Abstract
Yarrowia lipolytica is an oleaginous saccharomycetous yeast with a long history of industrial use. It aroused interest several decades ago as host for heterologous protein production. Thanks to the development of numerous molecular and genetic tools, Y. lipolytica is now a recognized system for expressing heterologous genes and secreting the corresponding proteins of interest. As genomic and transcriptomic tools increased our basic knowledge on this yeast, we can now envision engineering its metabolic pathways for use as whole-cell factory in various bioconversion processes. Y. lipolytica is currently being developed as a workhorse for biotechnology, notably for single-cell oil production and upgrading of industrial wastes into valuable products. As it becomes more and more difficult to keep up with an ever-increasing literature on Y. lipolytica engineering technology, this article aims to provide basic and actualized knowledge on this research area. The most useful reviews on Y. lipolytica biology, use, and safety will be evoked, together with a resume of the engineering tools available in this yeast. This mini-review will then focus on recently developed tools and engineering strategies, with a particular emphasis on promoter tuning, metabolic pathways assembly, and genome editing technologies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Overview of Yarrowia lipolytica Biology and Use
The saccharomycetous yeast Yarrowia lipolytica is an oleaginous species found in a large range of ecosystems (soils, marine waters, mycorrhizae, oil-polluted environments) and a variety of foods (notably meat and dairy products, including cheeses) [1]. It has aroused industrial interest since more than 50 years, due to its remarkable lipolytic activity and high capacity of enzyme secretion and production of organic acids [1,2,3]. This yeast is regarded as non-pathogenic and used in several industrial processes classified as GRAS (generally recognized as safe) [1, 4]. Since a few decades, Y. lipolytica has emerged as a powerful and versatile host for heterologous gene expression and recombinant protein secretion or surface display [5,6,7]. This non-conventional yeast is also a model organism in several research areas (notably secretion, dimorphism, salt tolerance, and lipid metabolism) and its biology and applications have been the subject of two volumes of the Microbiology Monographs series [8, 9]. Sequencing of several strains, increased knowledge of its metabolism, and development of innovative genetic tools offer now new perspectives for metabolic pathway engineering in this yeast, paving the way for its use as cell factory for various applications. A chronology of the most important achievements in developing Y. lipolytica engineering is presented in Table 1, which constitutes the backbone of this mini-review.
Development of Molecular and Genetic Tools for Y. lipolytica Engineering
Interest for Y. lipolytica was oriented at first towards producing biomass (single-cell protein: SCP) or valuable metabolites (citric acid) from wild-type or traditionally improved strains [1, 2]. In the 1980s, several patents on Y. lipolytica transformation and use for heterologous gene expression and recombinant protein secretion (cf. Table 1) were filed by Pfizer Inc. (USA) or INRA (French National Institute for Agricultural Research). They represented the first steps towards genetic engineering of this yeast, for both academical studies and industrial applications. As the major molecular and genetic tools used for heterologous expression in Y. lipolytica have been extensively described and compared in previous reviews [5,6,7], this mini-review will essentially focus on recent improvements in the domain of promoters, DNA assembly and genome editing.
INRA played at first a pivotal role in early development of Y. lipolytica as expression host, by providing elements of expression cassettes (promoters, signals), engineered recipient strains, and the first ready-to-use expression/secretion vectors (cf. Table 1, 2000) [18, 20]. Some of these tools, like the Suc+ derivatives [10] of the high-secretor wild-type strain W29 (Po1 series of recipient strains) [18, 19] or the multi-UASs (upstream activating sequences) recombinant promoter hp4d, are among the most widely used tools, worldwide, for Y. lipolytica engineering [6, 7]. Recently, the development of new assembly and editing tools for this yeast, by several American, Chinese and Danish laboratories, has allowed designing several toolboxes for easy and rapid engineering of Y. lipolytica (cf. Table 1, 2017 and 2018, and infra) [68, 72, 74].
Promoters
Among natural Y. lipolytica promoters that were evaluated for heterologous expression [5,6,7], the most used are the strong constitutive TEF promoter (abbreviated pTEF, etc.) (cf. Table 1, 1998) [17] and the inducible pPOX2 [77]. This latter, isolated from an acyl-CoA oxidase gene from fatty acids synthesis pathway, is highly inducible by fatty acids and alkanes, and repressed by glucose and glycerol. Hydrophobicity of these inducers and incomplete substrate repression, however, limit pPOX2 use in industrial processes [77]. Interestingly, heterologous gene expression from many Y. lipolytica promoters can be enhanced using an intron-mediated enhancement (IME) strategy, as reviewed previously [6]. IME consists in retaining an upstream intron (noted “in”) within a promoter, in order to benefit from its positive effect on mRNA stability, and can lead to spectacular increases: expression from pTEFin is 17-fold higher than that from intronless pTEF [78].
As an alternative to searching natural promoters, building recombinant ones, with characteristics precisely tailored for each intended application, can now be envisioned. This new strategy was initiated with patented hp4d promoter, constructed by inserting four tandem repeats of UAS1B from XPR2 gene (UAS1XPR2) upstream of minimal pLEU2 (cf. Table 1, 1995) [18]. This recombinant promoter drives a growth-phase-dependent gene expression, which increases when stationary phase begins [18, 20, 79], a characteristic particularly interesting for heterologous production since it allows a partial dissociation of growth and expression phases. Its functional elements derived from pXPR2, historically important during development of Y. lipolytica engineering but which complex regulation hindered industrial use [18].
The concept of multi-UASs recombinant promoter was generalized at the University of Texas at Austin (cf. Table 1, 2011): a large array of promoters carrying one to 32 copies of UAS1XPR2 inserted upstream of minimal pLEU2 or pTEF were evaluated [45]. Surprisingly, transcription factor availability did not appear to limit the increase in expression obtained with high UAS copy numbers: expression eightfold higher than with preferred natural promoters was reported, the strongest ever for Y. lipolytica [45]. This suggest that endogenous promoters are enhancer-limited, which can be alleviated through UASs (notably UAS1XPR2) addition. Combination of disparate UAS elements can also be generalized for de novo construction of synthetic promoters: a new UAS from pTEF was identified and, combined to UAS1XPR2, drove sevenfold higher expression than pTEF [46]. Engineering pPOX2 by addition of tandem repeats of UASPOX2, a newly identified fatty acid-inducible element, produced a strong inducible recombinant promoter with an unprecedented 48-fold induction potential [48]. Besides the interest of such a strong inducible promoter for controlling metabolism, it could also constitute a useful fatty acid sensor. Similarly, the newly described pEYK1 has been used to develop hybrid inducible promoters carrying added tandem repeats of either UAS1XPR2 or its own UAS1EYK1 [49]. This new promoter, from EYK1 erythrulose kinase gene, and its hybrid derivatives are strongly inducible by erythritol and erythrulose, which can be used as free (non-metabolized) inducer in a ΔEYK1 strain [49].
Aiming at developing Y. lipolytica as microbial factory, INRA recently tested a pool of vectors with variable-strength promoters for producing proteins of industrial interest [47]. Six recombinant promoters carrying 2–8 UAS1XPR2 inserted upstream of pTEF or minimal pLEU2 (hp4d, hp8d) were evaluated, comparatively to pTEF. If the strongest hybrid promoters 8UAS1-pTEF and hp8d allowed the best production of RedStar2 and secreted glucoamylase, this was surprisingly 2UAS1-pTEF that was associated with highest yield and activity of secreted xylanase C (despite mRNA levels correlated to UAS copy number) [54]. This revealed that using stronger promoters can sometime be counterproductive. Consequently, the authors developed a method for easily identifying the best promoter for a given protein of interest, by combining Gateway cloning into a pool of vectors with variable-strength promoters and activity screening, which was successfully assayed on YFP and secreted α-amylase [49].
Signal Sequences for Secretion, Surface Display, and Termination
Secretion signals (pre or prepro regions) and terminator sequences are generally derived from genes encoding abundantly secreted proteins, such as XPR2 (encoding extracellular AEP protease) or LIP2 (extracellular lipase). These tools were described and compared in previous reviews: XPR2 pre region and terminator are most widely used [5,6,7]. It is also interesting to note that short synthetic terminator sequences, initially designed at UT Austin for Saccharomyces cerevisiae and able to increase expression by a fourfold factor in this yeast, were also shown to be fully functional in Y. lipolytica [80].
