate these studies, showing a wide distribution of HBoV in Italian sewages, and consequently in the population living in Italy. We detected a large number of samples positive for HBoV (79 %), with subtype 2 and 3 more prevalent, and subtype 4 detected only in 1 sample.
As far as we know, no previous studies have been performed on HBoV in sewages or other water environments in Italy. However, few data are available from some clinical studies, where low percentages of positivity for HBoV were found in different types of clinical specimens. Principi and coworkers detected HBoV in 104 (5.7 %) nasopharyngeal samples collected from children attending an emergency room for a respiratory tract infection, all belonging to species 1 (Principi et al. 2015). In a study on the prevalence of viral infections in children with community-acquired pneumonia, HBoV was detected in 10 % of samples, mainly in association with other viruses (particularly Respiratory Syncytial virus and rhinovirus) (Mameli and Zuccotti 2013). Data on the species detected in these positive samples were not available. Medici and coworkers studied the prevalence of HBoV in children with acute gastroenteritis, and detected HBoV in 3.2 % of samples, characterized as types 1 (21.7 %), 2 (34.8 %), and 3 (43.5 %) (Medici et al. 2012). In another study, HBoV was detected in stool samples from hospitalized patients with gastrointestinal syndromes in only 2.0 % of samples (5/246) (Rovida et al. 2013).
A serological survey performed in Italy to determine the prevalence of antibodies against human bocavirus in an Apulian population, indicate HBoV infection is ubiquitous in the general population and early acquired from preschool children (Guido et al. 2012), with more than 90 % of subjects older than 4 years displaying previous exposure to HBoV. Our results support the notion that HBoV is widely circulating in the population with prevalence of human bocavirus 2 and 3, and a large number of samples containing both these species. This result might reflect enhanced human gut tropism for HBoV-2 and HBoV-3 relative to HBoV-1 as already suggested by other authors (Blinkova et al. 2009). Indeed, there is satisfactory evidence demonstrating an association between HBoV-1 and respiratory disease in children, and there is evidence that HBoV-2 (and possibly the HBoV-3 and HBoV-4 species) are associated with gastroenteritis (Berry et al. 2015; Broccolo et al. 2015). In the present study, one sample was classified as HBoV-4. According to our data, this is the first detection of this species of HBoV in Italy.
No seasonal prevalence was observed for HBoV, which is in agreement with a previous study (Bastien et al. 2006). However, other studies suggested that HboV infection had a higher detection rate in winter or in summer (Jiang et al. 2016).The true incidence and seasonality of this pathogen thus remain unknown.
The considerable presence of HBoV in sewage waters suggest that it may spread to other water environments. Indeed, Hamza and collaborators described the detection of HBoV in 41 % of river water samples (Hamza et al. 2009). Also, Li and coworkers recently detected HBoV along with other enteric viruses in sewage-contaminated fresh and marine waters (Li et al. 2016). Therefore, a potential role of water in the transmission of HBoV should not be neglected.
In conclusion, this study is the first to evaluate the prevalence and genetic diversity of HBoV in Italy. Our results show a wide circulation of HBoV in sewages and, therefore, in the Italian population. This could mean that the virus circulates silently, not necessarily associated with disease status, or it potentially causes undiagnosed infections. Further studies are needed to determine the exact clinical and epidemiological roles of HBoV. Moreover, a potential role of water in the transmission of HBoV should be investigated.
To our knowledge, this is the first investigation on HBoV in sewages in Italy and the first detection of Species 4 in this country. Our results show a wide circulation of HBoV in sewages with prevalence of species 2 and 3. Although there is no indication of waterborne transmission for HBoV, the significant presence in sewage waters suggests that HBoV may spread to other water environments, and therefore, a potential role of water in the HBoV transmission should not be neglected.
References
Akturk, H., Sik, G., Salman, N., Sutcu, M., Tatli, B., Akcay, C. M., et al. (2015). Atypical presentation of human bocavirus: severe respiratory tract infection complicated with encephalopathy. Journal of Medical Virology, 87, 1831–1838.
Allander, T., Tammi, M. T., Eriksson, M., Bjerkner, A., Tiveljung-Lindell, A., & Andersson, B. (2005). Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA, 102, 12891–12896.
