Abstract
Information about Human Enterovirus circulation in Uruguay is scarce. The aim of this study was to generate the first description about their circulation in the country through the study of sewage samples collected before and after the switch from Oral Poliovirus Vaccine to Inactivated Poliovirus Vaccine. Viruses were concentrated by an adsorption–elution to a negatively charged membrane, and real-time quantitative PCR and qualitative PCR methods were used to detect, quantify, and characterize enteroviruses. Positive samples were inoculated in RD cells and two passages were performed. Additionally, RD+ samples were subsequently passed onto L20B cells. Human Enteroviruses were detected in 67.6% of the samples, with concentrations between 4.9 and 6.6 Log10 genomic copies per liter. 10% of positive samples replicated in RD cells, of which none in L20B cells. Molecular characterization of Human Enterovirus strains directly detected from sewage sample concentrates allowed the identification of highly divergent members of species C such as Enterovirus C99 and Coxsackievirus A13, as well as the frequent detection of species A and B members (particularly Coxsackievirus A16 and Echovirus 6, respectively). Other detected types were Coxsackievirus A2, A22, B1, B5, Echovirus 5, and 9. The characterization of viruses isolated in cell culture revealed the presence of Echovirus 6 and Coxsackievirus B3. Despite the absence of poliovirus, a wide circulation of different enterovirus types was evidenced in Uruguayan sewage samples, highlighting that the local populations are exposed to different kinds of diseases originated by several human enterovirus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human Enteroviruses (HEVs) are classified into four viral species (HEV-A to HEV-D) inside the genus Enterovirus, Picornaviridae family (Knowles et al. 2011). The virions are non-enveloped, with an icosahedral symmetry of approximately 30 nm of diameter, containing a single-stranded positive-sense RNA genome about 7.5 kb in length with 5′ and 3′ Non-Coding Regions (NCRs) flanking a unique Open Reading Frame (ORF). A single polyprotein is translated from the ORF, followed by proteolytic cleavages generating capsid (VP1–VP4) and non-structural (2A–2C, and 3A–3D) proteins such as protease and RNA-dependent RNA polymerase (Pallansch et al. 2013).
While HEVs usually cause silent or subclinical infections (Witsø et al. 2007), they are associated with a wide spectrum of clinical manifestations. HEVs could be responsible from a common cold or minor febrile illness to severe diseases as hand foot and mouth (HFM) disease, acute hemorrhagic conjunctivitis, acute myocarditis, aseptic meningitis, encephalitis, cardiac disease, severe neonatal sepsis-like disease, or acute flaccid paralysis (AFP) (Blondel et al. 2005; Muehlenbachs et al. 2015). The ratio of number of diseases cases to number of infections depends on the HEV serotype (Nathanson and Kew 2010) and reported cases of infection represent a small proportion of infected people in a community (Hovi et al. 1986).
HEV classification was initially based on antigenicity, biological activity, and disease, grouping them into Coxsackievirus A (CVA), Coxsackievirus B (CVB), Poliovirus (PV), and Echovirus (E). Nevertheless this scheme collapsed when new emerging viruses with different serological characteristics to those already described were discovered, and as a consequence they were numbered starting from EV68 (Pallansch et al. 2013).
Afterwards, the molecular typing overcame the difficulties of the EV typing old scheme and helped to establish a new classification system. In fact, nucleotide sequence of VP1 coding segment correlates well with enterovirus serotype and members of the same serotype usually share more than 75% identity at nucleotide level (85% at amino acid level) in this region of the genome (Oberste et al. 1999; Oberste and Pallansch 2005). Up to date it has been identified 3 PV, 21 CVA, 6 CVB, 28 E, and 48 numbered EV types (The Pirbright Institute 2018).
Since HEVs replicate in the gastrointestinal tract and are shed in huge concentrations in feces from both asymptomatic and symptomatic individuals, the fecal–oral route is the main route of transmission (Hovi et al. 2012). This leads to human feces from sewage being the major source of HEVs in the environment, spreading to other environmental water matrixes like lakes, rivers, or seas (Connell et al. 2012; Allmann et al. 2013; Prevost et al. 2015).
Nevertheless, structural stability of HEVs in the environment allows the study of sewage samples with the aim to describe the epidemiology of them in the human population, which excretes feces to sanitary sewer network (Global Polio Eradication Initiative 2015).
Environmental surveillance is an effective approach investigating circulation of HEVs in several communities (Wieczorek et al. 2011; Battistone et al. 2014; Tao et al. 2016; Farías et al. 2018) and provides supplementary information when clinical cases reports are absent or are carried out under poor-quality standards. Several countries, both for early detection of wild PV (WPV) introduction and transmission as well as for detection of vaccine-derived neuro-virulent polioviruses (VDPVs) that emerge following the use of Oral Poliovirus Vaccine (OPVs), implemented this approach.
In Uruguay, the available information about the circulation of HEVs in the population is scarce and proceed from the study of meningitis and encephalitis of viral origin as well as the AFP, which constitute notifiable diseases to the public health authorities (Uruguayan Government 2012). Uruguay reported the last case with isolation of WPV in 1978 and certified the elimination of paralysis cases due the virus in 1994, jointly with the Americas (de Quadros et al. 1997). Since then, Uruguay never registered cases of paralysis by VDPVs, despite the utilization of the trivalent OPV up to May 2012, when the attenuated vaccine was completely replaced by the Inactivated Poliovirus Vaccine (IPV) in the National Calendar of Vaccination.