More recently (cf. Table 1, 2008), a surface display system was developed at the Ocean University of China (OUC): transcriptional fusion with C-terminal part of cell wall protein YlCWP1p was used for displaying heterologous proteins on the yeast cell surface [35]. This makes use of the GPI (glycosylphosphatidylinositol) anchor domain of YlCWP1p to target secreted fusion proteins to cell wall, where they covalently bind β-1,6 glucans. As reviewed previously [6, 7], other Y. lipolytica cell wall proteins have also been evaluated for surface display [37, 38], together with alternative strategies: fusion with homologous [38] or heterologous [36] flocculation domains or with a chitin binding module [37]. Arming Y. lipolytica cells can have multiple biotechnological applications, such as biosensors or live vaccines [35], and are particularly interesting as microbial factories for whole-cell biocatalysis [36,37,38].
Another interesting application of Y. lipolytica is using this oleaginous yeast as a platform for producing tunable arming oleosomes (cf. Table 1, 2013). Such functional nanostructures were designed, at the University of Hawaii at Manoa, by using transcriptional fusion with plant oleosins for displaying heterologous proteins on the surface of oleosomes [57]. Arming oleosomes can serve multiple purposes, notably cell-targeting/reporting functions (targeted drug delivery, pathogen detection) and in vivo self-assembly of protein nanofactories, by using high-affinity binding properties of cohesin/dockerin domains [57].
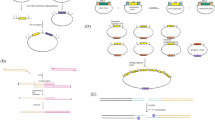
An overview of the possibilities offered by Y. lipolytica engineering tools, with their intended applications, is presented in the Fig. 1.
Overview of the possibilities offered by Y. lipolytica heterologous expression and engineering tools, with their intended applications. Single or multiple expression cassettes (promoter/ORF/terminator construct, with possibly secretion and/or targeting signals) are introduced into cells using integrative or replicative shuttle vectors. CRISPR-Cas9 tools for genome editing are available for targeted integration or for gene deletion, together with CRISPRi tools for transcriptional regulation. Heterologous genes can be expressed intracellularly for metabolic pathway engineering purposes, and their product can be targeted to the oleosomes (oleosin fusion) or, in presence of a signal peptide, directed to the secretory pathway. The recombinant protein can be secreted (or associated to the membrane), or, in presence of a GPI anchor domain, displayed on the cell surface (arming yeast). Some major applications of engineered Y. lipolytica strains are indicated in italics. Cf. details and references in the text. NHEJ non-homologous end-joining; ERT enzyme replacement therapy; PUFA polyunsaturated fatty acids
Main Recipient Strains and Selection Markers
Physiology and genealogy of Y. lipolytica laboratory strains were extensively described long ago [77], and recipient strains used for genetic engineering were listed in previous reviews [5,6,7]. The reference strain for Y. lipolytica species is E150 (CLIB122), which genome was fully sequenced and annotated [22]. Most used recipient strains are E129 (CLIB121) and, mainly, the Po1 series of strains (Po1d, f, g, and h—cf. Table 1, 2000) [18, 19], derived at INRA from the industrially relevant wild-type strain W29 (CLIB89, ATCC20460, CBS7504). Assembled genome sequences are available for W29 [59, 60], Po1f [58] (cf. Table 1, 2014), and a ku70 mutant of Po1g [68]. E150, E129, and the Po1 series are all engineered strains, able (in contrast to wild-type Y. lipolytica) to use sucrose as carbon source due to heterologous expression of SUC2 gene from S. cerevisiae [10]. This feature is particularly interesting for industrial applications, since it allows the use of molasses, from agro-industrial wastes, as cheap substrate [11, 82]. All these strains can be ordered from INRA’s CIRM-Levures Yeasts Library (https://www6.inra.fr/cirm_eng/Yeasts/Strain-catalogue).
A few wild-type Y. lipolytica isolates were also selected for remarkable features and engineered for peculiar applications, as reviewed previously [6]. For example, WSH-Z06 strain, a natural overproducer of α-ketoglutarate (α-KG), was engineered, at Jiangnan University, for increasing further α-KG production [83]. However, the most notable remains: H222 (DSM 27185) strain, available from DSMZ (Deutsche Sammlung für Mikroorganismen und Zellkulturen GmbH, https://www.dsmz.de/), from which a traditionally obtained α-KG-overproducing derivative was engineered, at Technische Universität Dresden (TUD), for the same purpose [84]. Like for the Po1 series of strains, some H222 derivatives have been engineered for sucrose utilization [85] or for increased homologous recombination efficiency (cf. infra), and are applied to developing industrial processes, notably organic acid production [85, 86].
Marker genes used for selection in Y. lipolytica are essentially auxotrophy complementation genes, notably LEU2 and URA3, as reviewed previously [5, 6]. The fact that URA3 marker gene can also be counter-selected (using 5-FOA medium [21]) makes it irreplaceable for marker rescue systems (cf. infra). Defective selection markers, with reduced promoters, are also available, which promote copy number amplification of integrated expression cassettes: ura3d4 allele [12] was extensively used for increasing heterologous expression [5,6,7], and similar defective leu2 alleles were developed (personal communication). As reviewed previously [5,6,7], Y. lipolytica is naturally resistant to most antibiotics, which limits the choice of dominant markers. This yeast is, however, sensitive to bleomycin/phleomycin, hygromycin B, and nourseothricin [74]. Hygromycin resistance hph gene is notably used in the Cre-lox based gene disruption/marker rescue system (cf. Table 1, 2003) [21] and in recently developed Y. lipolytica toolboxes (cf. infra) [68, 74]. The use of Escherichia coli guaB as dominant marker, for resistance to mycophenolic acid, was also described very recently [75]. The increased use of dominant markers in many recently developed engineering tools was prompted by the identification of the impact of some auxotrophic markers on the overall phenotype of producing strains [41]. Most notably, leucine biosynthesis and metabolism were shown to impact lipogenesis: leucine biosynthetic pathway was downregulated under lipid accumulation conditions and leucine supplementation of a leucine-auxotrophic lipid-producing strain resulted in increased lipogenesis [41]. This unexpected involvement of leucine biosynthesis in lipid accumulation in Y. lipolytica, firstly established at UT Austin, was more recently confirmed in a multifactorial study from a Sweden/USA consortium of laboratories [87].
Po1d strain was derived from W29 for heterologous production [12], and Po1f, g [18], and h [19] were improved further for this purpose: these high-secretor Suc+ strains are deleted for both extracellular proteases (AEP and AXP) and carry non-leaky non-reverting auxotrophies (Leu− and/or Ura−). In addition, the Leu− Po1g strain was equipped with an integrated pBR322 docking platform, for further targeting of LEU2-carrying pBR-based integrative vectors [18]. A triple auxotrophic derivative of Po1f, Po1j (Leu−, Ura−, Trp−), was very recently constructed at UT Austin [75]. Another derivative of Po1d was more specifically adapted for genetic engineering of lipid metabolic pathways: the Ura− JMY1212 strain is deleted for three main lipases (LIP2p, 7p, and 8p) and equipped with an integrated zeta docking platform, for further targeting integration of zeta-based integrative vectors [31]. Zeta sequences are long terminal repeats (LTRs) of Ylt1 retrotransposon (absent from W29 and derivatives) [88], used as targeting elements in some INRA vectors [15, 20] (cf. infra). JMY1212 is used in the high-throughput system, designed by INSA, CNRS, and INRA, for screening new biocatalysts through expression cloning and improving them through directed enzyme evolution (cf. Table 1, 2007) [31, 32].
Besides being widely used for heterologous production, Po1 strains have also notably served as basis for constructing “obese” strains, engineered for enhanced lipid storage (cf. Table 1, 2008, and infra). In addition, Po1g was recently chosen to construct Cell Atlas, a collection of seven strains with fluorescently tagged organelles (cf. Table 1, 2017) [68]. This set of isogenic strains, useful for cell biology studies (live assessment of gene expression, enzyme localization), is available from the Fungal Genetics Stock Center (http://www.fgsc.net).
Engineered Strains with Increased Homologous Recombination Efficiency
In contrast to S. cerevisiae, Y. lipolytica uses mainly non-homologous end-joining (NHEJ), and not homologous recombination (HR), for repairing DNA double-strand breaks (DSB). Consequently, targeted integration of exogenous DNA by single crossover can occur at acceptable rates (up to 80%, but seemingly locus and strain dependent) only if flanking homologous regions of at least 0.5 kb, and preferably 0.75–1 kb, are present [21, 55, 56, 81]. In order to increase HR efficiency during transformation, strains deleted for Ku70 and/or Ku80 gene(s) were independently constructed, from Po1d at INRA, and from H222 at TUD (cf. Table 1, 2013) [55, 56]. These NHEJ-deficient strains demonstrated increased HR efficiencies, despite notable differences between the French and German groups’ results (respectively, no effect versus a positive effect on HR, for ΔKu80 strain; 30–100 versus 4–5-fold decrease of transformation efficiency in ΔKu70 strain) [55, 56]. Reported HR frequencies, for different flanking homologous regions sizes, were also variable, possibility underlining influences from locus and strain background. The French group notably reported 100% of homologous integration in ΔKu70 strain, but without any effect of homology lengths from 50 to 250 bp, a result that the German group found at odds with their own results and those on other yeasts [56]. The German group observed a remaining 15% frequency of non-homologous recombination in ku70/ku80-deleted strains, which suggests that other recombination mechanisms may exist in Y. lipolytica, like microhomology-mediated end-joining (MMEJ) [56]. They also reported an increased sensitivity to UV light for ΔKu70 strain, implicating that NHEJ is partly required for cell defense against UV [56]. A series of ΔKu70 strains with different auxotrophies were also derived from Po1g strain, by a consortium of Richland laboratories, as part of their molecular genetic toolbox (cf. Table 1, 2017) [68].
