Abstract
Human bocavirus (HBoV) infections are related to respiratory and gastroenteric diseases. The aim of this study was to investigate the presence of HBoV in both sewage and surface waters in Uruguay. Sixty-eight sewage samples from the cities of Salto, Paysandú, Bella Unión, Fray Bentos, Treinta y Tres and Melo and 36 surface water samples from the cities of Salto, Florida and Santa Lucía were studied. HBoV was screened by multiplex qPCR for the detection of the four subtypes, followed by monoplex qPCRs for the independent quantification of each subtype. A qualitative PCR followed by DNA sequencing and phylogenetic analysis was carried out for molecular characterization of HBoV strains. HBoV was present in a high frequency (69%) in sewage and only one positive sample (3%) was found in surface water. Concerning sewage samples, HBoV1 was detected in 11 (23%) out of the 47 positives samples, with a mean concentration of 8.2 × 104 genomic copies/Liter (gc/L), HBoV3 was detected in 35 (74%) of the positive samples with a mean concentration of 4.1 × 106 gc/L and subtypes 2 and/or 4 were detected in 39 (83%) of the positive samples with a mean concentration of 7.8 × 106 gc/L. After the phylogenetic analysis performed by a Bayesian approach, the four HBoV subtypes were confirmed. This is the first study determining a high frequency of HBoV and the presence of the four HBoV subtypes in aquatic matrices in Latin America, mainly in sewage. Although HBoV was scarcely detected in surface water, a waterborne transmission is likely to occur if people enter in contact with polluted surface waters for recreational activities such as fishing or swimming since an elevated frequency of HBoV was detected in raw sewage which is usually directly discharged into surface waters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human bocaviruses (HBoV) are non-enveloped viruses with an icosahedral capsid belonging to the family Parvoviridae, subfamily Parvovirinae, genus Bocaparvovirus. The genome is composed by a negative sense single-stranded DNA of 5.3 kb with three open-reading frames (ORFs) which codify two non-structural (NS1 and NP1 in ORF1 and ORF2, respectively) and two structural (VP1 and VP2 in ORF3) proteins (Guido et al. 2016).
HBoV strains are genetically classified into four subtypes: HBoV1, 2, 3 and 4. HBoV1 was the first detected in 2005 from samples of the respiratory tract (Allander et al. 2005) and is commonly associated with respiratory infections and diseases. Subtypes 2, 3 and 4 were initially detected in human stool samples (Arthur et al. 2009; Kapoor et al. 2009, 2010) and are associated with gastrointestinal infections. Previous studies have reported the presence of HBoV in up to 63% of the stool specimens analyzed, estimating a mean global prevalence of 6% in gastrointestinal infections (Guido et al. 2016).
Concerning HBoV dissemination into the environment, there are studies documenting the presence of HBoV in surface water (37–40%) and sewage (60–93%) worldwide (Blinkova et al. 2009; Hamza et al. 2009; Bibby and Peccia 2013; Myrmel et al. 2015; Iaconelli et al. 2016; Hamza et al. 2017; La Rosa et al. 2017). The presence of HBoV in environmental waters in Latin America is unknown although several studies performed in this region evidenced the presence of HBoV in fecal specimens from children with gastroenteritis and suggested a causative role for HBoV in gastrointestinal manifestations observed in acute respiratory infection patients in Brazil (Albuquerque et al. 2007; Proenca-Modena et al. 2011).
Several enteric viruses (Aichivirus, Astrovirus, Norovirus and Rotavirus) have been studied in sewage in Uruguay (Victoria et al. 2014, 2016; Burutarán et al. 2015; Lizasoain et al. 2015; Tort et al. 2015). However, the presence of HBoV is unknown in our country. Therefore, the aim of this study was to determine the frequency, viral concentration and molecular characterization of HBoV through the analysis of both sewage and surface water from Uruguay, as a useful approach to describe HBoV strain circulation in the local population where the environmental samples were collected.
Materials and Methods
Collection of Samples
Sewage samples (42 mL each) were collected in four cities (Bella Unión, Salto, Paysandú and Fray Bentos) in the northwestern region and two (Melo and Treinta y Tres) in the eastern region of Uruguay. Salto, Paysandú and Fray Bentos discharge raw sewage directly to the Uruguay river which is the most important river of the country as previously mentioned by Victoria et al. (2014); on the other hand, Bella Unión treats its sewage by a stabilization pond. Both cities located in the eastern region present a Sewage Treatment Plant with activated sludge treatment followed by ultraviolet (UV) disinfection. In all the cities, raw sewage was collected with the exception of Bella Unión where effluent samples were collected.
