Abstract
Molecular evaluation of KRAS, BRAF, and PIK3CA mutation has become an important part in colorectal carcinoma evaluation, and their alterations may determine the therapeutic response to anti-EGFR therapy. The current study demonstrates the evaluation of KRAS, BRAF, and PIK3CA mutation using direct sequencing in 204 samples. The frequency of KRAS, BRAF, and PIK3CA mutations was 23.5, 9.8, and 5.9 %, respectively. Five different substitution mutations at KRAS codon 12 (G12S, G12D, G12A, G12V, and G12C) and one substitution type at codon 13 (G13D) were observed. KRAS mutations were significantly higher in patients who were >50 years, and were associated with moderate/poorly differentiated tumors and adenocarcinomas. All mutations in BRAF gene were of V600E type, which were frequent in patients who were ≤50 years. Unlike KRAS mutations, BRAF mutations were more frequent in well-differentiated tumors and right-sided tumors. PIK3CA–E545K was the most recurrent mutation while other mutations detected were T544I, Q546R, H1047R, G1049S, and D1056N. No significant association of PIK3CA mutation with age, tumor differentiation, location, and other parameters was noted. No concomitant mutation of KRAS and BRAF mutations was observed, while, interestingly, five cases showed concurrent mutation of KRAS and PIK3CA mutations. In conclusion, to our knowledge, this is the first study to evaluate the PIK3CA mutation in Indian CRC patients. The frequency of KRAS, BRAF, and PIK3CA was similar to worldwide reports. Furthermore, identification of molecular markers has unique strengths, and can provide insights into the pathogenic process and help optimize personalized prevention and therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the most common gastrointestinal malignancy occurring fourth in males and third in females across the globe associated with high mortality and morbidity [1]. The incidence of CRC is relatively high in the western countries. Furthermore, within Asia, the incidence of CRC was reported to vary from lower in South Asian developing countries and higher in developed Asian countries [2]. In India, CRC stands fourth in males and fifth in females of the number of patients living and diagnosed within the past 5 years [3]. While surgery is still the only curative method for patients with localized tumors [4], with advanced disease progression, several combinations of chemotherapeutic drugs are used to extend the rate of overall and disease-free survival.
Activation of multiple signaling pathways specifically RAS–RAF–MAPK and PI3K–PTEN–AKT pathway plays an important role in regulating cell proliferation, angiogenesis, cell motility, and apoptosis [5, 6]. Accumulation of several mutations in the tumor suppressor genes and proto-oncogenes in these signaling pathways plays key roles in promoting CRC. Among these, mutations of KRAS, NRAS, and BRAF genes have already been widely studied in the past few years in CRC [7]. Oncogenic mutations of KRAS, NRAS, and BRAF produce a constitutively active ras protein, leading in turn to EGFR-independent activation of the MAPK pathway [8, 9].
Various studies have reported that about 30–45 % of CRC tumors show the presence of KRAS mutation, indicating that approximately 55 % of CRC patients might respond to anti-EGFR therapy [10, 11]. On the other hand, nearly 40–60 % of KRAS-negative tumors shows the failure of the therapy [12], indicating that additional genetic alterations might be contributing to this non-responsiveness. Recent studies have suggested that the presence of BRAF mutations in about 5–20 % of CRC tumors can also affect the response to these agents, especially in patients whose tumors lack KRAS mutations [13, 14].
Initiation of the PI3K/AKT/mTOR pathway is frequently mediated by genetic alterations in the p110α subunit of PIK3CA, with most mutations (>80 %) occurring either in exon 9, which codes for the helical domain, or exon 20, which codes for the kinase domain [5]. Furthermore, the literature survey suggests that mutations of the PIK3CA are the most recurrent genetic alterations after KRAS/BRAF mutation, and has been reported in the range of 5–15 %, which may contribute to the non-responsiveness to anti-EGFR therapy [15, 16]. It is interesting to note that PIK3CA mutations may coexist with either KRAS or BRAF alterations within the same tumor [17], but KRAS and BRAF mutations appear to be mutually exclusive [18].
In the past decade, many groups have reported the frequency of these mutations and their association to the non-responsiveness to anti-EGFR therapy through extensive cell line studies, and various clinical trials across the globe [13, 14, 19]. In spite of so much of research, only limited studies have been reported about KRAS and BRAF mutations in Indian CRC patients [20–23], whereas no precise published data exist about the presence of PIK3CA mutations, despite the fact that colon cancer is one of the most common cancer associated with high morbidity and mortality in India. There is a need to investigate the presence of these markers in larger cohort, which will enhance our knowledge in further understanding the genetic heterogeneity of Indian CRC patients. Therefore, in the current study, we aimed to assess the frequency and distribution pattern of KRAS, BRAF, and PIK3CA mutations, and correlate these mutations with clinicopathological features. To the best of our knowledge, this is the first large study to evaluate the presence of PIK3CA mutation and its correlation with other candidate gene mutations in Indian CRC patients.