These ΔKu70 strains constitute interesting recipient strains for efficient gene deletion and homologous recombination, provided a reduced transformation efficiency is tolerable. However, an alternative strategy is suggested by the recent development of CRISPR interference (CRISPRi) in Y. lipolytica: Ku70 or Ku80 repression by CRISPRi could offer the same benefit of increased HR, without permanent genetic knockout (cf. Table 1, 2017, and infra) [73].
At last, besides genetic engineering, cell cycle synchronization has been used to improve gene targeting: hydroxyurea (HU)-mediated cell cycle arrest in S-phase was shown to allow enhancing HR in various yeasts, including Y. lipolytica [89]. A consortium of Korean laboratories combined and compared these chemical (HU) and biological (ΔKu70) approaches for HR enhancement: although HU treatment was efficient on both wild-type and ΔKu70 cells, the best gene targeting efficiency (90%) was obtained in HU-treated wild type [76]. HU treatment thus appears as the most simple and effective method for HR enhancement in Y. lipolytica. These authors, however, favored HU-treated ΔKu70 cells, for complex engineering projects, since they allowed them to obtain repeated insertion/excision steps of a URA3 marker gene by HR between 100-bp flanking homology regions (“URA3-blaster” cassette for marker rescue, cf. infra) [76].
Glyco-Engineered Strains for Therapeutic Applications
When producing recombinant therapeutic proteins, differences between N-glycosylation pathways in yeasts and mammals can become a source of problems: yeast glycoproteins display high mannose-type N-glycans, which can reduce in vivo protein half-life or be immunogenic in humans and other mammals [28, 29]. Consequently, many research groups have developed glyco-engineered (aka humanized) strains in S. cerevisiae and non-conventional yeasts currently used for heterologous production, in order to produce more human-compatible glycoproteins [28, 29]. N-glycan biosynthesis engineering works performed in Y. lipolytica, by two laboratory consortia from Belgium and South Korea (cf. Table 1, 2007), have been reviewed previously [6, 7]. Briefly, the South Korean consortium constructed a double mutant strain lacking yeast-specific hypermannosylation and mannosyl phosphorylation [27]. This glyco-engineered strain was modified further by surface display of a fungal mannosidase, which conferred it a mannose trimming activity [30]. The Belgian consortium constructed a double mutant strain, lacking both yeast-specific mannosyltransferases and expressing a fungal mannosidase, able to produce homogeneous Man5GlcNAc2 residues [28]. Another project, involving a mannosyltransferase mutant strain further engineered by overexpression of a glucosyltransferase and heterologous overexpression of a mannosidase and a glucosidase from fungi, provided a strain able to produce homogeneous Man3GlcNAc2 residues, a core common to all mammalian N-glycan structures and that can be modified further in vitro to yield any complex-type N-glycan [29]. These new Y. lipolytica expression platforms should be able to produce recombinant proteins carrying humanized N-linked oligosaccharides compatible with therapeutic applications.
The same consortium of Belgian research groups, including Oxyrane (Belgium), has also engineered Y. lipolytica N-glycosylation pathway by expressing a bacterial glycosidase, which increases the level of mannose-6-phosphate [54]. Oxyrane UK Ltd is applying this glyco-engineered strain to the production of recombinant human lysosomal enzymes for ERTs (enzyme replacement therapies) of lysosomal storage diseases (cf. Table 1, 2012). Notably, a recombinant α-glucosidase enriched in mannose-6-phosphate is under validation for treatment of Pompe disease: high levels of mannose-6-phosphate enable its efficient targeting to the lysosomes of diseased cells, via interaction with specific receptors [54].
Strategies for Genetic Engineering of Y. lipolytica
Integrative and Replicative Expression Vectors
Replicative vectors make use of ARSs (autonomously replicating sequences) isolated from Y. lipolytica chromosomes, in which centromeric (CEN) and replicative functions are co-localized (cf. Table 1, 1993) [13]. This feature limits their use for heterologous production (one or a few copies per cell, high loss frequency requiring selective pressure during cultivation [13]) but they are used for pathway engineering [72] and constitute the preferred tool for transient expression (e.g., marker rescue using Cre-lox recombination [21]) and for newly developed CRISPR/Cas9-based tools for targeted markerless gene integration (cf. infra). If only low-copy ARS/CEN elements are available in Y. lipolytica, it is however possible to engineer them for increased copy number, as demonstrated recently at UT Austin: different natural or recombinant promoters were fused upstream of the centromeric region, leading to a more than 80% increase in vector copy number [90]. Expression of a reporter heterologous gene was concomitantly increased, with a dynamic range effect of 2.7-fold [90]. Although moderate in its impact, this method for increasing expression levels from replicative vectors could, however, be combined to promoter engineering strategies (cf. supra), for a synergistic effect.
As reviewed previously [5,6,7], integrative vectors, targeted to a genomic locus or integrated docking platform by HR, constitute preferred tools for heterologous expression and genetic engineering in Y. lipolytica. Despite the prevalence of NHEJ recombination in Y. lipolytica, expression cassettes or linearized vectors can be effectively targeted when using large (0.5–1 kb) flanking homologous regions [21, 81]. Integrated cassettes are very stable: as reviewed previously [6], they are retained without rearrangement after more than 100 generations without selective pressure. Integrative vectors can target rDNA, different genomic loci (e.g., URA3, XPR2), zeta sequences (in Ylt1-bearing strains), or previously integrated docking platforms (e.g., pBR322, pHSS6, zeta in Ylt1-devoid strains) [6]. An example of easy-to-use vector/strain combination is pBR-based expression/secretion vectors, able to transform Po1g strain, when linearized in their pBR322 backbone region, with an efficiency in the 105 per µg range and a targeting efficiency of near 80% [18]. Integration of a unique copy at a known genomic site renders this system particularly adapted to genetic engineering of enzymes, since the mutations’ effect can be directly compared on transformants’ activity [23, 25]. This expression system is commercialized by Yeastern Biotech Co. as the YLEX kit (http://www.yeastern.com) (cf. Table 1, 2006).
Interestingly, a consortium of Richland laboratories designed, as part of their molecular genetic toolbox, a multipurpose vector which can either be used as a replicative vector or as an auto-cloning (cf. infra) integrative vector, when linearized. This vector, designed for expression of fluorescently tagged proteins, was used to construct Y. lipolytica Cell Atlas (cf. Table 1, 2017) [68].
Integrative vectors carrying two [43] or three [44, 69] expression cassettes, for co-expression of several genes from heterologous metabolic pathways, were employed by different research groups (cf. Table 1, 2010). When integrated into the yeast genome, these constructs appear to be fairly stable, despite the presence of direct repeats of promoter and terminator sequences [44]. Newly developed in vivo and in vitro methods for biosynthetic pathway assembly (cf. infra) push further this strategy, for example by assembling five expression cassettes on a large (nearly 19 kb) replicative vector (YaliBricks, cf. Table 1, 2017) [72].
Auto-Cloning Expression Vectors and Multicopy Integration
Integration of bacterial backbones from shuttle vectors, and especially of antibiotic resistance markers, into producing strains constitutes a drawback regarding acceptance by regulatory authorities for industrial or pharmaceutical applications. Auto-cloning vectors were designed to avoid this problem: bacterial moiety can be discarded before using purified integration cassette for transformation [20, 26]. Resulting recombinant strains bear no bacterial DNA, retaining their GRAS status. The most widely used auto-cloning vectors carry zeta sequences as expression cassette flanking regions [20]: this integration cassette can either be targeted to genomic zeta sequences in Ylt1-bearing strains (and strains equipped with an integrated zeta platform) or be integrated randomly in Ylt1-devoid strains [15]. This series of URA3-carrying zeta-based integrative vectors can be used in any Ura− strain, but are generally integrated at random into Ylt1-devoid Po1d, f, or h recipient strains [18,19,20]. Namely, NHEJ recombination is sufficiently effective in Y. lipolytica to allow integration of cassettes with non-homologous flanking regions, with only tenfold reduced transformation efficiency (cf. Table 1, 1998) [15]. This expression system was designed at INRA for both historical and practical reasons: avoiding using of HR for transformation allowed to circumvent a Pfizer patent (cf. Table 1, 1983) and more dispersed random multiple integrations were expected to be more stable than tandem ones obtained with HR [15].