Beyond meningitis, encephalitis, and acute flaccid paralysis, and up to our best knowledge, Uruguay lacks a HEV surveillance system and the etiological agent of most aforementioned diseases remains unknown.
This work attempts to generate a base line of information about the HEV circulation in Uruguay, conducting a retrospective study of sewage samples collected in different cities, mainly during the OPV vaccination period.
Methods
Environmental Samples
This is a retrospective study of two sets of environmental samples (42 ml each) collected in Uruguay between 2011 and 2013. Set One was composed of 96 sewage grab samples obtained bi-weekly between March 2011 and February 2012 from four cities located at the western region of the country (Bella Unión [BU], Salto [SA], Paysandú [PY], and Fray Bentos [FB] cities). In Bella Unión, samples were collected downstream of a stabilization pond, while in the other cities untreated sewage was collected. Set Two was composed of 20 composite sewage samples (24 h) collected bi-monthly between September 2011 and April 2013 from the influent of two sewage treatment plants located in two cities from the eastern region of the country: Melo (ME) and Treinta y Tres (TT). For Set One, samples from each month and each city were pooled reducing the 96 initial samples to 48 samples of 84 ml each. Samples were stored at − 20 °C from the collection data until the virological analysis.
Internal Process Control
Before viral concentration, 4.80 × 109 genomic copies of a PP7 bacteriophage were inoculated in each sample as an Internal Process Control (IPC) (Rajal et al. 2007).
Viral Concentration
Viral particles were concentrated using a protocol of adsorption/elution to a negatively charged membrane described by Katayama et al. (2002) with modifications (Haramoto et al. 2009). Briefly, MgCl2 was added to the sample to a final concentration of 25 mM before filtering through a type HA negatively charged membrane with a diameter of 47 mm and a pore size 0.45 µm. The membrane was rinsed with 200 ml of 0.5 mM H2SO4 (pH 3.0) and placed into a petri dish where the elution of viruses was performed by stirring with 4 ml of 1 mM NaOH (pH 10.8) for 10 min. To neutralize the solution, 40 µl of 50 mM H2SO4 and 40 µl of 100X TE Buffer (pH 8.00) were added.
The concentrated sample was centrifuged for 20 min at 4000×g to clarify, and the supernatant was stored at − 20 °C for further analysis.
RNA Extraction and cDNA Synthesis
The RNA extraction was performed from 200 µl of viral concentrate using the QIAamp cador Pathogen Mini Kit (Qiagen) according to the manufacturer’s instructions to obtain 60 µl of RNA extracted solution. cDNA synthesis with random hexamers (Thermo Fisher Scientific) and gene-specific primers were carried out, depending on the PCR protocols requirements, from 10 µl of extracted RNA using the RevertAid Reverse Transcriptase (Thermo Fisher Scientific).
For gene-specific cDNA synthesis, primers AN32, AN33, AN34, and AN35 (Nix et al. 2006) were used at a final concentration of 0.4 µM each and dNTPs were used in a concentration of 1000 µM each. Random hexamers at a final concentration of 2.0 ng/µl were used for random cDNA synthesis.
Real-Time PCR for the IPC and for Human Enteroviruses
Real-time PCRs were carried out with TaqMan technology and reactions were performed in duplicated with 2X Sensi Fast Probe No Rox Kit (Bioline Reagents) in a Rotor-Gene Q instrument (Qiagen) following manufacturer’s instructions.
The PP7 genomic detection was performed with 247F and 320R primers towards the helicase coding segment (Rajal et al. 2007).
Quantification of HEVs was achieved using rRT-R and rRT-F primers (Oberste et al. 2010) towards the 5′ NCR region of the enterovirus genome with a standard curve performed with serial dilutions (106–100) of a plasmid containing the fragment of the expected size as an insert.
Each reaction of PP7 detection and HEV quantification was performed from five microliters of random cDNA.
Human Enterovirus Molecular Characterization
The HEV molecular characterization was carried out by a qualitative PCR towards the VP1 capsid protein coding segment with the pair of primers 222/224 and AN88/AN89 described by Nix et al. (2006) in the first- and second-round PCRs, respectively. First-round PCR reaction was performed from 5.0 µl of gene-specific cDNA, and second-round PCR reaction was performed from 2.0 µl of first-round PCR product, with 2X Mango Mix (Bioline Reagents) following the manufacturer’s instructions.
Second-round PCR products were resolved in a 2% agarose gel electrophoresis and the expected size amplicons (~ 350 bp) were excised from the gel and purified with the Quick Gel Extraction and PCR Purification Combo Kit (Invitrogen™). Macrogen Inc. (Korea) performed DNA sequencing with primers AN88 and AN89. Sequences were curated with BioEdit Sequence Alignment Editor (Hall 1999) and aligned with Clustal W tool with reference sequences and sequences reported elsewhere retrieved from the GenBank database in Mega 6.0 software (Tamura et al. 2013).
Phylogenetic analyses were performed with Mega 6.0 software using the Neighbor-Joining method and Kimura two-parameter model (Tamura et al. 2013). As a measure of robustness of each node, the bootstrap method with 1,000 pseudo replicates was used. The sequences obtained in this study were submitted to the GenBank database with the accession numbers MF093652–MF093682 and MF537000–MF537003.