Arthur, J. L., Higgins, G. D., Davidson, G. P., Givney, R. C., & Ratcliff, R. M. (2009). A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathogens, 5, e1000391.
Bastien, N., Brandt, K., Dust, K., Ward, D., & Li, Y. (2006). Human bocavirus infection, Canada. Emerging Infectious Diseases, 12(5), 848–850.
Berry, M., Gamieldien, J., & Fielding, B. C. (2015). Identification of new respiratory viruses in the new millennium. Viruses, 7, 996–1019.
Bibby, K., & Peccia, J. (2013). Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environmental Science and Technology, 47, 1945–1951.
Blinkova, O., Rosario, K., Li, L., Kapoor, A., Slikas, B., Bernardin, F., et al. (2009). Frequent detection of highly diverse variants of cardiovirus, cosavirus, bocavirus, and circovirus in sewage samples collected in the United States. Journal of Clinical Microbiology, 47, 3507–3513.
Broccolo, F., Falcone, V., Esposito, S., & Toniolo, A. (2015). Human bocaviruses: possible etiologic role in respiratory infection. Journal of Clinical Virology, 72, 75–81.
Campe, H., Hartberger, C., & Sing, A. (2008). Role of human bocavirus infections in outbreaks of gastroenteritis. Journal of Clinical Virology, 43, 340–342.
Cantalupo, P. G., Calgua, B., Zhao, G., Hundesa, A., Wier, A. D., Katz, J. P., et al. (2011). Raw sewage harbors diverse viral populations. MBio., 2, e00180–e00211.
Chhabra, P., Payne, D. C., Szilagyi, P. G., Edwards, K. M., Staat, M. A., Shirley, S. H., et al. (2013). Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008–2009. Journal of Infectious Diseases, 208, 790–800.
Di Bonito, P., Libera, S. D., Petricca, S., Iaconelli, M., Accardi, L., Muscillo, M., & La Rosa, G. (2014). Frequent and abundant merkel cell polyomavirus detection in urban wastewaters in Italy. Food and Environmental Virology, 7, 1–6.
Fry, A. M., Lu, X., Chittaganpitch, M., Peret, T., Fischer, J., Dowell, S. F., et al. (2007). Human bocavirus: a novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. Journal of Infectious Diseases, 195, 1038–1045.
Guido, M., Zizza, A., Bredl, S., Lindner, J., De, D. A., Quattrocchi, M., et al. (2012). Seroepidemiology of human bocavirus in Apulia, Italy. Clinical Microbiology Infection, 18, E74–E76.
Hamza, I. A., Jurzik, L., Wilhelm, M., & Uberla, K. (2009). Detection and quantification of human bocavirus in river water. Journal of General Virology, 90, 2634–2637.
Jartti, T., Hedman, K., Jartti, L., Ruuskanen, O., Allander, T., & Soderlund-Venermo, M. (2012). Human bocavirus-the first 5 years. Reviews in Medical Virology, 22, 46–64.
Jiang, W., Yin, F., Zhou, W., Yan, Y., & Ji, W. (2016). Clinical significance of different virus load of human bocavirus in patients with lower respiratory tract infection. Scientific Reports, 6, 20246.
Kapoor, A., Simmonds, P., Slikas, E., Li, L., Bodhidatta, L., Sethabutr, O., et al. (2010). Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. Journal of Infectious Diseases, 201, 1633–1643.
Kapoor, A., Slikas, E., Simmonds, P., Chieochansin, T., Naeem, A., Shaukat, S., et al. (2009). A newly identified bocavirus species in human stool. Journal of Infectious Diseases, 199, 196–200.
Kim, S. (2014). Prevalence of human bocavirus 1 among people without gastroenteritis symptoms in South Korea between 2008 and 2010. Archives of Virology, 159, 2741–2744.
La Rosa, G., Della Libera, S., Iaconelli, M., Donia, D., Cenko, F., Xhelilaj, G., et al. (2015). Human bocavirus in children with acute gastroenteritis in Albania. Journal of Medical Virology, 88, 906–910.
La Rosa, G., Fratini, M., Accardi, L., D’Oro, G., Della Libera, S., Muscillo, M., & Di Bonito, P. (2013). Mucosal and cutaneous human papillomaviruses detected in raw sewages. PLoS One, 8, e52391.
