Abstract
Pea rust is a devastating disease of peas especially in the sub-tropical regions of the world and greatly influenced by the environmental conditions during disease development. Molecular markers associated with pea rust resistance would be useful in marker assisted selection (MAS). Utility of molecular markers associated with the pea rust resistance were evaluated in 30 diverse pea genotypes using four SSR markers (AA446 and AA505 flanking the major QTL Qruf; AD146 and AA416 flanking the minor QTL, Qruf1). QTL, Qruf flanking markers were able to identify all the resistant genotypes when used together, except Pant P 31. While, SSR markers AD146 and AA416 flanking the minor QTL, Qruf1 were able to identify all the pea resistant genotypes used for validation, except for HUDP-11 by AD146 and Pant P 31 by AA416. Similarly, SSR markers AA446 and AA505 were able to identify all the susceptible pea genotypes, except IPFD 99–13, HFP 9415 and S- 143. SSR markers AD146 and AA416 were together able to identify all the pea susceptible genotypes used for validation, except KPMR 526, KPMR 632 and IPFD 99–13. On the basis of marker allele analysis it may be concluded that SSR markers (AA446, AA505, AD146 and AA416) can be used in MAS of pea rust resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pea rust caused by Uromyces fabae Pers. de-Bary is a major disease of peas and is responsible for significant yield losses especially in the sub-tropical regions characterized by warm humid weather conditions (Kushwaha et al. 2006). These conditions usually coincide with the reproductive phase of pea and favour rust outbreak (Kushwaha et al. 2007). One of the best possible ways to stabilize the productivity of pea crop is to grow rust resistant varieties. Resistance to rust in pea is reported to be governed by single dominant gene (Tyagi and Srivastava 1999), a oligogene (Vijayalakshmi et al. 2005) showing partial dominance along with some minor genes and 2–3 additive genes (Singh et al. 2012). Resistance to pea rust has been reported to be post haustorial type, and none of the pea genotypes is reported to be free from rust infection (Singh and Srivastava 1985; Chand et al. 2006). Rust severity is greatly influenced by the environment during infection initiation and disease development. This is the major bottleneck in screening and selection for rust resistance. Use of molecular markers would allow indirect selection for rust resistance independent of environmental effects (Rai et al. 2011).
Information regarding association of molecular markers with rust resistance is available (Vijayalaxmi et al., 2005; Rai et al. 2011), which seems to be governed by one major and one minor QTL. Vijayalakshmi et al. (2005) identified two RAPD markers SC10-82 360 and SCRI- 71 1000 flanking the rust resistance gene (Ruf) with a distance of 10.8 cM (0.097 rF and LOD of 5.05) and 24.5 cM (0.194 rF and a LOD of 2.72), respectively by using a BC1F2 population. Rai et al. (2011) emphasized the quantitative nature of pea rust resistance and re-designated the ‘Ruf’ gene as Qruf. The following QTLs were identified for rust resistance: Qruf (22.4–58.8 % of phenotypic variation) on LG VII (flanked by SSR markers AA446 and AA505); Qruf1 (11.2–12.4 % of phenotypic variation) on LG VII (flanked by SSR markers AD146 and AA416). The closest markers to Ruf, Qruf and Qruf1 were SC10-82360 (10.8 cM), AA505 (1.0 cM) and AD146 (0.6 cM), respectively. To our knowledge, there is no other report on QTLs for U. fabae resistance in pea. However, Barilli et al. (2010) identified a major QTL for resistance to U. pisi in wild pea (P. fulvum L.). The reported QTL was flanked by RAPD markers OPY111316 and OPV171078; located on linkage group 3 and explained 63 % of the total phenotypic variation. The present study was conducted with an objective to validate the associated SSR markers in a diverse set of rust resistant pea genotypes so that these markers may be used directly in marker assisted selection (MAS) for rust resistance breeding in pea.
Materials and methods
Plant material
For validating the efficacy of molecular markers associated with rust resistance, 15 rust resistant (Table 1) and 15 rust susceptible (Table 2) pea genotypes along with two parental genotypes i.e., HUVP 1 (rust susceptible) and FC 1 (rust resistant) were used in the present study. These 30 genotypes were selected based on their rust reaction at multi location screening sites under AICRP on MULLaRP (All India Coordinated Research Project on Mung, Urd, Lentil Lathyrus, Rajmash and Pea) of ICAR, New Delhi, India.
Assessment of rust severity
Test genotypes, including all the 30 pea genotypes and two checks viz., HUVP 1 (susceptible check) and FC 1 (resistant check), were planted in the polyhouse facility at Institute of Agricultural Sciences, Banaras Hindu University, Varanasi, India during crop seasons 2011–12 and 2012–13. Each genotype was planted in two pots filled with garden soil, each pot representing a replication. Throughout the experiment, plants were maintained in vegetative phase by bud clipping and the plants were inoculated by the method of Chand et al. (2004). After inoculation, 10th nodal leaf before the expected appearance of the disease was tagged from five plants of each genotype in each replication. Data on rust severity was recorded three times at weekly intervals. Estimation of rust severity was based on 0–9 scale (Sokhi et al. 1984) after initiation of rust infection on susceptible check, HUVP 1.