Sewage samples were collected fortnightly between March 2011 and February 2012 in the cities located in the northwestern region of the country as described by Victoria et al. (2014). For this study, these samples were pooled to obtain one sample per month in each city. Sewage samples in Melo and Treinta y Tres were collected bi-monthly between September 2011 and April 2013.
Surface water samples (500 mL each) were collected monthly in Uruguay river, downstream to Salto and in Santa Lucía river (southern region of the country), downstream to Florida and Santa Lucía cities between June 2015 and May 2016. It is worth mentioning that Santa Lucía river is the source of drinking water for Montevideo (1,318,755 inhabitants), the capital of Uruguay located downstream of this basin.
Viral Concentration and Nucleic Acid Extraction
Sewage samples were subjected to ultracentrifugation method to perform the viral concentration as previously described in Victoria et al. (2014) and Tort et al. (2015).
Concentration of viral particles from surface water samples was performed by adsorption and elution to a negatively charged membrane (Katayama et al. 2002; Haramoto et al. 2009).
Nucleic acid extraction was performed with the QIAmp Cador Pathogen mini kit (QIAGEN®, Hilden, Germany) according to the manufacturer’s instructions.
Screening and Quantification of HBoV
Screening and quantification of HBoV was performed by a multiplex quantitative PCR (qPCR) with TaqMan® technology, which detects the four HBoV subtypes (Kantola et al. 2010). Positive samples by this multiplex qPCR were analyzed for each subtype by monoplex qPCRs for subtypes 1 and 3 and by a multiplex qPCR for subtypes 2 and 4 (Kantola et al. 2010). The nucleic acid of negative HBoV samples was diluted tenfold and subjected to qPCR reactions to overcome the presence of inhibitors. Primers and probes were directed towards the 5′ untranslated region (UTR) and the 5′ region of the NS1 gene of HBoV genome (Kantola et al. 2010). Reactions were performed in duplicate according to manufacturer’s recommendations with SensiFastTM II Probe Kit (Bioline Reagents Ltd.) and a Rotor-Gene Q instrument (QIAGEN®, Hilden, Germany) (Table 1).
One HBoV genome fragment amplified by quantitative PCR for each subtype was cloned into a plasmid and confirmed by sequencing to generate four standard curves to quantify all HBoV subtypes. Standard curves were constructed with eight points of tenfold serial dilutions of plasmid (107 to 100 genomic copies/reaction) that yielded slopes of − 3.66, − 3.21, − 3.34, − 3.73 and reaction efficiencies of 0.88, 1.05, 0.99, 0.85 for multiplex qPCR, monoplex of HBoV1, monoplex of HBoV3 and multiplex of HBoV2/4, respectively. The coefficient of correlation (R2) was 0.99 for all qPCR reactions.
HBoV Molecular Characterization
Positive samples detected and quantified by qPCR were subjected to amplification by a qualitative nested PCR for molecular characterization of HBoV Uruguayan strains as described by Iaconelli et al. (2016) and La Rosa et al. (2015). Both sets of primers target the VP1–VP2 region of the HBoV genome with an expected amplicon of 382 bp (Table 1).
Amplicons were purified using PureLink™ Quick Gel Extraction and PCR Purification Combo kit (Invitrogen, Carlsbad, California, United States) and sequenced by Macrogen Platform (South Korea) in an ABI3730XL Genetic Analyzer (Applied Biosystems, CA, USA). Sequences were edited with SeqMan® Software (DNAstar Lasergene®) and aligned using MUSCLE program (Edgard 2004) with reference sequences obtained from the NCBI Database. Model that best adjusts to the dataset was calculated using j-Model test program (Guindon and Gascuel 2003; Darriba et al. 2012). Phylogenetic trees were constructed with a Bayesian approach using Mr. Bayes program (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) with 400,000 generations and a sample frequency of 200.
Statistical Analyses
Statistical analyses were performed using the R software (R Core Team 2013). Associations between positive HBoV samples and location, seasonality, and detected subtypes were carried out using the Chi-squared test. p values were used as the measure of association between the studied variables.