As reviewed previously [5,6,7], zeta-based auto-cloning vectors with fully functional or defective selection marker were widely used worldwide for heterologous protein production. Using a defective marker promotes an amplification process leading to delayed appearance of colonies with increased copy numbers (generally around ten) of the expression cassette, at the expense of transformation efficiency, reduced by two orders of magnitude [12]. The flip side of the coin with randomly integrating auto-cloning vectors is a strong heterogeneity among transformants: since integration locus can impair cell growth or influence gene expression, careful selection of best producers is required [6]. Despite these drawbacks, multicopy auto-cloning vectors have been successfully used for increasing expression levels of numerous homologous [26] or heterologous genes [12, 15, 20]. However, as reviewed previously [6], long-term stability of randomly integrated multiple copies does not comply with the high standard of GMP (good manufacturing practices) guidelines, which limits their use for industrial applications. In addition, several newly developed engineering strategies can now offer more rapid and reliable alternatives for amplifying gene expression, like using multi-UASs promoters (cf. supra) and/or targeting copies at different selected genomic loci with genome editing technologies (cf. infra).
In Vivo and In Vitro DNA Assembly Methods
In recent years, the use of DNA assembly methods has considerably advanced genetic engineering of complex metabolic pathways in Y. lipolytica. A DNA assembler method allowed one-step integration of an entire β-carotene synthesis pathway, via in vivo HR, by a consortium of Shanghai laboratories (cf. Table 1, 2014) [61]. DNA fragments were at first assembled by overlap extension PCR (OE-PCR) into four expression cassettes (three overexpressed or heterologous genes and a selection marker), which were then used to co-transform yeast cells. Despite the fact that efficient HR in Y. lipolytica requires large flanking regions (cf. supra), the total efficiency of in vivo one-step assembly of the four DNA fragments was around 20% with overlaps between cassettes as small as 65 bp [61]. Flanking homologous sequences used for targeting integration at rDNA locus were, however, larger (0.6 kb). The orange/red color of successfully engineered colonies allowed visual screening and selection of best producers. Unexpected additional integration of partial cassettes was observed in the transformant with deepest color, probably due to NHEJ [61]. Simultaneous gene integration by in vivo HR thus appears as an efficient and rapid method (assembly of an 11-kb pathway in one week), in contrast to sequential integration that requires roughly one week per gene. The same consortium used recently the same DNA assembler method to integrate another 10-kb β-carotene synthesis pathway, based on a heterologous multifunctional carotene synthase, and showed that efficiency was greatly enhanced (63%) by double ku70/ku80 deletion [62]. A similar strategy of one-step in vivo assembly (OE-PCR followed by in vivo HR and rDNA targeting) was used at Nanjing Tech University to assemble an arachidonic acid (ARA) synthesis pathway (three genes, 10 kb) [63]. Overlapping region length was shown to influence efficiency, which reached 23% with 1-kb overlaps. The engineered strain exhibited robust growth and long-term genetic stability [63].
A Golden Gate Assembly (GGA) platform for complex engineering of Y. lipolytica has been recently designed at INRA, by constructing a library of donor plasmids bearing interchangeable building blocks, for one-step in vitro assembly [69]. GGA uses Type IIS restriction endonucleases, cutting outside of recognition sequence, to directionally assemble multiple DNA fragments. GGA platform operability was demonstrated on the same β-carotene synthesis pathway previously assembled in vivo [61]. GGA destination vector was a zeta-based auto-cloning vector, either integrated at random into Po1d or targeted at JMY1212 zeta docking platform (cf. supra). Global efficiency for GGA and yeast transformation (orange/red colonies) was 90% for random integration and 67% for platform targeting [69]. However, high variability in carotenoid-producer phenotype was observed in the former case, probably due to influence of random integration on gene expression (as discussed supra), when less variability was found among targeted transformants [69]. GGA efficiency (67–90% of desired phenotype) was thus much higher than when using in vivo DNA assembly (20%), for same carotenoid pathway genes [61, 69]. Moreover, GGA versatility (virtually limitless library of interchangeable building blocks, from endogenous or heterologous origin) makes it a tool of choice for fast assembly of any complex pathway. This GGA platform was also used to optimize expression of β-carotene pathway genes, in a promoter-shuffling experiment that allowed to enhance production by a sixfold factor [70]. An “obese” Y. lipolytica strain, carrying two copies of this promoter-optimized GGA-constructed β-carotene pathway, was reported to be the best microbial producer ever for this compound, in flask culture [70].
Another rapid in vitro assembly method is YaliBricks system, designed at the University of Maryland, Baltimore County [72]. A set of 12 YaliBrick vectors makes use of four compatible restriction sites to combine modular parts, complying with BioBrick standards, for rapid assembly of multigene pathways on replicative vectors. As proof of concept, the 12-kb five-gene violacein biosynthetic pathway was assembled in one week [72]. The library of YaliBrick vectors was also expanded to include CRISPR/Cas9 genome editing features (cf. infra) and is publicly available from Addgene website (https://www.addgene.org/).
New Genome Editing and Marker Rescue Tools
Since a few years, CRISPR/Cas9-based methods for genome editing and transcriptional regulation were developed in many organisms [64, 65]. Streptococcus pyogenes Cas9 endonuclease, when complexed with a targeting single-guide RNA (sgRNA), generates DSB at precise genomic loci, which repair by NHEJ causes indel mutations disrupting gene function. In presence of a homologous donor sequence, Cas9-induced DSB can be repaired by homology-directed repair (HDR), resulting in site-specific integration [64, 65]. Alternatively, mutated inactive Cas9 (dCas9), still able to bind sgRNA-complementary DNA but unable to generate DSB, can be targeted to a chosen promoter in order to suppress transcription by CRISPR interference (CRISPRi) [73]. Additionally, a transcriptional activator can be fused to dCas9 in order to promote transcription [73]. Collectively, these tools enable to disrupt and control genes of interest or to target new sequences into the genome, but their adaptation for each new species remains challenging. CRISPR–Cas9 tools for markerless gene disruption/integration in Y. lipolytica have been independently developed by American and Chinese research groups (cf. Table 1, 2016).
The University of California, Riverside (UCR), and the Clemson University used recombinant promoters, combining native RNA-PolIII promoters with a tRNA, for transcription of sgRNA, thus exploiting endogenous tRNA processing to produce mature sgRNA for Cas9 targeting. A codon-optimized Cas9 gene was expressed from an 8UAS1-pTEF promoter (cf. supra), and the two functional elements were combined on a single pCRISPRyl replicative vector [64]. Co-transformation into Po1f with a HDR donor plasmid resulted in markerless HR integration with 64% efficiency. This HR efficiency reached 100% in a NHEJ-disrupted ΔKu70 derivative [64]. This CRISPR/Cas9-based tool was further adapted for easy markerless integration of new metabolic pathways into Y. lipolytica genome [69]. After screening/selection of five genomic loci for accepting gene integration without impact on cell growth, a standardized tool comprising five pairs of plasmids (homologous donor and CRISPR/Cas9 expression plasmids), each targeting one of selected sites, was designed. It was applied to rapid integration of four genes from a semisynthetic lycopene biosynthesis pathway, at four loci (cf. Table 1, 2016) [66]. UCR also used this Cas9-expressing tool for developing a CRISPRi system in Y. lipolytica: dCas9 was targeted to Ku70 and Ku80 promoters, using multiplex sgRNA, in order to repress NHEJ, and HR efficiency was increased further when Mxi1 repressor was fused to dCas9 (cf. Table 1, 2017) [73]. The corresponding optimized CRISPRi-NHEJ plasmid, together with a ready-to-clone CRISPRi vector for use of alternative sgRNA, was deposited at Addgene.
Similarly, a consortium of Shanghai laboratories designed a set of two vectors for CRISPR/Cas9 genome editing in Y. lipolytica, which they assayed on different genomic loci: pCAS1yl and pCAS2yl express both Cas9 and sgRNA from a pTEFin promoter, and pCAS2yl additionally bears homologous donor DNA [65]. Maximal disruption efficiency was more than 85% when using pCAS1yl in Po1f (by NHEJ), and more than 94% when using pCAS2yl in a ΔKu70ΔKu80 derivative (by HDR only). Simultaneous multigene disruption was shown to be possible: a pCAS1yl carrying two tandem sgRNA expression cassettes provided double disruption with efficiency similar to that for single gene, and triple gene disruption was also obtained with less than twofold lower efficiency. At last, multiple rounds of genome editing were shown to be possible, following plasmid curing on non-selective medium [65].