PCR procedure
The genomic DNAs from the test genotypes were extracted using DNeasyTM Plant Mini Kit (Quiagen, Hilden, Germany). About 100 mg of young leaf tissue was excised from aseptically grown seedlings of each genotype. PCR amplification using SSR markers was performed in 10 mM Tris–HCl pH 9.0; 1.5 mM MgCl2; 50 mM KCl and 0.01 % gelatin), 0.5 mM MgCl2, 200 mM dNTPs, 1.25 μM of primer, 20 to 25 ng of DNA and 1 unit of Taq polymerase per 25Kl reaction volume. After initial denaturation at 94 °C for 5 min, the following cycle was repeated 40 times: denaturing at 94 °C for 1 min, annealing as per primer (Loridon et al. 2005) for 1 min, extension at 72 °C for 2 min, the final extension segment being held for 7 min. Polymerase chain reactions were carried out in a Thermal Cycler (Touchgene Gradient, Techne, UK). The PCR products were separated electrophoretically in 2.5 % (w/v) agarose gel using 0.5× TBE [54.0 g Tris-base; 27.5 g boric acid; 0.5 M EDTA (pH 8.0) in 1000 ml volume] buffer. Ethidium bromide solution at a final concentration of 0.5 μg/ml was added to the agarose solution. Finally, the gels were visualized under a UV light source in a gel documentation system (AlphaImager 2200, Alpha Innotech Corporation, San Leandro, USA).
Results and discussion
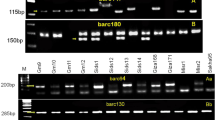
In order to determine the utility of molecular markers associated with the pea rust resistance, 30 pea genotypes (15 rust resistant and 15 susceptible) were screened with four SSR markers associated with rust resistance. The first 15 pea genotypes (Table 1) were highly resistant to rust and most of them are expected to carry resistance genes from FC 1, the rust resistant parent used in the study of Vijayalakshmi et al. (2005) and Rai et al. (2011). SSR markers AA446 and AA505 flanking the major QTL Qruf amplified a fragment of 450 and 140 bp, respectively, in FC 1. SSR marker AA446 failed to amplify the fragment of 450 bp in four genotypes Pant P 11, HUDP-17, Pant P 25 and Pant P 31, whereas marker AA505 was unable to amplify the band of 140 bp in HUDP-8 and Pant P 31 (Table 1, Fig. 1). However these two markers will be able to identify all the genotypes when used together, except Pant P 31. SSR markers AD146 and AA416 flanking the minor QTL, Qruf1 were able to identify all the pea resistant genotypes used for validation, except for HUDP-11 by AD146 and Pant P 31 by AA416 (Table 1, Fig. 2). SSR markers AD146 and AA416 flanking the QTL, Qruf1 amplified a fragment of 430 and 280 bp, respectively in FC 1.
PCR banding pattern of the SSR markers AA446 and AA505 flanking the major QTL ‘Qruf’. R: resistant parent (FC 1), S: susceptible parent (HUVP 1), 1–15 (pea genotypes as listed in table 1)
PCR banding pattern of the SSR markers AD146 and AA416 flanking the minor QTL ‘Qruf1’. R: resistant parent (FC 1), S: susceptible parent (HUVP 1), 1–15 (pea genotypes as listed in table 1)
SSR markers flanking the rust resistance loci were also validated in 15 rust susceptible pea genotypes (Table 2). SSR marker AA446 able to identify 10 out of 15 susceptible genotypes as amplifying a band similar to susceptible check, HUVP 1 (Table 2, Fig. 3). Whereas, marker AA505 was able to identify 12 out of 15 susceptible genotypes (Table 2, Fig. 3). So, SSR markers AA446 and AA505 flanking the major QTL, Qruf were able to identify all the susceptible pea genotypes used for validation, except IPFD 99–13, HFP 9415 and S- 143. SSR markers AD146 and AA416 flanking the minor QTL, Qruf1 were together able to identify all the pea susceptible genotypes used for validation, except KPMR 526, KPMR 632 and IPFD 99–13 (Table 2). However, individually SSR markers AD146 and AA416 are not so efficient to identify susceptible genotypes. It is interesting to note that, DDR and KPMR series susceptible pea genotypes are having ‘HFP 4’ as a parent in their pedigree.