Materials and methods
Samples
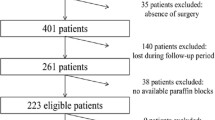
The current study was performed at the Research and Development Division of SRL Ltd., Mumbai, India. A total of 204 formalin-fixed paraffin-embedded (FFPE) CRC tissue samples were evaluated in the current study. These samples were resected at various cancer hospitals and sent to our center during October 2011 to May 2013. Tumor sections from FFPE tissue samples were hematoxylin–eosin (H&E)-stained and reviewed by two experienced histopathologist for histopathological examination and ensuring the presence of at least 50 % tumor cells at our center. From each of the 204 FFPE tumor blocks, 4–5 sections of 5 μm thickness (depending on the biopsy size) were obtained and processed for genomic DNA extraction using QIAamp® DNA Mini Kit (QIAGEN, Germany) as per the manufacturer’s instructions with slight modification. Treatment and outcome were not evaluated, and the study was approved by the institutional ethical committee. The tumors were classified following the Sixth edition of the AJCC/UICC TNM staging systems, and the details of the clinical characteristics of all patients are depicted in Table 1.
Mutation analysis for KRAS codons 12 and 13
Genomic DNA was amplified for the identification of KRAS mutations by nested PCR as reported earlier with slight modification [24]. The primer sequences for first-round PCR were as follows: KRAS Outer Forward 5′-AGGCCTGCTGAAAATGACTGAATA-3′; KRAS Outer Reverse 5′-CTGTATCAAAGAATGGTCCTGCAC-3′, and for nested: KRAS Inner Forward 5′-AAAATGACTGAATATAAACTTGTGG-3′; KRAS Inner Reverse 5′-CTCTATTGTTGGATCATATTCGTC-3′. The total volume of the reaction mixture for both first and nested PCR was 25 µL containing 50 ng of starting genomic DNA, 1.5 mmol/L MgCl2, 0.2 mM dNTPs, 10 pmol of forward and reverse primers, and 1.5 units of Taq polymerase (Sigma). The cycling conditions consisted of an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. In the nested PCR, 1 µL of the first-round PCR product was added and the same cycling conditions were applied, followed by the analysis of nested product on 2 % agarose gel.
BRAF mutation analysis for V600E mutation
Molecular evaluation of BRAF V600E mutation analysis was done by PCR and direct sequencing as per previous report [25]. The primer sequences were as follows: BRAF Forward 5′-GTGATTTTGGTCTAGCTACAGT-3′ and BRAF Reverse 5′-GGCCAAAATTTAATCAGTGG-3′. PCR was performed in 25 µL reaction volume containing 50 ng of genomic DNA, 1.5 mmol/L MgCl2, 0.2 Mm dNTPs, 10 pmol of forward and reverse primers, and 1.5 units of Taq polymerase (Sigma). The PCR conditions were initial heating step at 95 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. PCR products were verified on 2 % agarose gel for amplification.
PIK3CA mutation analysis for exons 9 and 20
Mutation analysis of PIK3CA exon 9 and exon 20 was performed using hemi-nested and nested PCR approach, respectively, using primers as per previous report [26–28]. The primer sequences were as follows: PIK3CA Exon 9 Outer Forward 5′-TTGCTTTTCTGTAAATCATCTGTG-3′, Inner Forward 5′- GGGAAAAATATGACAAAGAAAGC-3′ and Common Reverse 5′-GAATCTCCATTTTAGCACTTACCTGTGACT-3′, and for PIK3CA exon 20, the primers were as follows: Outer Forward 5′-TTTTCTCAATGATGCTTGGC-3′, Outer Reverse 5′-GGATTGTGCAATTCCTATGC-3′ and Inner Forward 5′-AATCTTTTGATGACATTGCATACATTCG-3′, Inner Reverse 5′-TCAGTTATCTTTTCAGTTCAATGCATG-3′. The total volume for both exon 9 and exon 20 PCR was 25 µL containing 50 ng of starting genomic DNA, 1.5 mmol/L MgCl2, 0.2 Mm dNTPs, 10 pmol of forward and reverse primers, and 1.5 units of Taq polymerase (Sigma). The PCR conditions were initial heating step at 95 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, annealing for exon 9 for both PCR rounds were 60 °C for 30 s and for exon 20, the first round was 55 °C, and the second was 64 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. Nested PCR products were verified on 2 % agarose gel for amplification.