These new CRISPR/Cas9 genome editing features were included in two recently developed Y. lipolytica toolboxes, YaliBrick vector library (cf. supra) [72], and EasyCloneYALI toolbox (cf. Table 1, 2018) [74]. Multipurpose EasyCloneYALI toolbox was designed at the Novo Nordisk Foundation Center for Biosustainability (Technical University of Denmark) and allows three different engineering strategies: marker-mediated integration, markerless (CRISPR/Cas9-based) integration, and markerless gene deletion [74]. It comprises a set of 27 standardized vectors (Biobrick elements) for integration of expression cassettes at defined genomic loci (11 selected intergenic sites allowing high expression levels and where integration did not affect growth), or for integration of mutation/knockout cassettes at loci of interest. The EasyCloneYALI toolbox present interesting innovative features: expression vectors are auto-cloning vectors accommodating two divergent expression cassettes; marker-mediated integration tools include dominant markers (hygromycin and nourseothricin resistance); markerless integration/deletion can use linear DNA fragments (e.g., double-stranded oligonucleotides or PCR products) as donor templates for HDR, instead of episomal vectors [74]. EasyCloneYALI integration vectors allowed CRISPR/Cas9 genome editing efficiencies above 80% with a transformation protocol using non-replicating DNA fragments as donor templates, and no loss of previously integrated cassettes could be detected after multiple engineering rounds (integrating 5–11 vectors) [74]. These tools can be obtained via AddGene.
In order to address the problem of inefficient sgRNA expression that limits CRISPR-Cas9 implementation in new fungal hosts, the UT Austin proposed to use a mutated version of a T7 polymerase to obtain sgRNA expression from a T7 promoter (cf. Table 1, 2016) [67]. Initially developed in S. cerevisiae, this methodology was further adapted for other yeasts, allowing genome editing of Kluyveromyces lactis and Y. lipolytica with, respectively, 96 and 60% efficiencies [67]. This T7-based sgRNA expression strategy is expected to enhance CRISPR systems efficiency in various fungal systems.
Alongside these CRISPR/Cas9-based methods, another genome editing strategy was also used in Y. lipolytica: TALEN (transcription activator-like effector nucleases)-based technology was applied to protein engineering by a research group from INSA (Toulouse University). TALEN are recombinant restriction enzymes, engineered to cut specific DNA sequences, obtained by fusing a TAL effector DNA-binding domain to a nuclease. TALEN-based genome editing tools were used to introduce targeted mutations into Y. lipolytica giant multifunctional fatty acid synthase (FAS), a key enzyme in lipid biosynthesis, for obtaining shorter fatty acid chain lengths (cf. Table 1, 2017) [71].
At last, in vivo piggyBac transposition was very recently demonstrated at UT Austin, using a codon-optimized hyperactive piggyBac transposase (hyPBase) [75]. Any cargo DNA sequence (i.e., selection marker or integration cassette), when flanked by piggyBac inverted terminal repeats (ITRs), can be mobilized (cut and pasted into Y. lipolytica genome) by hyPBase transposase expressed from a replicative vector. This transposition system, based on a TTAA-specific transposon (originally isolated from cabbage looper Trichoplusia ni) was developed into a platform, for constructing genome-wide insertional mutagenesis libraries and introducing scarless genomic modifications in Y. lipolytica, using a series of existing and new auxotrophic or dominant selection markers (cf. Table 1, 2018) [75]. In addition, the piggyBac-born integrated cargo sequence can be precisely excised from the genome using an engineered excision+/integration− mutant transposase, thus providing a scarless marker rescue system [75]. In contrast to the previously described Tn3 transposon-generated mutant library (cf. Table 1, 1998) that required multiple rounds of bacterial transformation/conjugation followed by yeast transformation with a library of Tn3-mutated DNA fragments [16], the piggyBac approach is achieved by direct transformation of Y. lipolytica followed by in vivo transposition [75]. However, contrary to some other transposons, piggyBac integration does not occur fully at random in the genome, but targets TTAA sequences (found in less than two-thirds of annotated Y. lipolytica coding sequences) and favors actively transcribed regions, which limits the representativeness of piggyBac-generated mutant libraries. Besides genome-wide insertional mutagenesis applications, the authors also propose using piggyBac-based tools for increasing transformation efficiency of randomly integrating cassettes or for enabling easy marker rescue following CRISPR/Cas9-directed integration of expression cassettes [75].
Besides marker rescue methods (Cre-lox, piggyBac) requiring the heterologous expression of a recombinase [21] or a transposase [75], a consortium of Korean laboratories proposed to use directly the high HR frequency of ΔKu70 cells for this purpose (cf. Table 1, 2018) [76]. These authors obtained 100% rescue of a URA3 marker gene in a ΔKu70 strain, by using HR between the 100-bp homology (3 tandem repeats of an HA tag) of the flanking regions of an “URA3-blaster” cassette [76]. When combined to HU-mediated cell cycle synchronization for improved gene targeting in ΔKu70 strains (cf. supra), this simple marker rescue strategy for repeated insertion/excision steps of an “URA3-blaster” cassette could considerably ease complex engineering projects, by allowing URA3 marker reuse, in repetitive rounds of transformations, for sequential multiple genetic modifications.
Applications of Y. lipolytica Engineered Strains
An extensive description of Y. lipolytica biotechnological applications can be found in a recent review [81], as well as in more general publications [1, 2, 9] or in more specific ones: environmental and industrial use [3]; food-processing applications [4]. Y. lipolytica has a long history of production of organic acids (notably citric acid) and SCP [1,2,3, 91] and has aroused interest more recently as an efficient single-cell oil (SCO) producer [39, 41, 42, 91]. Among organic acids for which Y. lipolytica is a recognized producer, the development of genetic engineering has greatly benefited in particular to alpha-ketoglutaric (cf. supra [83, 84]) and to succinic acid production [86]. Notably, process optimization of succinic acid production from a H222-derived engineered Y. lipolytica strain established this yeast as a competitive producer of this organic acid used as food additive, dietary supplement, and building block for bio-plastics [86]. A resume of some major applications of engineered Y. lipolytica strains is schematized in Fig. 1, but only a few examples will be briefly evoked here.
Y. lipolytica is an oleaginous yeast of particular interest since it can accumulate lipids up to 40% of its dry cell weight (DCW) and is the species with the highest proportion of linoleic acid (more than 50% of fatty acids) [39]. These properties served as basis for further engineering of lipid storage capacity (cf. Table 1, 2008) an “obese” strain, derived from Po1d at INRA, accumulated lipids up to 75% of DCW [40] and another, derived from Po1f at UT Austin, 90% [41]. At last, the Massachusetts Institute of Technology reported the highest lipid yield, titer (55 g/L), and productivity to date, for YL-ad9 “obese” strain, in which carbon to lipid conversion yield was 85% of theoretical maximal one [42]. YL-ad9, obtained notably by overexpressing a rate-limiting enzyme identified by reverse engineering of mammalian cell obese phenotype, exhibits a threefold growth advantage over its parent LEU2-complemented Po1g strain [42]. This work represents an important step towards efficient and cost-effective Y. lipolytica processes for biodiesel production or other oil-derived compounds from renewable resources.
Indeed, commercial scale production of microbial oil-based biodiesel from various economical/waste substrates constitutes a challenge for which Y. lipolytica is one of the most promising microorganisms [92, 93]. Microbial oils are also gaining importance since genetic engineering can enrich them in unusual desired fatty acids [71, 94], such as building blocks for bio-based chemistry (e.g., long-chain dicarboxylic acids) [85] and nutraceuticals (e.g., polyunsaturated fatty acids, PUFAs) [33, 34, 43]. Production of PUFA-rich SCO is the first process using engineered Y. lipolytica that reached the stage of commercialization (cf. Table 1, 2007): EPA-rich oil [34] was used as dietary supplement, and EPA-rich yeast cells as feed for pisciculture [1, 2, 91].
Numerous other high-value products can also be derived from engineered Y. lipolytica strains, such as medium chain-length polyhydroxyalkanoates (PHAs), biopolymers that constitutes renewable and biodegradable bio-plastics [96], polyketides, secondary metabolites that serve as building blocks for chemical catalysis/polymerization [97], and carotenoids for use in food industry, as exemplified supra [61, 62, 66, 69, 70]. At last, one of the key factors for establishing economic viability of biotechnological processes is the use of cheap substrates and preferably of renewable resources. Y. lipolytica constitutes a particularly interesting host in this regard, since its metabolic engineering allows the use of inexpensive carbon sources, such as agricultural or industrial wastes (e.g., molasses [82], oily food waste [96], or lignocellulosic biomass [98,99,100]), for producing biofuels and chemicals, as reviewed recently [91, 101].