Results
HBoV in Sewage Samples
HBoV was detected in 69% (47/68) of the sewage samples analyzed. In the six studied cities, HBoV detection ranged from 67 to 90%, except for Bella Unión city, where a frequency of 33% was observed in treated sewage, which was significantly different (p = 0.02). Concerning HBoV subtypes, HBoV2/4 were the prevalent, observed in 58% of the analyzed samples, followed by HBoV3, which was detected in 51% and HBoV1 which was detected in 16% (Table 2). It is important to highlight that the frequency of detection of gastroenteric subtypes (HBoV2/4 and 3) were higher than the frequency of the respiratory subtype (HBoV1) (p = 0.003). All negative samples that were tenfold diluted and amplified for the detection of HBoV were negative, which confirms the absence of inhibitors for the enzymatic reactions.
Different subtypes were co-detected in 62% of the samples. In 2% of them subtypes 1 and 3 were co-detected while 20% presented subtypes 1, 3 and 2/4, and 40% presented subtypes 3 and 2/4 (data not shown).
Concerning to the temporal distribution of HBoV in sewage from the northwestern region of Uruguay, it was observed that HBoV was detected throughout the year with no clear pattern of seasonality (p = 0.43) (Fig. 1). Similar results were observed from the analysis of the sewage samples collected in the eastern region (data not shown).
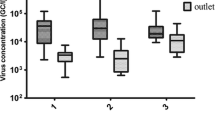
HBoV concentrations determined by qPCR identified HBoV1 as the subtype with the lowest concentration (mean of 8.2 × 104 genomic copies/L (gc/L), ranging from 1.9 × 104 gc/L to 2.2 × 105 gc/L). Higher concentrations were observed for subtypes frequently associated with gastroenteritis: HBoV3 presented a mean of 4.1 × 106 gc/L, ranging from 4.9 × 104 to 2.5 × 107 gc/L and HBoV 2/4 presented a mean concentration of 7.8 × 106 gc/L ranging from 1.2 × 105 to 8.1 × 107gc/L (Table 3).
HBoV in Surface Water Samples
HBoV was detected in 3% of the analyzed surface water samples (1/36). The HBoV-positive sample corresponding to the Santa Lucía river was collected in Florida and belongs to subtype 3 (Fig. 2), with a concentration of 2.7 × 104 genomic copies/L.
Phylogenetic tree constructed by the Bayesian method of HBoV detected in sewage and surface water in six Uruguayan cities. The sequences used corresponded to the partial VP1/VP2 gene (382 nts). The model that best adjusted to the dataset was HKY with gamma distribution. Reference strains are shown with the corresponded accession number and subtype. Names of Uruguayan strains are coded as followed: city code-month and year of collection-primers used in nested PCR-accession number-preceded by a black circle. BU Bella Unión, SA Salto, PY Paysandú, FB Fray Bentos, TyT Treinta y Tres, CL Cerro Largo (Melo) and FL Florida. Only posterior values over 90% are shown
Molecular Characterization of HBoV
Six out of eleven (54%) samples amplified by qPCR for HBoV1 and 25 out of 47 (53%) samples that were amplified by qPCR for HBoV2/4 and/or 3 were successfully sequenced. The most frequent subtype confirmed by phylogenetic analysis was HBoV3 with 12 sequences, followed by HBoV2 with 11 sequences, 6 sequences were classified as HBoV1 and 2 as HBoV4 (Fig. 2).
Discussion
As far as we know, this is the first report concerning the detection, quantification and molecular characterization of HBoV in Uruguay, as well as the first performed in sewage and surface water in Latin America.
The results obtained in this study showed that HBoV is frequently excreted by infected persons since a high frequency (69%) was observed in the analyzed sewage samples. This was in agreement with previous studies performed in other regions of the world that reported frequencies ranging between 59 and 93% (Blinkova et al. 2009; Bibby and Peccia 2013; Myrmel et al. 2015; Iaconelli et al. 2016; Hamza et al. 2017).
HBoV was detected in all cities, with frequencies ranging from 67 to 90%. Only Bella Unión presented a significant lower frequency of detection (33%), this could be because these samples were obtained from the effluent of a sewage treatment plant (stabilization pond). This kind of treatment allows a moderate remotion of enteric viruses (generally less than one log10-unit). Nevertheless, in our study, we were unable to determine the removal efficiency of this pond since no raw sewage sample was obtained (Verbyla and Mihelcic 2015).