Human Enterovirus Isolation by Cell Culture
In order to know the viability of the detected viral particles, viral concentrates corresponding to positive samples were assayed in cell culture, performing the infection of RD (human rhabdomyosarcoma) cells. Each inoculum was vortexed with chloroform (1:1 v/v) by 2 min followed by a centrifugation step at 4000×g for 10 min. 0.3 ml of the upper phase was inoculated onto 25 cm2 flask with 80% confluent RD cell monolayer. After 5 days of incubation (36 °C, 5% CO2) and daily observation, second passages were performed. Those HEVs which replicate in RD cells were subsequently passed (two passages) onto L20B cells (mouse fibroblast cells that express the human poliovirus receptor) in order to allow the isolation of possible PV present in the sample.
RD (NIBSC Accession Number 081003) and L20B (NIBSC Accession Number 081102) cell lines were provided by the WHO Global Polio Laboratory Network (GPLN). Cell culture procedures were performed according to WHO guidelines (World Health Organization 2004).
Molecular Characterization of Human Enterovirus Isolates
One hundred and forty microliters of supernatant (after three cycles of freezing/thawing) of samples showing cytopathic effect were subject to RNA extraction with QiAmp Viral RNA Kit (Qiagen). The molecular characterization of isolates was performed following the protocol designed by Nix et al. (2006) as described above.
Results
Human Enterovirus Detection and Quantification
The IPC was detected in all samples through a TaqMan real-time PCR assay.
HEVs were detected in 42 samples (61.8%), of which four samples (9.52%) were determined as positive by qPCR towards 5′ NCR segment, 11 samples (26.2%) by qualitative PCR towards VP1 coding segment, and 27 (64.3%) were determined as positive by both methods.
Viral concentration ranged between 4.9 and 6.6 Log10 genomic copies per liter (Log10 gcl−1) presenting average values of 5.3 in Treinta y Tres, 5.5 in Fray Bentos, 5.6 in Salto, 5.8 in Melo, and 6.0 in Paysandú. In Bella Unión, three samples were positive by qualitative PCR, although none of them was positive by qPCR (Fig. 1).
Presence and concentration of Human Enteroviruses in sewage samples collected in cities from the western (a) and eastern (b) regions of Uruguay between March 2011 and April 2013. BU Bella Unión, SA Salto, PY Paysandú, FB Fray Bentos, ME Melo, and TT Treinta y Tres. Thick squares in the grid indicate samples in which Human Enteroviruses were detected by qualitative PCR towards VP1 segment (Nix et al. 2006). Values of concentration are expressed as Log10 of genomic copies per liter and are result of a qPCR towards the 5′ NCR in the enterovirus genome (Oberste et al. 2010)
Human Enterovirus Isolation in Cell Culture
All the positive samples (n = 42) were assayed in cell culture. HEVs were isolated in RD cells in four (~ 10%) samples from Salto (March 2011), Treinta y Tres (November 2011), Paysandú (February 2012), and Melo (April 2012). None of these four samples presented characteristic cytopathic effect of poliovirus in L20B cells.
Sequencing and Phylogenetic Analysis
Sequences were obtained from 31 strains out of 42 qualitative PCR-positive samples and from the four strains isolated in RD cells, and were used in phylogenetic reconstructions.
As Fig. 2 shows, 11 strains were identified as belonging to HEV-A, 20 strains to HEV-B, and 4 strains to HEV-C, while HEV species D was not detected.
Phylogenetic analysis of Human Enterovirus strains detected in Uruguayan sewage samples based on partial sequences of the VP1 coding segment. Phylogenetic trees for Human Enterovirus species A, B, and C are shown in a–c, respectively. Black circles or squares represent Uruguayan strains when characterized directly from sewage or from an isolate, respectively. The GenBank accession number followed by the three-letter country code (ISO 3166-1 standard) and the year of detection is used to indicate strains reported elsewhere. Reference strains are in bold. Only bootstrap values higher than 70% are shown. The bars at the bottom denote genetic distance
Regarding HEV-A (Fig. 2a), CVA2 and CVA16 were detected in three and eight samples, respectively. CVA16 Uruguayan strains clustered all together with a bootstrap support of 92% and a high degree of similarity among them (Table 1). On the other hand, CVA2 Uruguayan strains clustered into two distinct groups (~ 70% bootstrap support) with up to 15% of divergence at nucleotide level among them and with strains reported elsewhere (Table 1). The 20 strains of HEV-B (Fig. 2b) correspond to CVB1 (two strains), CVB3 (one strain), CVB5 (three strains), E6 (11 strains), E5 (two strains), and E9 (one strain) types. CVB5 and E6 Uruguayan strains were the most divergent HEV-B types and strains of each type clustered in two distinct groups. An Uruguayan CVB5 strain (MF093668) clustered outside of the other Uruguayan CVB5 strains (100% bootstrap support) presenting up to 8.6 and 3.9% of nucleotide and amino acid diversity with them (Table 1). The divergence among the Uruguayan E6 strains reached values up to 4.8 and 2.9% at nucleotide and amino acid level, respectively, and clustered into two distinct groups (100% bootstrap support). The most numerous group also contain E6 strains reported in 2009–2011 in Finland, Denmark, and Netherlands.