Conclusion
Engineering Y. lipolytica for production of biofuel or high-value products from renewable resources is a very rapidly expanding research area, and the addition of DNA assembly and genome editing technologies to the global Y. lipolytica toolbox is expected to revolutionize this field by enabling fast combinatorial assembly of complex synthetic pathways. Metabolic engineering strategies will also afford benefit of detailed knowledge of key biological processes involved in lipid accumulation or organic acid production, brought by genomic, transcriptomic, metabolomic, and fluxomic analyses from several research groups throughout the world [102,103,104]. Bioinformatics and applied mathematics have also a role to play by allowing building of genome-scale models of Y. lipolytica metabolic networks (cf. Table 1, 2012) [50,51,52,53]. All these tools are expected to concur for establishing this yeast as a workhorse for biotechnological applications.
Note Added in Proof
A new in vivo transposition system, for Hermes transposon, has just been used at UCI to construct a library of insertion mutants that has been applied to studying Y. lipolytica metabolism (https://doi.org/10.1016/j.ymben.2018.05.008).
References
Groenewald, M., Boekhout, T., Neuvéglise, C., Gaillardin, C., van Dijck, P. W., & Wyss, M. (2014). Yarrowia lipolytica: Safety assessment of an oleaginous yeast with a great industrial potential. Critical Reviews in Microbiology, 40(3), 187–206.
Sibirny, A., Madzak, C., & Fickers, P. (2014). Genetic engineering of non-conventional yeast for the production of valuable compounds. In F.D. Harzevili, H. Chen (Eds.), Microbial biotechnology: Progress and trends (pp. 63–111). Boca Raton: CRC Press.
Bankar, A. V., Kumar, A. R., & Zinjarde, S. S. (2009). Environmental and industrial applications of Yarrowia lipolytica. Applied Microbiology and Biotechnology, 84(5), 847–865.
Zinjarde, S. S. (2014). Food-related applications of Yarrowia lipolytica. Food Chemistry, 152, 1–10.
Madzak, C., Gaillardin, C., & Beckerich, J. M. (2004). Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: A review. Journal of Biotechnology, 109(1–2), 63–81.
Madzak, C., & Beckerich, J. M. (2013). Heterologous protein expression and secretion, in Yarrowia lipolytica. In G. Barth (Ed.), Yarrowia lipolytica: Biotechnological applications (Vol. 25, pp. 1–76). Heidelberg: Springer.
Madzak, C. (2015). Yarrowia lipolytica: Recent achievements in heterologous protein expression and pathway engineering. Applied Microbiology and Biotechnology, 99(11), 4559–4577.
Barth, G. (2013). Microbiology monographs, Vol. 24: Yarrowia lipolytica: Genetics, genomics, and physiology. Heidelberg: Springer.
Barth, G. (2013). Microbiology monographs, Vol. 25: Yarrowia lipolytica: Biotechnological applications. Heidelberg: Springer.
Nicaud, J. M., Fabre, E., & Gaillardin, C. (1989). Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker. Current Genetics, 16(4), 253–260.
Lazar, Z., Rossignol, T., Verbeke, J., Crutz-Le Coq, A. M., Nicaud, J. M., & Robak, M. (2013). Optimized invertase expression and secretion cassette for improving Yarrowia lipolytica growth on sucrose for industrial applications. Journal of Industrial Microbiology and Biotechnology, 40(11), 1273–1283.
Le Dall, M. T., Nicaud, J. M., & Gaillardin, C. (1994). Multiple-copy integration in the yeast Yarrowia lipolytica. Current Genetics, 26(1), 38–44.
Fournier, P., Abbas, A., Chasles, M., Kudla, B., Ogrydziak, D. M., et al. (1993) Colocalization of centromeric and replicative functions on autonomously replicating sequences isolated from the yeast Yarrowia lipolytica. Proceedings of the National Academy of Sciences of the United States of America, 90(11), 4912–4916.
Chen, D. C., Beckerich, J. M., & Gaillardin, C. (1997). One-step transformation of the dimorphic yeast Yarrowia lipolytica. Applied Microbiology and Biotechnology, 48(2), 232–235.
Juretzek, T., Le Dall, M. T., Mauersberger, S., Gaillardin, C., Barth, G., & Nicaud, J. M. (2001). Vectors for gene expression and amplification in the yeast Yarrowia lipolytica. Yeast, 18(2), 97–113.
Neuvéglise, C., Nicaud, J. M., Ross-Macdonald, P., & Gaillardin, C. (1998). A shuttle mutagenesis system for tagging genes in the yeast Yarrowia lipolytica. Gene, 213(1–2), 37–46.
Müller, S., Sandal, T., Kamp-Hansen, P., & Dalbøge, H. (1998). Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast, 14(14), 1267–1283.
Madzak, C., Tréton, B., & Blanchin-Roland, S. (2000). Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. Journal of Molecular Microbiology and Biotechnology, 2(2), 207–216.
Madzak, C. (2003). New tools for heterologous protein production in the yeast Yarrowia lipolytica. In S. G. Pandalai (Ed.), Recent research developments in microbiology (Vol. 7, pp. 453–479). Trivandrum: Research Signpost.
Nicaud, J. M., Madzak, C., van den Broek, P., Gysler, C., Duboc, P., et al. (2002). Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Research, 2(3), 371–379.
Fickers, P., Le Dall, M. T., Gaillardin, C., Thonart, P., & Nicaud, J. M. (2003). New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. Journal of Microbiological Methods, 55(3), 727–737.
Dujon, B., Sherman, D., Fischer, G., Durrens, P., Casaregola, S., et al. (2004). Genome evolution in yeasts. Nature., 430(6995), 35–44.
Madzak, C., Mimmi, M. C., Caminade, E., Brault, A., Baumberger, S., et al. (2006). Shifting the optimal pH of activity for a laccase from the fungus Trametes versicolor by structure-based mutagenesis. Protein Engineering, Design & Selection, 19(2), 77–84.
Bordes, F., Cambon, E., Dossat-Létisse, V., André, I., Croux, C., et al. (2009). Improvement of Yarrowia lipolytica lipase enantioselectivity by using mutagenesis targeted to the substrate binding site. Chembiochem, 10(10), 1705–1713.
Galli, C., Gentili, P., Jolivalt, C., Madzak, C., & Vadalà, R. (2011). How is the reactivity of laccase affected by single-point mutations? Engineering laccase for improved activity towards sterically demanding substrates. Applied Microbiology and Biotechnology, 91(1), 123–131.
Pignède, G., Wang, H. J., Fudalej, F., Seman, M., Gaillardin, C., & Nicaud, J. M. (2000). Autocloning and amplification of LIP2 in Yarrowia lipolytica. Applied and Environment Microbiology, 66(8), 3283–3289.
Park, J. N., Song, Y., Cheon, S. A., Kwon, O., Oh, D. B., et al. (2011). Essential role of YlMPO1, a novel Yarrowia lipolytica homologue of Saccharomyces cerevisiae MNN4, in mannosylphosphorylation of N- and O-linked glycans. Applied and Environment Microbiology, 77(4), 1187–1195.
De Pourcq, K., Vervecken, W., Dewerte, I., Valevska, A., Van Hecke, A., & Callewaert, N. (2012). Engineering the yeast Yarrowia lipolytica for the production of therapeutic proteins homogeneously glycosylated with Man8GlcNAc2 and Man5GlcNAc2. Microbial Cell Factories, 11, 53.
De Pourcq, K., Tiels, P., Van Hecke, A., Geysens, S., Vervecken, W., & Callewaert, N. (2012) Engineering Yarrowia lipolytica to produce glycoproteins homogeneously modified with the universal Man3GlcNAc2 N-glycan core. PLoS ONE, 7(6), e39976.
Moon, H. Y., Van, T. L., Cheon, S. A., Choo, J., Kim, J. Y., & Kang, H. A. (2013). Cell-surface expression of Aspergillus saitoi-derived functional α-1,2-mannosidase on Yarrowia lipolytica for glycan remodeling. Journal of Microbiology, 51(4), 506–514.
Bordes, F., Fudalej, F., Dossat, V., Nicaud, J. M., & Marty, A. (2007). A new recombinant protein expression system for high-throughput screening in the yeast Yarrowia lipolytica. Journal of Microbiological Methods, 70(3), 493–502.