La Rosa, G., Della Libera, S., Iaconelli, M., Ciccaglione, A. R., Bruni, R., Taffon, S., et al. (2014). Surveillance of hepatitis A virus in urban sewages and comparison with cases notified in the course of an outbreak, Italy 2013. BMC Infectious Diseases, 14, 419.
La Rosa, G., & Muscillo, M. (2013). Molecular detection of viruses in water and sewage. In C. Nigel (Ed.), Viruses in food and water: risks, surveillance and control. Cambridge: Woodhead Publishing.
La Rosa, G., Pourshaban, M., Iaconelli, M., & Muscillo, M. (2010). Quantitative real-time PCR of enteric viruses in influent and effluent samples from wastewater treatment plants in Italy. Annali dell Istituto Superiore di Sanita, 46, 266–273.
Lau, S. K., Yip, C. C., Que, T. L., Lee, R. A., Au-Yeung, R. K., Zhou, B., et al. (2007). Clinical and molecular epidemiology of human bocavirus in respiratory and fecal samples from children in Hong Kong. Journal of Infectious Diseases, 196, 986–993.
Li, X., Harwood, V. J., Nayak, B., & Weidhaas, J. (2016). Ultrafiltration and microarray detect microbial source tracking marker and pathogen genes in riverine and marine systems. Applied and Environment Microbiology, 82, 1625–1635.
Mameli, C., & Zuccotti, G. V. (2013). The impact of viral infections in children with community-acquired pneumonia. Current Infectious Disease Reports, 15, 197–202.
Medici, M. C., Tummolo, F., Albonetti, V., Abelli, L. A., Chezzi, C., & Calderaro, A. (2012). Molecular detection and epidemiology of astrovirus, bocavirus, and sapovirus in Italian children admitted to hospital with acute gastroenteritis, 2008–2009. Journal of Medical Virology, 84, 643–650.
Muscillo, M., Fratini, M., Graffeo, R., Sanguinetti, M., Martella, V., Green, K. Y., et al. (2013). GIV noroviruses in wastewaters and in stool specimens from hospitalized patients. Food and Environmental Virology, 5, 194–202.
Myrmel, M., Lange, H., & Rimstad, E. (2015). A 1-year quantitative survey of noro-, adeno-, human boca-, and hepatitis e viruses in raw and secondarily treated sewage from two plants in Norway. Food and Environmental Virology, 7, 213–223.
Nawaz, S., Allen, D. J., Aladin, F., Gallimore, C., & Iturriza-Gomara, M. (2012). Human bocaviruses are not significantly associated with gastroenteritis: results of retesting archive DNA from a case control study in the UK. PLoS One, 7, e41346.
Ong, D. S., Schuurman, R., & Heikens, E. (2016). Human bocavirus in stool: a true pathogen or an innocent bystander? Journal of Clinical Virology, 74, 45–49.
Peltola, V., Soderlund-Venermo, M., & Jartti, T. (2013). Human bocavirus infections. The Pediatric Infectious Disease Journal, 32, 178–179.
Pozo, F., Garcia-Garcia, M. L., Calvo, C., Cuesta, I., Perez-Brena, P., & Casas, I. (2007). High incidence of human bocavirus infection in children in Spain. Journal of Clinical Virology, 40, 224–228.
Principi, N., Piralla, A., Zampiero, A., Bianchini, S., Umbrello, G., Scala, A., et al. (2015). Bocavirus infection in otherwise healthy children with respiratory disease. PLoS One, 10, e0135640.
Rovida, F., Campanini, G., Piralla, A., Adzasehoun, K. M., Sarasini, A., & Baldanti, F. (2013). Molecular detection of gastrointestinal viral infections in hospitalized patients. Diagnostic Microbiology and Infectious Disease, 77, 231–235.
Sadeghi, M., Kantola, K., Finnegan, D. P., McCaughey, C., Hedman, L., Soderlund-Venermo, M., & Hedman, K. (2013). Possible involvement of human bocavirus 1 in the death of a middle-aged immunosuppressed patient. Journal of Clinical Microbiology, 51, 3461–3463.
Schildgen, O. (2013). Human bocavirus: lessons learned to date. Pathogens, 2, 1–12.