The strains characterized as HEV-C were CVA22 (two strains), CVA13 and EVC99 (one strain each) (Fig. 2c). The Uruguayan CVA22 strains were up to 6.2 and 2.9% divergent at nucleotide and aminoacid level, respectively, with CVA22 strains reported elsewhere (Table 1). On the other hand, the Uruguayan CVA13 strain was up to 22.5 and 12% divergent at nucleotide and amino acid levels, respectively, with strains reported in Argentina and Madagascar (Fig. 1c; Table 1). Although the Uruguayan EVC99 strain was also divergent around 20% at nucleotide level with strains reported elsewhere, it presented a lower degree of divergence at amino acid level with these strains compared with CVA13 (Table 1).
Temporal and Geographical Distribution of HEV Types
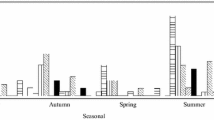
While types as CVB1, CVB3, CVA13, E5, and E9 were detected only in the eastern region of the country (Melo and Treinta y Tres), CVA16 or EVC99 were detected exclusively in the western region (Salto, Paysandú, and Fray Bentos). On the other hand, some types (E6, CVA2, CVA22, and CVB5) were detected in both regions of the country. Regarding the temporal fluctuation of the HEV types, although most of them were detected sporadically, CVA16 and E6 were consecutively detected in the periods of May–October and November–March, respectively, in the western region of the country (Fig. 3).
Temporal and geographical distribution of Human Enterovirus types detected in sewage samples from Uruguay between March 2011 and April 2013. The upper scheme represents cities of the western region of Uruguay (SA Salto, PY Paysandú, FB Fray Bentos) and the lower one, cities of the eastern region (ME Melo and TT Treinta y Tres)
Discussion
We have described herein the occurrence of several HEV types in the environment from Uruguay, which constitutes, up to our knowledge, the first report from this country concerning the environmental molecular diversity of HEVs.
Two methods designed originally for detection and molecular characterization of HEVs from clinical specimens (Nix et al. 2006; Oberste et al. 2010) were applied here in parallel, for HEV screening in sewage sample concentrates. A higher frequency of detection for HEVs was obtained by the use of these two methods in parallel, than using one of them only, as suggested by Oberste et al. (2010).
As the IPC was detected by real-time PCR in all samples, we discarded that negative results in qPCR for HEVs have been due to the presence of inhibitors. Several positive samples by qualitative PCR for HEVs were negative when the qPCR was applied, which could be attributed to viral concentrations under the limit of detection for qPCR. This probably happened for BU samples, since they were collected after a treatment in a stabilization pond.
In general, the information about HEV circulation in the environment is a sub-product of the PV environmental surveillance (Battistone et al. 2014; Ndiaye et al. 2014; Wahjuhono et al. 2014; Wang et al. 2014; Tao et al. 2016). This is carried out by inoculation of sewage specimens in PV-specific cell lines—which do not allow the isolation of all HEV types (World Health Organization 2004), generating an incomplete description of the real HEV diversity. The HEV detection directly from sewage concentrates using molecular methods could help to obtain a more realistic view of the HEV diversity, avoiding a bias by isolation in cell lines specific for PV. In this work, the direct sequencing of PCR amplicons allowed the identification of 10 different types of three HEV species in 31 samples. The primers proposed by Nix et al. (2006) were designed for broad target specificity and could amplify all recognized and proposed HEV serotypes when they tested originally with RNA of clinical specimens. Moreover, this method has already been used to characterize HEVs isolated from sewage (Shukla et al. 2013; Battistone et al. 2014; Nakamura et al. 2015), and to amplify and typing HEVs directly from sewage sample concentrates (Ibrahim et al. 2014). Contrarily to Ibrahim et al. (2014) who characterized a low percentage (~ 12%) of HEVs from positive sewage samples using the PCR designed by Nix et al. (2006), we have characterized 67% of the positive sewage samples using this method. Moreover, we were able to identify variants seldom isolated in cell culture, as CVA22 (Brown et al. 2003). These results highlight the importance in selecting appropriate methods for the study of enterovirus in environmental samples.
It is worth mentioning the detection of HEV-C types. Although sporadically detected, EVC99 and CVA13 were highly divergent regarding the worldwide circulating strains. Previous studies reveal that strains of CVA13 and neuro-virulent VDPVs that circulated contemporary in a same geographic area were highly related in the non-structural region (Rakoto-Andrianarivelo et al. 2007; Delpeyroux et al. 2013). Besides, recombination between OPV strains and other HEV-C contributes as a favorable factor in the emergence of pathogenic VDPVs (Jegouic et al. 2009; Bessaud et al. 2016).
In our study, there was no isolation of OPV strains, regardless of a vaccination scheme with OPV or with IPV. Previous studies conducted in other countries reported PV and/or OPV strain detection in sewage (Mueller et al. 2009; Tao et al. 2012b; Alam et al. 2014; de Oliveira Pereira et al. 2016), indeed after the switch to IPV (Battistone et al. 2014; Wahjuhono et al. 2014). The absence of PV isolation from samples collected during routinely OPV vaccination and the low percentage of Non-Polio Enterovirus (NPEV) isolation in this study could be associated to different factors such as population size, sanitation level, and population density, as well as to geographic or climatic factors (Global Polio Eradication Initiative 2015). Other important aspect to highlight is that samples remained frozen at − 20 °C from the sampling up to early 2016 when this study began, and viruses could lose viability during this period.