PCR banding pattern of the SSR markers AA446 and AA505 flanking the major QTL ‘Qruf’. R: resistant parent (FC 1), S: susceptible parent (HUVP 1), 1–15 (pea genotypes as listed in table 2)
The level of polymorphism for microsatellite markers in pea is high, with an average of 3.8 alleles per polymorphic locus spanning 1430 cM of composite genetic map covered by 239 SSR marker loci (Loridon et al. 2005). Aubert et al. (2006) have developed a functional map of pea comprising 1,458 cM total length and depicting locations of 363 loci, including 111 markers anchored to specific genes having known functions. To allow a dependable marker assisted selection (MAS) for rust resistance, a marker should be tightly linked to the gene of interest. For MAS, it is usually imperative to saturate the appropriate regions of the genome with markers in order to obtain those ones that are sufficiently tightly linked to genes of interest to permit a reliable indirect selection for them. When, flanking markers are used together, the effectiveness of MAS would be improved considerably (Tanksley 1983). Ek et al. (2005) reported that pea powdery mildew resistance gene ‘er1’ was flanked by the markers PSMPSAD60 and PSMPS5 and use of both markers for selection of resistant plants resulted in only 1.6 % error in selection. Since, the flanking SSR markers AA446 and AA505 are tightly associated with the major QTL (Qruf) and AD146 and AA416 with the minor QTL (Qruf1) (Rai et al. 2011), they are assumed to cosegregate with their corresponding QTL. SSR markers associated with Qruf and Qruf1 together successfully identified all the resistant as well as susceptible pea genotypes used to validate them except, a susceptible genotype IPFD 99–13. If two flanking markers are used for MAS, the chance of both the markers becoming unlinked to the concerned QTL, Qruf and Qruf1 would be only 0.1 and 0.04 %, respectively (Rai et al. 2011). On the basis of marker allele analysis it may be concluded that SSR markers (AA446, AA505, AD146 and AA416) can be used in marker assisted selection (MAS) of pea rust resistance.
References
Aubert G, Morin J, Jacquin F, Loridon K, Quillet MC, Petit A, Rameau C, Lejeune- Henaut I, Huguet T, Burstin J (2006) Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor Appl Genet 112:1024–1041
Barilli E, Satovic Z, Rubiales D, Torres AM (2010) Mapping of quantitative trait loci controlling partial resistance against rust incited by Uromyces pisi (Pers.) Wint. in a Pisum fulvum L. intraspecific cross. Euphytica 175:151–159
Chand R, Srivastava CP, Kushwaha C (2004) Screening technique for pea (Pisum sativum) genotypes against rust disease (Uromyces fabae). Ind J Agric Sci 74:166–167
Chand R, Srivastava CP, Singh BD, Sarode SB (2006) Identification and characterization of slow rusting components in pea (Pisum sativum L.). Genet Resour Crop Evol 53:219–224
Ek M, vonPost R, Dayteg C, Henriksson T, Weibull P, Ceplitis A, Isaac P, Tuvesson S (2005) Microsatellite markers for powdery mildew resistance in pea (Pisum sativum L.). Hereditas 142:86–91
Kushwaha C, Chand R, Srivastava CP (2006) Role of aeciospores in outbreak of pea (Pisum sativum L) rust (Uromyces fabae). Euro J Plant Pathol 115:323–330
Kushwaha C, Srivastava CP, Chand R, Singh BD (2007) Identification and evaluation of a critical time for assessment of slow rusting in pea against Uromyces fabae. Field Crop Res 103:1–4
Loridon K, McPhee K, Morin J, Dubreuil P, Pilet-Nayel ML, Aubert G, Rameau C, Baranger A, Coyne C, Lejeune-Henaut I, Burstin J (2005) Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor Appl Genet 111:1022–1031
Rai R, Singh AK, Singh BD, Joshi AK, Chand R, Srivastava CP (2011) Molecular mapping for resistance to pea rust caused by Uromyces fabae (Pers.) de-Bary. Theor Appl Genet 123:803–813
Singh AK, Rai R, Srivastava CP, Singh BD, Kushwaha C, Chand R (2012) A quantitative analysis of rust (Uromyces fabae) resistance in pea (Pisum sativum) using RILs. Ind J Agric Sci 82:190–192
Singh RM, Srivastava CP (1985) Evaluation classification and usefulness of pea germplasm for quantitative characters. Legume Res 8:68–72
Sokhi SS, Singh SJ, Munshi GD (1984) Parameters of rating pea varieties/lines against rust. Ind Phytopathol 37:252–255
Tanksley S (1983) Molecular markers in plant breeding. Plant Mol Biol Rep 1:3–8
Tyagi MK, Srivastava CP (1999) Inheritance of powdery mildew and rust resistance in pea. Annals Biol 15:13–16
Vijayalakshmi S, Yadav K, Kushwaha C, Sarode SB, Srivastava CP, Chand R, Singh BD (2005) Identification of RAPD markers linked to the rust (Uromyces fabae) resistance gene in pea (Pisum sativum.). Euphytica 144:265–274
Acknowledgments
The First author is thankful to UGC, New Delhi, India for providing financial assistance in form of UGC Research Fellowship during Ph.D. program and AICRP on MULLaRP, IIPR, Kanpur, India for providing plant material.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, A.K., Rai, R., Singh, B.D. et al. Validation of SSR markers associated with rust (Uromyces fabae) resistance in pea (Pisum sativum L.). Physiol Mol Biol Plants 21, 243–247 (2015). https://doi.org/10.1007/s12298-015-0280-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-015-0280-8