Sequence analysis
The PCR products were sequenced bidirectionally by automated ABI Prism 3100 Avant Genetic Analyzer (Applied Biosystems, Foster City, Calif.) using ABI prism BigDye Terminator version 3.1 Cycle Sequencing Kit. The abnormal results were reconfirmed by at least two repeated analysis right from PCR amplification.
Statistical analysis
The data were analyzed by chi-square or Fisher’s exact tests to study the correlation between the clinicopathological features and the occurrence of a particular mutation. All the P values were two-tailed, and the statistical significance was set at P < 0.05.
Result
Clinical characteristics of CRC patients
In the current study, we analyzed 204 tumor tissues from Indian CRC patients. The frequency of CRC was found to be elevated in males (58.8 %) as compared to females (41.2 %) (Table 1). Also, the number of cases gradually increased with the increase in the age of the patients with a peak in the age group of 51–60 years of age (below 50 years: 32.8 %, 51–60 years: 29.4 %, above 60 years: 36.3 %). The median age of the patient cohort was 58 years, ranging from 6 to 85 years. Distribution across various histological subtypes was as follows: adenocarcinoma: 152 (74.5 %), mucinous adenocarcinoma: 34 (16.7 %), and signet ring cell adenocarcinoma: 18 (8.8 %). The tumors were graded following the WHO criteria and categorized according to the clinicopathological features described as rectum, left-sided (including sigmoid colon and splenic flexure), and right-sided (cecum, hepatic flexure, transverse colon). Sixty-nine samples were located in the left colon, eighty in the right colon, and forty-nine were in the rectum. For eight samples, the tumor location within the colon was unknown (Table 1).
KRAS mutation type and its correlation with clinicopathological findings
Among the 204 samples analyzed by direct sequencing, 48 cases (23.5 %) harbored KRAS mutations while the remaining 156 cases were normal for both the codons. Of these 48 cases, codon 12 mutations were more frequently observed in 30 (62.5 %) cases, while codon 13 mutations were detected in 18 (37.5 %) cases (Table 2). We identified five different amino acid substitutions at codon 12 (G12S, G12D, G12A, G12V, and G12C) and one substitution type at codon 13 (G13D) (Table 3) (Fig. 1a–f). As shown in Table 3, G12D was the most recurrent codon 12 mutation (5.9 %) followed by G12V (4.9 %), G12A (1.9 %), and 1.0 % each for G12S and G12C. All patients with different mutations were heterozygous and retained a wild-type allele. None of the cases harbored concomitant mutations at both the codons.
Correlation of the KRAS mutation with clinicopathological data indicated that KRAS mutations were predominantly observed in females than their male counterparts (29.8 vs. 19.2 %, P = 0.079) (Table 2). Interestingly, KRAS mutations were significantly higher in patients who were older than 50 years in comparison with patients <50 years of age (32.1 vs. 6 %, P < 0.0001), indicating that KRAS mutations are uncommon in younger patients. On correlation with histopathological data, KRAS mutation was found to be significantly associated with moderate/poorly differentiated tumors in comparison with well-differentiated tumors (27.0 %, 41/152 vs. 13.5 %, 7/52; P = 0.047). Likewise, KRAS mutations were more prevalent in adenocarcinomas when compared to some of a special histological subtype such as mucinous/signet ring cell type of histology (27.0 %, 41/152 vs. 13.5 %, 7/52; P = 0.047). No significant association with tumor site was noted, though a higher tendency of KRAS mutation toward the left side of the colon was observed. Similarly, no significant correlation of KRAS mutations with the TNM staging system was observed, though a trend toward more frequent KRAS mutation in N0 designated cases in comparison with others (N1–NX) (29.7 %, 22/74 vs. 20.0 %, 26/130; P = 0.053) was observed (Table 2).
BRAF mutation type and its correlation with clinicopathological findings
In our sample cohort, a total of 20 (9.8 %) BRAF codon 600 mutations were detected out of 204 tumor tissues. Sequencing analysis revealed that GTG>GAG (85 %, 17/20) was the most frequent alteration observed affecting codon 600 of the BRAF gene, followed by GTG>GAA (15 %, 3/20) (Table 3; Fig. 1g, h). Both of these nucleotide changes resulted in the V600E mutation, thereby suggesting that other codon 600 variants were not present in this cohort. None of the BRAF mutation-positive cases showed concomitant KRAS mutation, indicating that both of them were mutually exclusive. It is interesting to note that a trend toward increased frequency of BRAF mutation was observed in patients who were <50 years (14.9 %, 10/67) as compared to the older-aged patients (7.5 %, 10/134) (P = 0.096) (Table 2). This indicates that the mutation of BRAF occurs at a lower rate in older patients. Additionally, right-sided tumors possessed more BRAF mutations (12.5 %, 10/80) than the tumors on the left side (5.9 %, 4/68) or rectum (10.4 %, 5/48) (P = 0.39). However, these differences were not significant. Furthermore, no significant association of BRAF mutation was present with any clinicopathological features like gender, tumor differentiation, tumor type, and TNM staging.