Despite the absence of PV detection in this study, the circulation of HEV-C strains should be carefully monitored in a country with IPV vaccination scheme such as Uruguay. The low intestinal immunity generated by the inactivated vaccine (Salas-Peraza et al. 2010) could allow the importation and circulation of OPV strains in a silent manner (Manor et al. 2014), with opportunity to recombine with some of the HEV-C present in the country and to circulate among population without being detected. Uruguay employs the IPV vaccine since 2012 and has not yet been generated data concerning the epidemiology of HEV-C.
The most frequent types detected during this study correspond to species A (CVA16) and species B (E6).
The CVA16 was detected in the western region of the country between May and October 2011, being all the strains highly related among them. This type has been well associated to HFM disease (Iwai et al. 2009; Xu et al. 2015) and although there is a lack of information about the incidence of CVA16 in Uruguayan children, the Public Health authorities have warned about the occurrence of HFM disease in the country (Public Health Ministry of Uruguay 2014).
Although the HFM disease occurs worldwide, information concerning the epidemiology of CVA16 in the Americas is scarce (Carrion et al. 2016). Major studies have been conducted in the Asiatic region, where this type is endemic and several sub-types have been characterized (Zhang et al. 2010; Yu et al. 2016; Zhao et al. 2016).
Our results would indicate a primary role of this viral pathogen in the account of HFM disease cases in Uruguay, since neither CVA10 nor CVA71 (the others main HEV types related to the disease) were detected in sewage samples in this study. The high similarity among the CVA16 Uruguayan strains and the sustained circulation at the western region of the country (mainly in Fray Bentos where CVA16 was detected for five consecutive months) suggest a recent introduction of this variant in the country. Unfortunately, the molecular epidemiology of the HEVs involved in the HFM disease in Uruguay remains unknown for more than 40 years after the first national clinical report (Aguirrezabal et al. 1973).
E6 was the other frequently detected type in this study and was present in both regions of the country. This type is mainly associated with outbreaks and sporadic cases of aseptic meningitis (Mao et al. 2010; Kim et al. 2012; Benschop et al. 2016) and several reports documented its presence in sewage samples in different countries (Sedmak et al. 2003; Tao et al. 2016; Wieczorek et al. 2017).
Despite E6 detection both in cold and warm months, which is in agreement with previous studies (Tao et al. 2012a, 2016), this type was exclusively detected in those warmest months of the year in the western region of Uruguay. Since infantile meningitis and encephalitis cases due to E6 in Uruguay have occurred in summer (Hernández 2014), it is worrying that viable E6 were detected in sewage samples collected in summer months in places without sewage treatment plants like Salto and Paysandú. In these cities, the Uruguay River (the main recreational course of water for their populations) daily receives thousands of liters of sewage without treatment, which means a possible risk of infection for susceptible people exposed to this environmental matrix, when our results are considered.
The pattern of occurrence of different HEV types, exclusively circulating at each region of the country, is similar to the previously reported distribution of Norovirus and Rotavirus types in both eastern and western regions of Uruguay (Tort et al. 2015; Victoria et al. 2016). This scenario could be attributed to the influence of the epidemiology of Brazil and Argentina on the Uruguayan eastern and western regions, respectively, and to the low population flux between both regions. Nevertheless, more epidemiological information is needed in order to confirm this hypothesis.
This study conducted an environmental approach to describe the circulation of HEVs in Uruguay, encompassing the period of OPV–IPV switch. Although OPV was not detected, the occurrence of several types belonging to HEV species A, B, and C, was reported, contributing to unveil the molecular epidemiology of those variants which could affect the population and for which no information had been generated in the country.
References
Aguirrezabal, M. P., Somma, M. P., Russi, J., Campione, J., Hortal, M., & Tosi, H. C. (1973). Maculopapular vesicular disease of hand, foot and mouth. First national observation. Archivos de Pediatría del Uruguay, 44, 111–113 (Spanish).
Alam, M. M., Shaukat, S., Sharif, S., Angez, M., Khurshid, A., Malik, F., et al. (2014). Detection of multiple cocirculating wild poliovirus type 1 lineages through environmental surveillance: Impact and progress during 2011–2013 in Pakistan. The Journal of the Infectious Diseases, 210(1), 324–332.
Allmann, E., Pan, L., Li, L., Li, D., Wang, S., & Lu, Y. (2013). Presence of enteroviruses in recreational water in Wuhan, China. Journal of Virological Methods, 193, 327–331.
Battistone, A., Buttinelli, G., Fiore, S., Amato, C., Bonomo, P., Patti, A. M., et al. (2014). Sporadic isolation of sabin-like polioviruses and high-level detection of non-polio enteroviruses during sewage surveillance in seven Italian cities, after several years of inactivated poliovirus vaccination. Applied and Environmental Microbiology, 80, 4491–4501.
Benschop, K. S., Geeraedts, F., Beuvink, B., Spit, S. A., Fanoy, E. B., Claas, E. C., et al. (2016). Increase in ECHOvirus 6 infections associated with neurological symptoms in the Netherlands, June to August 2016. Euro Surveillance: European Communicable Disease Bulletin. https://doi.org/10.2807/1560-7917.ES.2016.21.39.30351.