Sewage samples collected in the northwestern region of Uruguay have been previously analyzed for several enteric viruses evidencing frequencies ranging from 45 to 72%. However, HBoV presented one of the highest frequencies of detection (Burutarán et al. 2015; Tort et al. 2015; Lizasoain et al. 2015; Victoria et al. 2016). In this study, HBoV was frequently detected in sewage samples but only once detected in surface water. On the other hand, La Rosa et al. (2017) observed a high frequency of HBoV in surface water suggesting that this virus could be used as a viral indicator of fecal contamination. Further studies are needed to determine the frequency of this virus in both sewage and surface water in other temperate regions of the world to confirm HBoV as a viral indicator of fecal contamination (Lau et al. 2007; Campe et al. 2008; Chhabra et al. 2013).
In this study, both the frequency of detection and concentration were higher for HBoV3, 2/4 than HBoV1. This is likely to occur since subtypes 2, 3 and 4 are associated with gastrointestinal infections, thus they could be replicating in the epithelium of the gastrointestinal tract, while HBoV1 infects epithelial cells of the respiratory tract. It has been suggested that HBoV1 once replicated in the respiratory tract can be swallowed together with nasopharyngeal secretion and can be detected in stools or sewage with lower titer than that observed in nasopharyngeal swabs (Proenca-Modena et al. 2013). There is scarce information concerning the concentration of HBoV in sewage worldwide. In our study, we observed concentration values one log higher for HBoV1, and two log higher for HBoV 2/3/4 when compared with quantifications described by other studies (Myrmel et al. 2015; Iaconelli et al. 2016; Hamza et al. 2017). Although the waterborne transmission has not been proven for HBoV, its elevated frequency and concentration in sewage samples suggest that this transmission route should not be ruled out. Moreover, previous studies performed with these sewage samples showed similar concentrations of enteric viruses clearly associated with waterborne transmission such as rotavirus and norovirus (Victoria et al. 2014).
Despite a seasonal pattern has been described for HBoV obtained from nasopharyngeal aspirates and stool samples (Choi et al. 2006; Nawaz et al. 2012), we have not detected a seasonal pattern in this study. This observation was confirmed statistically and is in agreement with results obtained by other authors (Hamza et al. 2009; Iaconelli et al. 2016; Hamza et al. 2017).
The presence of HBoV was also investigated in surface water samples collected in two important rivers of Uruguay. A low frequency of HBoV was observed with only one positive sample (3%) out of 36 analyzed. There are only two studies where HBoV was analyzed in river samples which indicate higher frequencies than that observed in our study (Hamza et al. 2009; La Rosa et al. 2017). These differences in the frequencies of HBoV detection can be observed since the previous studies used higher volume of surface water to perform the viral concentration and the basin of these rivers are located in areas with high population densities. The positive sample observed in our study corresponds to HBoV3, however, results from a previous study indicated that HBoV2 is the prevalent in surface waters collected in rivers from Italy (La Rosa et al. 2017).
Phylogenetic analysis was performed with 25 sequences out of 47 positives samples for HBoV2, HBoV3 or HBoV4, and 6 sequences out of 11 positives samples for HBoV1. We were unable to obtain the sequence of 27 positive samples which can be due to the higher sensitivity of qPCR when compared with qualitative conventional PCR. However, studies comparing the sensitivity between qPCR and Nested PCR for HBoV are lacking. The phylogenetic tree showed that 12 sequences obtained in this study clustered in the clade of HBoV3, 11 in the clade of HBoV2, 6 in HBoV1 clade, and 2 corresponded to HBoV4. These results were statistically supported since these clades present posterior values higher than 90%. Phylogenetic analysis confirmed that samples that were positive by qPCR for HBoV2 or 4 corresponded mostly to subtype 2 with only 2 sequences belonging to subtype 4. These results were in accordance with previous studies confirming that HBoV4 is rarely detected both in environmental (Myrmel et al. 2015; Iaconelli et al. 2016; Hamza et al. 2017) and clinical samples (Kapoor et al. 2010; Kantola et al. 2010).
To the best of our knowledge, this is the first work where the presence of HBoV was analyzed in Uruguay, confirming an elevated frequency in sewage from all the six studied cities. Although all the HBoV subtypes described where detected in sewage samples, those subtypes associated with gastrointestinal infections where observed in higher concentration than HBoV1, which is associated with respiratory infections. Taken together, HBoV is frequently excreted by infected persons and is present in sewage which suggests that a waterborne transmission of this virus is likely to occur if sewage is discharged in recreational waters, consumption water, water for irrigation of crops and bivalve cultivation.