Sinclair, R. G., Choi, C. Y., Riley, M. R., & Gerba, C. P. (2008). Pathogen surveillance through monitoring of sewer systems. Advances in Applied Microbiology, 65, 249–269.
Smith, D. B., Paddy, J. O., & Simmonds, P. (2015). The use of human sewage screening for community surveillance of hepatitis E virus in the UK. Journal of Medical Virology, 88, 915–918.
Tozer, S. J., Lambert, S. B., Whiley, D. M., Bialasiewicz, S., Lyon, M. J., Nissen, M. D., & Sloots, T. P. (2009). Detection of human bocavirus in respiratory, fecal, and blood samples by real-time PCR. Journal of Medical Virology, 81, 488–493.
Vicente, D., Cilla, G., Montes, M., Perez-Yarza, E. G., & Perez-Trallero, E. (2007). Human bocavirus, a respiratory and enteric virus. Emerging Infectious Diseases, 13, 636–637.
Victoria, M., Tort, L. F., Lizasoain, A., Garcia, M., Castells, M., Berois, M., et al. (2016). Norovirus molecular detection in Uruguayan sewage samples reveals a high genetic diversity and GII.4 variant replacement along time. J Appl Microbiol. doi:10.1111/jam.13058.
Acknowledgments
We thank Professor Herbert W. Virgin, Washington University (St. Louis, Missouri, United States) for providing the murine norovirus strain used as sample process control.
We thank the assistance of the following members of the Italian Wastewater Network for wastewater sample collection: E. Lorenzi, M. De Ceglia (SMAT Spa, Castiglione T.se, Torino), L. Meucci, D. Giacosa, A. Poncino (Centro Ricerche SMAT, Torino); N. Mesiano (SMAT, Collegno); W. Bodini, C. Amadasi (Vettabbia Spa, Milano); F. Montanelli, L. Boscolo, P. Viola, F. Pizza (Amiacque Spa, Milano); M. Tomasoni, D. Monteverdi, L. Massafra (A2A Spa, Brescia); Werner Strobl, M. Poli; E. Scarperi, (APPA, Bolzano), M. Poli, M. Dekas (Eco Center Spa AG, Bolzano); L. Bruni, G. Gatti, L. Tomasi, G. Cimadon (Provincia Autonoma di Trento, Trento); I. Pellegrini, A. Ventura, C. Zorzetto (ARPA FVG, Trieste); P. Parati, E. Dell’Andrea, G. Gambillara (Arpa Veneto, Venezia); C. Galas, R. Tomassini (Arpalazio, Roma), G. Ranalletta (ACEA ATO2 S.p.A., Roma); V. Perrino, M. Mariani (ARPA Puglia, Bari); G. Manassero (Arpa Valle D’Aosta, Aosta); A. Martello (Corpo Forestale Aldostano); S. Gaiter, L. Sola (ARPA Genova); M.A. Corvaglia, P. Albertelli (ARPA Emilia Romagna, Bologna); C. Mengarelli, A. Trimboli (ARPA Marche, Ancona); A. Gambaccioni, M. Razzolini (Publiacqua S.p.A. Firenze); E. Renna, G. Saltalamacchia, M. Lucarini (ARPA Umbria, Perugia); A. Renzi, D. Rosoni, C. Spatola Mayo (ARTA Abruzzo, Pescara); A. Lucci, V. Colecchia, C. Iorio (ARPA Molise, Campobasso); E. Rufolo, R. Martino (ARPA Campania, Napoli); R. Vita, R. Masotti, R. Martoccia (ARPA Basilicata, Potenza); F. Pedulla, G. Belmusto (ARPA Calabria, Reggio Calabria); L. Librici, G. Abbate (ARPA Sicilia, Palermo); A.M. Mereu, M. Secci (ARPA Sardegna, Cagliari); R. Bucci, A. Schito, T. Carpinelli (ARPA Puglia, Lecce).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Iaconelli, M., Divizia, M., Della Libera, S. et al. Frequent Detection and Genetic Diversity of Human Bocavirus in Urban Sewage Samples. Food Environ Virol 8, 289–295 (2016). https://doi.org/10.1007/s12560-016-9251-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-016-9251-7