Bessaud, M., Joffret, M. L., Blondel, B., & Delpeyroux, F. (2016). Exchanges of genomic domains between poliovirus and other cocirculating species C enteroviruses reveal a high degree of plasticity. Scientific Reports. https://doi.org/10.1038/srep38831.
Blondel, B., Colbère-Garapin, F., Couderc, T., Wirotius, A., & Guivel-Benhassine, F. (2005). Poliovirus, pathogenesis of poliomyelitis, and apoptosis. Current Topics in Microbiology and Immunology, 289, 25–56.
Brown, B., Oberste, M. S., Maher, K., & Pallansch, M. A. (2003). Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. Journal of Virology, 77, 8973–8984.
Carrion, G., Huaman, J. L., Silva, M., Ampuero, J. S., Paz, I., Ocaña, V. R., et al. (2016). Molecular epidemiology of coxsackievirus A16 strains from four sentinel surveillance sites in Peru. International Journal of Infectious Diseases, 52, 83–85.
Connell, C., Tong, H. I., Wang, Z., Allmann, E., & Lu, Y. (2012). New approaches for enhanced detection of enteroviruses from Hawaiian environmental waters. PLoS ONE. https://doi.org/10.1371/journal.pone.0032442.
de Oliveira Pereira, J. S., da Silva, L. R., de Meireles Nunes, A., de Souza Oliveira, S., da Costa, E. V., & da Silva, E. E. (2016). Environmental Surveillance of Polioviruses in Rio de Janeiro, Brazil, in support to the activities of Global Polio Eradication Initiative. Food and Environmental Virology, 8(1), 27–33.
de Quadros, C. A., Hersh, B. S., Olivé, J. M., Andrus, J. K., da Silveira, C. M., & Carrasco, P. A. (1997). Eradication of wild poliovirus from the Americas: Acute flaccid paralysis surveillance, 1988–1995. The Journal of the Infectious Diseases, 175(1), 37–42.
Delpeyroux, F., Colbère-Garapin, F., Razafindratsimandresy, R., Sadeuh-Mba, S., Joffret, M. L., Rousset, D., et al. (2013). Eradication of poliomyelitis and emergence of pathogenic vaccine-derived polioviruses: From Madagascar to Cameroon. Médecine/Sciences, https://doi.org/10.1051/medsci/20132911021. (French).
Farías, A. A., Mojsiejczuk, L. N., Pisano, M. B., Flores, F. S., Aguilar, J. J., Jean, A. N., et al. (2018). Environmental Surveillance of enteroviruses in Central Argentina: First detection and evolutionary analyses of E14. Food and Environmental Virology, 10(1), 121–126.
Global Polio Eradication Initiative. (2015). Guidelines on environmental surveillance for detection of polioviruses. Working draft—March 2015. Retrieved February 21, 2018 from http://polioeradication.org/wp-content/uploads/2016/07/GPLN_GuidelinesES_April2015.pdf.
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
Haramoto, E., Katayama, H., Utagawa, E., & Ohgaki, S. (2009). Recovery of human norovirus from water by virus concentration methods. Journal of Virological Methods, 160(1–2), 206–209.
Hernández, E. (2014). Typing of community non-polio enteroviruses responsible of neurovirosis in Uruguayan children. Biochemistry Bachelor´s Thesis. Faculty of Sciences, University of the Republic, Uruguay. Spanish. Retrieved July 24, 2017 from http://www.bib.fcien.edu.uy/files/etd/pasan/uy24-17275.pdf.
Hovi, T., Cantell, K., Huovilainen, A., Kinnunen, E., Kuronen, T., Lapinleimu, K., et al. (1986). Outbreak of paralytic poliomyelitis in Finland: Widespread circulation of antigenically altered poliovirus type 3 in a vaccinated population. Lancet, 1, 1427–1432.
Hovi, T., Shulman, L. M., van der Avoort, H., Deshpande, J., Roivainen, M., & De Gourville, E. M. (2012). Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiology and Infection, 140(1), 1–13.
Ibrahim, W., Ouerdani, I., Pillet, S., Aouni, M., Pozzetto, B., & Harrath, R. (2014). Direct typing of human enteroviruses from wastewater samples. Journal of Virological Methods, 207, 215–219.
Iwai, M., Masaki, A., Hasegawa, S., Obara, M., Horimoto, E., Nakamura, K., et al. (2009). Genetic changes of coxsackievirus A16 and enterovirus 71 isolated from hand, foot, and mouth disease patients in Toyama, Japan between 1981 and 2007. Japanese Journal of Infection Diseases, 62, 254–259.
Jegouic, S., Joffret, M. L., Blanchard, C., Riquet, F. B., Perret, C., Pelletier, I., et al. (2009). Recombination between polioviruses and co-circulating Coxsackie A viruses: Role in the emergence of pathogenic vaccine-derived polioviruses. PLoS Pathogens. https://doi.org/10.1371/journal.ppat.1000412.
Katayama, H., Shimasaki, A., & Ohgaki, S. (2002). Development of a virus concentration method and its application to detection of enterovirus and norwalk virus from coastal seawater. Applied and Environmental Microbiology, 68, 1033–1039.
Kim, H. J., Kang, B., Hwang, S., Hong, J., Kim, K., & Cheon, D. S. (2012). Epidemics of viral meningitis caused by echovirus 6 and 30 in Korea in 2008. Virology Journal. https://doi.org/10.1186/1743-422X-9-38.