References
Albuquerque, M. C. M., Rocha, L. N., Benati, F. J., Soares, C. C., Maranhão, A. G., Ramirez, M. L., et al. (2007). Human bocavirus infection in children with gastroenteritis, Brazil. Emerging Infectious Diseases, 13, 1756–1758.
Allander, T., Tammi, M. T., Eriksson, M., Bjerkner, A., TiveljungLindell, A., & Andersson, B. (2005). Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proceedings of the National academy of Sciences of the United States of America, 102, 12891–12896.
Arthur, J. L., Higgins, G. D., Davidson, G. P., Givney, R. C., & Ratcliff, R. M. (2009). A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathogens, 5, e1000391.
Bibby, K., & Peccia, J. (2013). Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environmental Science and Technology, 47, 1945–1951.
Blinkova, O., Rosario, K., Li, L., Kapoor, A., Slikas, B., Bernardin, F., et al. (2009). Frequent detection of highly diverse variants of cardiovirus, cosavirus, bocavirus, and circovirus in sewage samples collected in the United States. Journal of Clinical Microbiology, 47, 3507–3513.
Burutarán, L., Lizasoain, A., García, M., Tort, L. F., Colina, R., & Victoria, M. (2015). Detection and molecular characterization of aichivirus 1 in wastewater samples from Uruguay. Food and Environmental Virology, 8, 13–17.
Campe, H., Hartberger, C., & Sing, A. (2008). Role of human bocavirus infections in outbreaks of gastroenteritis. Journal of Clinical Virology, 43, 340–342.
Chhabra, P., Payne, D. C., Szilagyi, P. G., Edwards, K. M., Staat, M. A., Shirley, S. H., et al. (2013). Etiology of viral gastroenteritis in children < 5 years of age in the United States, 2008–2009. Journal of Infectious Diseases, 208, 790–800.
Choi, E. H., Lee, H. J., Kim, S. J., Eun, B. W., Kim, N. H., Lee, J. A., et al. (2006). The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000–2005. Clinical Infectious Diseases, 43(5), 585–592.
Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9(8), 772.
Edgard, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–1795.
Guido, M., Tumolo, M. R., Verri, T., Romano, A., Serio, F., De Giorgi, M., et al. (2016). Human bocavirus: current knowledge and future challenges. World Journal of Gastroenterology, 22, 8684–8697. https://doi.org/10.3748/wjg.v22.i39.8684.
Guindon, S., & Gascuel, O. (2003). A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology, 52, 696–704.
Hamza, I. A., Jurzik, L., Wilhelm, M., & Uberla, K. (2009). Detection and quantification of human bocavirus in river water. Journal of General Virology, 90, 2634–2637.
Hamza, H., Leifels, M., Wilhelm, M., & Hamza, I. A. (2017). Relative abundance of human bocaviruses in urban sewage in Greater Cairo, Egypt. Food and Environmental Virology. https://doi.org/10.1007/s12560-017-9287-3.
Haramoto, E., Katayama, H., Utagawa, E., & Ohgaki, S. (2009). Recovery of human norovirus from water by virus concentration methods. Journal of Virological Methods, 160(1–2), 206–209.
Huelsenbeck, J. P., & Ronquist, F. (2001). MR BAYES: bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755.
Iaconelli, M., Divizia, M., Della Libera, S., Di Bonito, P., & La Rosa, G. (2016). Frequent detection and genetic diversity of human bocavirus in urban sewage samples. Food and Environmental Virology, 8, 289–295.
Kantola, K., Sadeghi, M., & Antikainen, J. (2010). Real-time quantitative PCR detection of four human bocaviruses. Journal of Clinical Microbiology, 48, 4044–4050.
Kapoor, A., Simmonds, P., Slikas, E., Li, L., Bodhidatta, L., Sethabutr, O., et al. (2010). Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. Journal of Infectious Diseases, 201, 1633–1643.
Kapoor, A., Slikas, E., Simmonds, P., Chieochansin, T., Naeem, A., Shaukat, S., et al. (2009). A newly identified bocavirus species in human stool. Journal of Infectious Diseases, 199, 196–200.