PIK3CA mutation type and its correlation with clinicopathological findings
The presence of PIK3CA mutations was noted in 12 cases (5.9 %), while the remaining 192 cases demonstrated normal wild-type alleles. PIK3CA mutations were predominantly found in exon 9 (9/12,) as compared to exon 20 (3/12) (Table 3). All mutations in exon 9 were substitution mutation with E545K (GAG>AAG) as the most recurrent type observed in seven cases, followed by T544I (ACT>ATT) (n = 1) and Q546R (CAG>CGG) (n = 1) (Table 3; Fig. 2a–c). Similarly, all the three mutations of exon 20 were substitution type, one case each of H1047R (CAT>CGT), G1049S (GGT>AGT), and D1056N (GAT>AAT) (Table 3; Fig. 2d–f). In addition to point mutations in exon 20, silent mutations (H1047H) and polymorphisms (T1025T) were also found (Fig. 2g, h).
It is worth noting that PIK3CA mutation was present in 5 of 48 (10.4 %) patients with KRAS mutations, when compared with only 7 of 156 (4.5 %) patients with KRAS wild type (P = 0.12). Similarly, KRAS mutation was detected in 5 of 12 (41.7 %) patients with PIK3CA mutations in comparison with 43 of 192 (22.3 %) patient with PIK3CA wild type. Interestingly, PIK3CA exon 9 mutations were significantly associated with KRAS mutations (10.4 %, 5/48 vs. 2.6 %, 4/156; P = 0.02), thereby suggesting that PIK3CA and KRAS gene mutation represents partially overlapping subgroups in colon cancer. In contrast, no concurrent mutation of BRAF mutation was observed, thereby suggesting that PIK3CA and BRAF mutations were mutually exclusive. No significant association between PIK3CA mutations and patients’ characteristics such as age, gender, tumor location, and tumor differentiation was observed (Table 2).
Discussion
Colorectal cancer (CRC) is one of the most common cancer responsible for cancer-related mortality and morbidity worldwide [29]. The 5-year survival rate of patients with CRC remains up to 90 % if diagnosed at an early stage [30]. The survival rate declines as the disease spreads, i.e., 35–60 %, when the lymph nodes are involved, and less than 10 %, when metastatic [31]. Therefore, the survival rate of CRC patients depends on the clinical and pathological staging of the disease at diagnosis.
Recently, significant improvements have been made in the treatment for metastatic CRC patients with the aid of monoclonal antibody therapy that targets the EGFR. Despite the advancements in therapeutics, a considerable amount of patients do not benefit from the anti-EGFR therapy because of the frequent activation of EGFR signaling pathway by oncogenic mutations of KRAS, BRAF, and PIK3CA that activates both the RAS/RAF/MAPK signaling pathway and the PIK3/AKT/mTOR pathway [8, 9].
In the present study, we have evaluated the frequency of KRAS, BRAF, and PIK3CA somatic mutations in a cohort of 204 Indian CRC patients. Additionally, we also assessed the correlation of these gene mutations with clinicopathological features. The total frequency of the cases with at least one mutation present was 36.8 % and the remaining 63.2 % were wild type for all the genes analyzed.
KRAS
It is evident from the previous studies that there lies a strong correlation between the KRAS mutation and non-responsiveness to anti-EGFR therapies [32–34]. The frequency of KRAS mutations varies considerably across the globe ranging between 13 and 53 % (Table 4) [35–68]. The frequency of KRAS mutation (23.3 %) in the current study was in accordance with the previous published data from Japan, France, Portugal, Korea, Italy, Malaysia, Thailand, and Morocco (20–25 %), higher than those from China, USA, Pakistan, and Portugal (12–19 %) as well as lower than USA, United Kingdom, Germany, France, Belgium, China, Greece, Taiwan, Japan, Egypt, and Australia (31–53 %) (Table 4). In comparison with Indian studies, the frequency of KRAS mutation tallies with previous report from our group [20] as well as study from Patil and colleagues (2013) [21] (20–24 %), although few studies have stated a higher occurrence of 26–66 % [22, 23], which may be attributed to the small sample set, and or clinically selected cases. However, the variations in the frequency of KRAS mutation across the globe can be credited to the ethnicity, geographical distribution as well as use of sensitive techniques in some studies [37, 41, 46, 53].