Knowles, N. J., Hovi, T., Hyypiä, T., King, A. M. Q., Lindberg, A. M., Pallansch, M. A., et al. (2011). Picornaviridae. In A. M. Q. King, M. J. Adams, E. B. Carstens & E. J. Lefkowitz (Eds.), Virus taxonomy: Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses (pp. 855–880). San Diego, CA: Elsevier.
Manor, Y., Shulman, L. M., Kaliner, E., Hindiyeh, M., Ram, D., Sofer, D., et al. (2014). Intensified environmental surveillance supporting the response to wild-type poliovirus type 1 silent circulation in Israel, 2013. Euro Surveillance: European Communicable Disease Bulletin, 19(7), 20708.
Mao, N., Zhao, L., Zhu, Z., Chen, X., Zhou, S., Zhang, Y., et al. (2010). An aseptic meningitis outbreak caused by echovirus 6 in Anhui province, China. Journal of Medical Virology, 82, 441–445.
Muehlenbachs, A., Bhatnagar, J., & Zaki, S. R. (2015). Tissue tropism, pathology and pathogenesis of enterovirus infection. The Journal of Pathology, 235, 217–228.
Mueller, J. E., Bessaud, M., Huang, Q. S., Martinez, L. C., Barril, P. A., Morel, V., et al. (2009). Environmental poliovirus surveillance during oral poliovirus vaccine and inactivated poliovirus vaccine use in Córdoba Province, Argentina. Applied and Environmental Microbiology, 75, 1395–1401.
Nakamura, T., Hamasaki, M., Yoshitomi, H., Ishibashi, T., Yoshiyama, C., Maeda, E., et al. (2015). Environmental surveillance of poliovirus in sewage water around the introduction period for inactivated polio vaccine in Japan. Applied and Environmental Microbiology, 81, 1859–1864.
Nathanson, N., & Kew, O. M. (2010). From emergence to eradication: The epidemiology of poliomyelitis deconstructed. American Journal of Epidemiology, 172, 1213–1229.
Ndiaye, A. K., Diop, P. A., & Diop, O. M. (2014). Environmental surveillance of poliovirus and non-polio enterovirus in urban sewage in Dakar, Senegal (2007–2013). Pan African Medical Journal. https://doi.org/10.11604/pamj.2014.19.243.3538.
Nix, W. A., Oberste, M. S., & Pallansch, M. A. (2006). Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. Journal of Clinical Microbiology, 44, 2698–2704.
Oberste, M. S., Maher, K., Kilpatrick, D. R., & Pallansch, M. A. (1999). Molecular evolution of the human enteroviruses: Correlation of serotype with VP1 sequence and application to picornavirus classification. Journal of Virology, 73, 1941–1948.
Oberste, M. S., & Pallansch, M. A. (2005). Enterovirus: Molecular detection and typing. Reviews in Medical Microbiology, 16(4), 163–171.
Oberste, M. S., Peñaranda, S., Rogers, S. L., Henderson, E., & Nix, W. A. (2010). Comparative evaluation of Taqman real-time PCR and semi-nested VP1 PCR for detection of enteroviruses in clinical specimens. Journal of Clinical Virology, 49(1), 73–74.
Pallansch, M. A., Oberste, M. S., & Whitton, J. L. (2013). Enteroviruses: Polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In B. N. Fields, D. M. Knipe & P. M. Howley (Eds.), Fields virology (6th ed., pp. 490–530). Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins Health.
Prevost, B., Lucas, F. S., Goncalves, A., Richard, F., Moulin, L., & Wurtzer, S. (2015). Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environment International, 79, 42–50.
Public Health Ministry of Uruguay. (2014). Communication: Hand-foot-mouth syndrome. 22nd, August 2014. Spanish. Retrieved June 21, 2017 from http://www.msp.gub.uy/comunicado/comunicado-s%C3%ADndrome-mano-pie-boca.
Rajal, V. B., McSwain, B. S., Thompson, D. E., Leutenegger, C. M., Kildare, B. J., & Wuertz, S. (2007). Validation of hollow fiber ultrafiltration and real-time PCR using bacteriophage PP7 as surrogate for the quantification of viruses from water samples. Water Research, 41, 1411–1422.
Rakoto-Andrianarivelo, M., Guillot, S., Iber, J., Balanant, J., Blondel, B., Riquet, F., et al. (2007). Co-circulation and evolution of polioviruses and species C enteroviruses in a district of Madagascar. PLoS Pathogens. https://doi.org/10.1371/journal.ppat.0030191.
Salas-Peraza, D., Avila-Agüero, M. L., & Morice-Trejos, A. (2010). Switching from OPV to IPV: Are we behind the schedule in Latin America? Expert Review of Vaccines, 9, 475–483.
Sedmak, G., Bina, D., & MacDonald, J. (2003). Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from Milwaukee, Wisconsin, collected August 1994 to December 2002. Applied and Environmental Microbiology, 69, 7181–7187.
Shukla, D., Kumar, A., Srivastava, S., Idris, M. Z., & Dhole, T. N. (2013). Environmental surveillance of enterovirus in Northern India using an integrated shell vial culture with a semi-nested RT PCR and partial sequencing of the VP1 gene. Journal of Medical Virology, 85, 505–511.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Tao, Z., Song, Y., Wang, H., Zhang, Y., Yoshida, H., Ji, S., et al. (2012a). Intercity spread of echovirus 6 in Shandong Province, China: Application of environmental surveillance in tracing circulating enteroviruses. Applied and Environmental Microbiology, 78, 6946–6953.