Leplat, C., Nicaud, J. M., & Rossignol, T. (2015) High-throughput transformation method for Yarrowia lipolytica mutant library screening. FEMS Yeast Research, 15(6), fov052.
Xue, Z., Sharpe, P. L., Hong, S. P., Yadav, N. S., Xie, D., et al. (2013). Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nature Biotechnology, 31(8), 734–740.
Xie, D., Jackson, E. N., & Zhu, Q. (2015). Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: From fundamental research to commercial production. Applied Microbiology and Biotechnology, 99(4), 1599–1610.
Yue, L., Chi, Z., Wang, L., Liu, J., Madzak, C., Li, J., & Wang, X. (2008) Construction of a new plasmid for surface display on cells of Yarrowia lipolytica. Journal of Microbiological Methods, 72(2), 116–123
Yang, X. S., Jiang, Z. B., Song, H. T., Jiang, S. J., Madzak, C., & Ma, L. X. (2009). Cell-surface display of the active mannanase in Yarrowia lipolytica with a novel surface-display system. Biotechnology and Applied Biochemistry, 54(3), 171–176.
Duquesne, S., Bozonnet, S., Bordes, F., Dumon, C., Nicaud, J. M., & Marty, A. (2014) Construction of a highly active xylanase displaying oleaginous yeast: Comparison of anchoring systems. PLoS ONE, 9(4), e95128.
Yuzbasheva, E. Y., Yuzbashev, T. V., Laptev, I. A., Konstantinova, T. K., & Sineoky, S. P. (2011). Efficient cell surface display of Lip2 lipase using C-domains of glycosylphosphatidylinositol-anchored cell wall proteins of Yarrowia lipolytica. Applied Microbiology and Biotechnology, 91(3), 645–654.
Beopoulos, A., Chardot, T., & Nicaud, J. M. (2009). Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie, 91(6), 692–696.
Dulermo, T., & Nicaud, J. M. (2011). Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metabolic Engineering, 13(5), 482–491.
Blazeck, J., Hill, A., Liu, L., Knight, R., Miller, J., et al. (2014). Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nature Communications, 5, 3131.
Qiao, K., Abidi, I., Liu, S. H., Zhang, H., Chakraborty, H., S. et al (2015). Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metabolic Engineering, 29, 56–65.
Chuang, L. T., Chen, D. C., Nicaud, J. M., Madzak, C., Chen, Y. H., & Huang, Y. S. (2010). Co-expression of heterologous desaturase genes in Yarrowia lipolytica. New Biotechnology, 27(4), 277–282.
Celińska, E., & Grajek, W. (2013). A novel multigene expression construct for modification of glycerol metabolism in Yarrowia lipolytica. Microbial Cell Factories, 12, 102.
Blazeck, J., Liu, L., Redden, H., & Alper, H. (2011). Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Applied and Environment Microbiology, 77(22), 7905–7914.
Blazeck, J., Reed, B., Garg, R., Gerstner, R., Pan, A., et al. (2013). Generalizing a hybrid synthetic promoter approach in Yarrowia lipolytica. Applied Microbiology and Biotechnology, 97(7), 3037–3052.
Dulermo, R., Brunel, F., Dulermo, T., Ledesma-Amaro, R., Vion, J., et al. (2017). Using a vector pool containing variable-strength promoters to optimize protein production in Yarrowia lipolytica. Microbial Cell Factories, 16(1), 31.
Shabbir-Hussain, M., Wheeldon, I., & Blenner, M. A. (2017). A strong hybrid fatty acid inducible transcriptional sensor built from Yarrowia lipolytica upstream activating and regulatory sequences. Biotechnology Journal, 12(10), 1700248.
Trassaert, M., Vandermies, M., Carly, F., Denies, O., Thomas, S., et al. (2017). New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica. Microbial Cell Factories, 16(1), 141.
Loira, N., Dulermo, T., Nicaud, J. M., & Sherman, D. J. (2012). A genome-scale metabolic model of the lipid-accumulating yeast Yarrowia lipolytica. BMC Systems Biology, 6, 35.
Pan, P., & Hua, Q. (2012) Reconstruction and in silico analysis of metabolic network for an oleaginous yeast, Yarrowia lipolytica. PLoS ONE 7(12), e51535.
Kavšček, M., Bhutada, G., Madl, T., & Natter, K. (2015). Optimization of lipid production with a genome-scale model of Yarrowia lipolytica. BMC Systems Biology, 9, 72.
Trébulle, P., Nicaud, J. M., Leplat, C., & Elati, M. (2017). Inference and interrogation of a coregulatory network in the context of lipid accumulation in Yarrowia lipolytica. NPJ Systems Biology and Applications, 3, 21.
Tiels, P., Baranova, E., Piens, K., De Visscher, C., Pynaert, G., et al. (2012). A bacterial glycosidase enables mannose-6-phosphate modification and improved cellular uptake of yeast-produced recombinant human lysosomal enzymes. Nature Biotechnology, 30(12), 1225–1231.
Verbeke, J., Beopoulos, A., & Nicaud, J. M. (2013). Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnology Letters, 35(4), 571–576.
Kretzschmar, A., Otto, C., Holz, M., Werner, S., Hübner, L., & Barth, G. (2013). Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Current Genetics, 59(1–2), 63–72.
Han, Z., Madzak, C., & Su, W. W. (2013). Tunable nano-oleosomes derived from engineered Yarrowia lipolytica. Biotechnology and Bioengineering, 110(3), 702–710.
Liu, L., & Alper, H. S. (2014) Draft genome sequence of the oleaginous yeast Yarrowia lipolytica PO1f, a commonly used metabolic engineering host. Genome Announcements, 2(4), e00652–e006614.
Pomraning, K. R., & Baker, S. E. (2015) Draft genome sequence of the dimorphic yeast Yarrowia lipolytica strain W29. Genome Announcements, 3(6), e01211–e01215.
Magnan, C., Yu, J., Chang, I., Jahn, E., Kanomata, Y., et al. (2016) Sequence assembly of Yarrowia lipolytica strain W29/CLIB89 shows transposable element diversity. PLoS ONE, 11(9), e0162363.
Gao, S., Han, L., Zhu, L., Ge, M., Yang, S., et al. (2014). One-step integration of multiple genes into the oleaginous yeast Yarrowia lipolytica. Biotechnology Letters, 36(12), 2523–2528.
Gao, S., Tong, Y., Zhu, L., Ge, M., Jiang, Y., et al. (2017). Production of β-carotene by expressing a heterologous multifunctional carotene synthase in Yarrowia lipolytica. Biotechnology Letters, 39(6), 921–927.
Liu, H. H., Madzak, C., Sun, M. L., Ren, L., Song, P., et al. (2017). Engineering Yarrowia lipolytica for arachidonic acid production through rapid assembly of metabolic pathway. Biochemical Engineering Journal, 119, 52–58.
Schwartz, C. M., Hussain, M. S., Blenner, M., & Wheeldon, I. (2016). Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica. ACS Synthetic Biology, 5(4), 356–359.
Gao, S., Tong, Y., Wen, Z., Zhu, L., Ge, M., et al. (2016). Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. Journal of Industrial Microbiology and Biotechnology, 43(8), 1085–1093.
Schwartz, C., Shabbir-Hussain, M., Frogue, K., Blenner, M., & Wheeldon, I. (2017). Standardized markerless gene integration for pathway engineering in Yarrowia lipolytica. ACS Synthetic Biology, 6(3), 402–409.
Morse, N. J., Wagner, J. M., Reed, K. B., Gopal, M. R., Lauffer, L. H., & Alper, H. S. (2018). T7 polymerase expression of guide RNAs in vivo allows exportable CRISPR-Cas9 editing in multiple yeast hosts. ACS Synthetic Biology, 7(4), 1075–1084.
Bredeweg, E. L., Pomraning, K. R., Dai, Z., Nielsen, J., Kerkhoven, E. J., & Baker, S. E. (2017). A molecular genetic toolbox for Yarrowia lipolytica. Biotechnology for Biofuels, 10, 2.
Celińska, E., Ledesma-Amaro, R., Larroude, M., Rossignol, T., Pauthenier, C., & Nicaud, J. M. (2017). Golden Gate Assembly system dedicated to complex pathway manipulation in Yarrowia lipolytica. Microbial Biotechnology, 10(2), 450–455.
Larroude, M., Celinska, E., Back, A., Thomas, S., Nicaud, J. M., & Ledesma-Amaro, R. (2018). A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnology and Bioengineering, 115(2), 464–472.
Rigouin, C., Gueroult, M., Croux, C., Dubois, G., Borsenberger, V., et al. (2017). Production of medium chain fatty acids by Yarrowia lipolytica: Combining molecular design and TALEN to engineer the fatty acid synthase. ACS Synthetic Biology, 6(10), 1870–1879.