Katayama, H., Shimasaki, A., & Ohgaki, S. (2002). Development of a virus concentration method and its application to detection of enterovirus and norwalk virus from coastal seawater. Applied and Environmental Microbiology, 68, 1033–1039.
La Rosa, G., Della Libera, S., Iaconelli, M., Donia, D., Cenko, F., Xhelilaj, G., et al. (2015). Human bocavirus in children with acute gastroenteritis in Albania. Journal of Medical Virology, 88, 906–910.
La Rosa, G., Sanseverino, I., Della Libera, S., Iaconelli, M., Ferrero, V. E. V., Barra Caracciolo, A., et al. (2017). The impact of anthropogenic pressure on the virological quality of water from the Tiber river, Italy. Lett Appl Microbiol. https://doi.org/10.1111/lam.12774.
Lau, S. K., Yip, C. C., Que, T. L., Lee, R. A., Au-Yeung, R. K., Zhou, B., et al. (2007). Clinical and molecular epidemiology of Human Bocavirus in respiratory and fecal samples from children in Hong Kong. Journal of Infectious Diseases, 196, 986–993.
Lizasoain, A., Tort, L. F., García, M., Gómez, M. M., Cristina, J., Leite, J. P., et al. (2015). Environmental assessment of classical human astrovirus in Uruguay. Food and Environmental Virology, 7, 142.
Myrmel, M., Lange, H., & Rimstad, E. (2015). A 1-year quantitative survey of noro-, adeno-, human boca-, and hepatitis e viruses in raw and secondarily treated sewage from two plants in Norway. Food and Environmental Virology, 7, 213–223.
Nawaz, S., Allen, D. J., Aladin, F., Gallimore, C., & Iturriza-Gomara, M. (2012). Human bocaviruses are not significantly associated with gastroenteritis: results of retesting archive DNA from a case control study in the UK. PLoS ONE, 7(7), e41346.
Proenca-Modena, J. L., Gagliardi, T. B., Escremim de Paula, F., Iwamoto, M. A., Criado, M. F., Camara, A. A., et al. (2011). Detection of human bocavirus mRNA in respiratory secretions correlates with high viral load and concurrent diarrhea. PLoS ONE, 6(6), e21083.
Proenca-Modena, J. L., Martinez, M., Amarilla, A. A., Espínola, E. E., Galeano, M. E., Fariña, N., et al. (2013). Viral load of Human Bocavirus-1 in stools from children with viral diarrhoea in Paraguay. Epidemiology and Infection, 141(12), 2576–2580.
Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12), 1572–1574.
R Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL. http://www.R-project.org/.
Tort, L. F., Victoria, M., Lizasoain, A., García, M., Berois, M., Cristina, J., et al. (2015). Detection of common, emerging and uncommon VP4, and VP7 human group A rotavirus genotypes from urban sewage samples in Uruguay. Food and Environmental Virology, 7, 342–353.
Verbyla, M. E., & Mihelcic, J. R. (2015). A review of virus removal in wastewater treatment pond systems. Water Research, 71, 107–124.
Victoria, M., Tort, L. F., García, M., Lizasoain, A., Maya, L., Leite, J. P., et al. (2014). Assessment of gastroenteric viruses from wastewater directly discharged into Uruguay river, Uruguay. Food and Environmental Virology, 6, 116–124.
Victoria, M., Tort, L. F., Lizasoain, A., García, M., Castells, M., Berois, M., et al. (2016). Norovirus molecular detection in Uruguayan sewage samples reveals a high genetic diversity and GII.4 variant replacement along time. Journal of Applied Microbiology, 120, 1427–1435.
Acknowledgments
We would like to thank the financial support given by the program ‘‘Polo de Desarrollo Universitario (PDU), Universidad de la República (UdelaR), Uruguay’’; project CSIC I+D 2010 and project CSIC I+D 2014, UdelaR. We also thank OSE and the staff of the Virology Section, School of Sciences, UdelaR, Uruguay for their technical assistance. MS is a master student at “Programa de Desarrollo de las Ciencias Básicas - PEDECIBA” and has a scholarship from “Agencia Nacional de Investigación e Innovación - ANII”, Uruguay.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Salvo, M., Lizasoain, A., Castells, M. et al. Human Bocavirus: Detection, Quantification and Molecular Characterization in Sewage and Surface Waters in Uruguay. Food Environ Virol 10, 193–200 (2018). https://doi.org/10.1007/s12560-017-9334-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-017-9334-0