Tao, Z., Wang, Z., Lin, X., Wang, S., Wang, H., Yoshida, H., et al. (2016). One-year Survey of human enteroviruses from sewage and the factors affecting virus adsorption to the suspended solids. Scientific Reports. https://doi.org/10.1038/srep31474.
Tao, Z., Zhang, Y., Liu, Y., Xu, A., Lin, X., Yoshida, H., et al. (2012b). Isolation and characterization of a type 2 vaccine-derived poliovirus from environmental surveillance in China, 2012. PLoS ONE. https://doi.org/10.1371/journal.pone.0083975.
The Pirbright Institute. (2018). Enterovirus. Retrieved March 23, 2018 from http://www.picornaviridae.com/enterovirus/enterovirus.htm.
Tort, L. F., Victoria, M., Lizasoain, A., García, M., Berois, M., Cristina, J., et al. (2015). Detection of common, emerging and uncommon VP4, and VP7 human Group A rotavirus genotypes from urban sewage samples in Uruguay. Food and Environmental Virology, 7, 342–353.
Uruguayan Government. (2012). Decree-Law Number 41/012 (2012, Feb 28). National Code of Sanitary Events and Diseases of Mandatory Notification. An update of the Decree 64/004. Spanish. Retrieved February 21, 2018 from https://www.impo.com.uy/bases/decretos/41-2012.
Victoria, M., Tort, L. F., Lizasoain, A., García, M., Castells, M., Berois, M., et al. (2016). Norovirus molecular detection in Uruguayan sewage samples reveals a high genetic diversity and GII.4 variant replacement along time. Journal of Applied Microbiology, 120, 1427–1435.
Wahjuhono, G., Revolusiana, Widhiastuti, D., Sundoro, J., Mardani, T., Ratih, W. U., et al. (2014). Switch from oral to inactivated poliovirus vaccine in Yogyakarta Province, Indonesia: Summary of coverage, immunity, and environmental surveillance. The Journal of Infectious Diseases, 210(1), 347–352.
Wang, H., Tao, Z., Li, Y., Lin, X., Yoshida, H., Song, L., et al. (2014). Environmental surveillance of human enteroviruses in Shandong Province, China, 2008 to 2012: Serotypes, temporal fluctuation, and molecular epidemiology. Applied and Environmental Microbiology, 80, 4683–4691.
Wieczorek, M., Ciąćka, A., Witek, A., Kuryk, Ł, & Żuk-Wasek, A. (2011). Environmental Surveillance of Non-polio Enteroviruses in Poland, 2011. Food and Environmental Virology, 7, 224–231.
Wieczorek, M., Krzysztoszek, A., Ciąćka, A., & Figas, A. (2017). Molecular characterization of environmental and clinical echovirus 6 isolates from Poland, 2006–2014. Journal of Medical Virology, 89, 936–940.
Witsø, E., Palacios, G., Rønningen, K. S., Cinek, O., Janowitz, D., Rewers, M., et al. (2007). Asymptomatic circulation of HEV71 in Norway. Virus Research, 123(1), 19–29.
World Health Organization. (2004). Polio Laboratory Manual. Fourth edition. Geneva. Switzerland. Retrieved February 21, 2018 from http://apps.who.int/iris/bitstream/10665/68762/1/WHO_IVB_04.10.pdf.
Xu, M., Su, L., Cao, L., Zhong, H., Dong, N., Dong, Z., et al. (2015). Genotypes of the enterovirus causing hand foot and mouth disease in Shanghai, China, 2012–2013. PLoS ONE. https://doi.org/10.1371/journal.pone.0138514.
Yu, W., Xu, H., & Yin, C. (2016). Molecular epidemiology of human coxsackievirus A16 strains. Biomedical Reports, 4, 761–764.
Zhang, Y., Wang, D., Yan, D., Zhu, S., Liu, J., Wang, H., et al. (2010). Molecular evidence of persistent epidemic and evolution of subgenotype B1 coxsackievirus A16-associated hand, foot, and mouth disease in China. Journal of Clinical Microbiology, 48, 619–622.
Zhao, G., Zhang, X., Wang, C., Wang, G., & Li, F. (2016). Characterization of VP1 sequence of Coxsackievirus A16 isolates by Bayesian evolutionary method. Virology Journal. https://doi.org/10.1186/s12985-016-0578-3.
Funding
Funding was provided provided by Universidad de la República Uruguay (Grant No. PDU Virología), Comisión Sectorial de Investigación Científica (Grant Nos. CSIC I+D 2010, Scholarship, Beca doctorado-Comisión Académica de Postgrado) and Programa de desarrollo de las Ciencias Básicas PEDECIBA (Grant No. Beca de movilidad).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Lizasoain, A., Burlandy, F.M., Victoria, M. et al. An Environmental Surveillance in Uruguay Reveals the Presence of Highly Divergent Types of Human Enterovirus Species C and a High Frequency of Species A and B Types. Food Environ Virol 10, 343–352 (2018). https://doi.org/10.1007/s12560-018-9351-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-018-9351-7