Wong, L., Engel, J., Jin, E., Holdridge, B., & Xu, P. (2017). YaliBricks, a versatile genetic toolkit for streamlined and rapid pathway engineering in Yarrowia lipolytica. Metabolic Engineering Communications, 5, 68–77.
Schwartz, C., Frogue, K., Ramesh, A., Misa, J., & Wheeldon, I. (2017). CRISPRi repression of nonhomologous end-joining for enhanced genome engineering via homologous recombination in Yarrowia lipolytica. Biotechnology and Bioengineering, 114(12), 2896–2906.
Holkenbrink, C., Dam, M. I., Kildegaard, K. R., Beder, J., Dahlin, J., et al. (2018) EasyCloneYALI: CRISPR/Cas9-based synthetic toolbox for engineering of the yeast Yarrowia lipolytica. Biotechnology Journal. https://doi.org/10.1002/biot.201700543.
Wagner, J. M., Williams, E. V., & Alper, H. S. (2018). Developing a piggyBac transposon system and compatible selection markers for insertional mutagenesis and genome engineering in Yarrowia lipolytica. Biotechnology Journal, 13(5), e1800022.
Jang, I. S., Yu, B. J., Jang, J. Y., Jegal, J., & Lee, J. Y. (2018) Improving the efficiency of homologous recombination by chemical and biological approaches in Yarrowia lipolytica. PLoS ONE 13(3), e0194954.
Juretzek, T., Wang, H. J., Nicaud, J. M., Mauersberger, S., & Barth, G. (2000). Comparison of promoters suitable for regulated overexpression of β-galactosidase in the alkane-utilizing yeast Yarrowia lipolytica. Biotechnology and Bioprocess Engineering, 5, 320–326.
Tai, M., & Stephanopoulos, G. (2013). Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metabolic Engineering, 15, 1–9.
Kopecný, D., Pethe, C., Sebela, M., Houba-Hérin, N., Madzak, C., et al. (2005). High-level expression and characterization of Zea mays cytokinin oxidase/dehydrogenase in Yarrowia lipolytica. Biochimie, 87(11), 1011–1022.
Curran, K. A., Morse, N. J., Markham, K. A., Wagman, A. M., Gupta, A., & Alper, H. S. (2015). Short synthetic terminators for improved heterologous gene expression in yeast. ACS Synthetic Biology, 4(7), 824–832.
Barth, G., & Gaillardin, C. (1996) Yarrowia lipolytica. In K. Wolf (Ed.), Nonconventional yeasts in biotechnology: A handbook. Springer, Heidelberg, pp. 313–388.
Yan, J., Han, B., Gui, X., Wang, G., Xu, L., et al. (2018). Engineering Yarrowia lipolytica to simultaneously produce lipase and single cell protein from agro-industrial wastes for feed. Science Report, 8(1), 758.
Guo, H., Su, S., Madzak, C., Zhou, J., Chen, H., & Chen, G. (2016). Applying pathway engineering to enhance production of alpha-ketoglutarate in Yarrowia lipolytica. Applied Microbiology and Biotechnology, 100(23), 9875–9884.
Yovkova, V., Otto, C., Aurich, A., Mauersberger, S., & Barth, G. (2014). Engineering the α-ketoglutarate overproduction from raw glycerol by overexpression of the genes encoding NADP+-dependent isocitrate dehydrogenase and pyruvate carboxylase in Yarrowia lipolytica. Applied Microbiology and Biotechnology, 98, 2003–2013.
Förster, A., Aurich, A., Mauersberger, S., & Barth, G. (2007). Citric acid production from sucrose using a recombinant strain of the yeast Yarrowia lipolytica. Applied Microbiology and Biotechnology, 75(6), 1409–1417.
Jost, B., Holz, M., Aurich, A., Barth, G., Bley, T., & Müller, R. A. (2015). The influence of oxygen limitation for the production of succinic acid with recombinant strains of Yarrowia lipolytica. Applied Microbiology and Biotechnology, 99(4), 1675–1686.
Kerkhoven, E. J., Kim, Y. M., Wei, S., Nicora, C. D., Fillmore, T. L., et al. (2017). Leucine biosynthesis is involved in regulating high lipid accumulation in Yarrowia lipolytica. MBio, 8(3), e00857–e00817.
Schmid-Berger, N., Schmid, B., & Barth, G. (1994). Ylt1, a highly repetitive retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. Journal of Bacteriology, 176, 2477–2482.
Tsakraklides, V., Brevnova, E., Stephanopoulos, G., & Shaw, A. J. (2015) Improved gene targeting through cell cycle synchronization. PLoS ONE 10(7), e0133434.
Liu, L., Otoupal, P., Pan, A., & Alper, H. S. (2014). Increasing expression level and copy number of a Yarrowia lipolytica plasmid through regulated centromere function. FEMS Yeast Research, 14(7), 1124–1127.
Liu, H. H., Ji, X. J., & Huang, H. (2015). Biotechnological applications of Yarrowia lipolytica: Past, present and future. Biotechnology Advances, 33(8), 1522–1546.
Xie, D. (2017). Integrating cellular and bioprocess engineering in the non-conventional yeast Yarrowia lipolytica for biodiesel production: A review. Frontiers in Bioengineering and Biotechnology, 5, 65.
Yan, J., Yan, Y., Madzak, C., & Han, B. (2017). Harnessing biodiesel-producing microbes: From genetic engineering of lipase to metabolic engineering of fatty acid biosynthetic pathway. Critical Reviews in Biotechnology, 37(1), 26–36.
Ledesma-Amaro, R., & Nicaud, J. M. (2016). Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Progress in Lipid Research, 61, 40–50.
Abghari, A., Madzak, C., & Chen, S. (2016). Combinatorial engineering Yarrowia lipolytica as a promising cell biorefinery platform for the production of multi-purpose long chain dicarboxylic acid building blocks from glycerol. Fermentation, 3(3), 40.
Gao, C., Qi, Q., Madzak, C., & Lin, C. S. (2015). Exploring medium-chain-length polyhydroxyalkanoates production in the engineered yeast Yarrowia lipolytica. Journal of Industrial Microbiology and Biotechnology, 42(9), 1255–1262.
Markham, K. A., Palmer, C. M., Chwatko, M., Wagner, J. M., Murray, C., et al. (2018) Rewiring Yarrowia lipolytica toward triacetic acid lactone for materials generation. Proceedings of the National Academy of Sciences of the United States of America, 115(9), 2096–2101.
Wei, H., Wang, W., Alahuhta, M., Wall, V., Baker, T., Taylor, J.O. et al. (2014). Engineering towards a complete heterologous cellulase secretome in Yarrowia lipolytica reveals its potential for consolidated bioprocessing. Biotechnology for Biofuels, 7(1), 148.
Guo, Z., Duquesne, S., Bozonnet, S., Cioci, G., Nicaud, J. M., et al. (2015). Development of cellobiose-degrading ability in Yarrowia lipolytica strain by overexpression of endogenous genes. Biotechnology for Biofuels, 8, 109.
Guo, Z. P., Robin, J., Duquesne, S., O’Donohue, M. J., Marty, A., & Bordes, F. (2018). Developing cellulolytic Yarrowia lipolytica as a platform for the production of valuable products in consolidated bioprocessing of cellulose. Biotechnology for Biofuels, 11, 141.
Ledesma-Amaro, R., & Nicaud, J. M. (2016). Metabolic engineering for expanding the substrate range of Yarrowia lipolytica. Trends in Biotechnology, 34(10), 798–809.
Morin, N., Cescut, J., Beopoulos, A., Lelandais, G., Le Berre, V., et al. (2011) Transcriptomic analyses during the transition from biomass production to lipid accumulation in the oleaginous yeast Yarrowia lipolytica. PLoS ONE. 6(11), e27966.
Pomraning, K. R., Wei, S., Karagiosis, S. A., Kim, Y. M., Dohnalkova, A. C., et al. (2015) Comprehensive metabolomic, lipidomic and microscopic profiling of Yarrowia lipolytica during lipid accumulation identifies targets for increased lipogenesis. PLoS ONE. 10(4), e0123188.
Sabra, W., Bommareddy, R. R., Maheshwari, G., Papanikolaou, S., & Zeng, A. P. (2017). Substrates and oxygen dependent citric acid production by Yarrowia lipolytica: Insights through transcriptome and fluxome analyses. Microbial Cell Factories, 16(1), 78.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Madzak, C. Engineering Yarrowia lipolytica for Use in Biotechnological Applications: A Review of Major Achievements and Recent Innovations. Mol Biotechnol 60, 621–635 (2018). https://doi.org/10.1007/s12033-018-0093-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-018-0093-4