Studies from western nations reported KRAS G12D as the most recurrent transitions followed by G12 V, G12C, G12S, and G12A [69, 70]. However, in our study, the corresponding order is G12D, G12 V, G12A, G12S, and G12C. Among KRAS codon 13 mutations, the majority of them were G13D, followed by G13C and G13R in western populations [69, 70]. However, in the current study, predominantly only G13D and rarely G13C mutations were observed, while none of the cases showed G13R. These data suggest that there may be some racial discrimination in the patterns of KRAS mutations. Furthermore, in agreement with the previous findings, none of the KRAS mutation cases harbored concurrent BRAF mutations, indicating that these mutations were mutually exclusive which is in line with the previous findings [13, 18].
Our findings suggested that KRAS mutations occurred more frequently in females when compared to their male counterpart, which is in agreement with the previous report [26] Similarly, patients above 50 years of age tend to show significantly higher KRAS mutations 32.1 % (43/134) than patients below 50 years of age, while tumors with KRAS mutations were significantly associated with moderate/poorly differentiated tumors as compared to well-differentiated tumors (moderate/poor 27.0 % vs. well 13.5 %, P = 0.04). Interestingly, tumors with a KRAS mutation tend to occur more in adenocarcinoma as compared to the special histological subtypes such as mucinous/signet ring cell type, which further supports previous findings [71]. Other clinicopathological findings such as tumor location and staging system did not show any association with the KRAS mutation, which is in agreement with a recent report of China [26].
BRAF
The BRAF V600E mutation is the most commonly observed genetic alterations after KRAS mutation in human cancers including CRC. Similar to KRAS, even BRAF gene encodes proteins that act in the EGFR signaling pathway. The distribution of BRAF mutations varies largely from 2 % to 25 % across the globe (Table 4). In the present study, we identified frequency of 9.8 % (V600E) mutation, which remains consistent when compared to European studies (7–9 %) [48, 56] and inconsistent with some of the Asian studies [59–61]. However, the variations in the mutation frequency point toward that geographical distribution, genetic makeup, and diverse ethnicity play and participate in deciding the gene mutation pattern. Several studies have reported that V600E mutation existed only in KRAS wild-type tumors, which is similar to our findings [13, 18]. In contrast, a recent Chinese study reported concurrent mutation of BRAF and KRAS mutation in as high as 24 % of their study group [26] Furthermore, V600E was the only mutation type that was observed in this large cohort of patients, which is in contrast to a recent small Chinese study wherein apart from V600E, other mutation type such as V600Q, V600L, and V600 V was reported [26]. The exact reason for this variation is still not known; however, it can be attributed to racial difference and environmental factors that may exist between these populations. In agreement with the previous findings, no significant association of BRAF mutation with clinicopathological data was observed [26, 71]. Furthermore, a holistic consensus on the predictive role of BRAF mutations in the anti-EGFR therapy of metastatic colon carcinoma is yet to be establish, since some of the study found that V600E mutation was associated with worse outcome in metastatic CRC patients [72], while others demonstrated that V600E was just a general poor prognostic marker rather than a predictive markers for anti-EGFR therapy, because its relationship with poor prognosis is independent from the given treatment [73].
PIK3CA
Several studies have reported the somatic mutations of phosphoinositide-3-kinase catalytic alpha; PIK3CA gene plays an important role in carcinogenesis and disease progression in many cancers including CRC [74]. Literature survey suggests a wide range of variation in PIK3CA mutation rates, ranging from 7 to 30 % [75, 76] of CRC cases presenting a mutated PIK3CA. In the present study, the frequency of PIK3CA mutation was found to be 5.9 %, which is consistent with Asian (4.7–4.9 %) [60, 68] and European studies (4–5.9 %) [39, 55], while it was lesser than some western studies (11–18 %) [17, 41, 43]. This discrepancy may be attributed to several factors like different diagnostic techniques (HRM, SSCP, direct sequencing, arms PCR, pyrosequencing, etc. [41, 44, 53], in addition to the difference in lifestyle pattern, ethnicity, genetic factors, geographical distribution.
Interestingly, we observed increased mutations in exon 9 (kinase domain) than exon 20 (helicase domain), which is consistent with western studies [51] but quite different from the results of Chinese studies, wherein exon 20 was more frequently mutated [26, 71]. It is important to note that exon 9 and exon 20 mutations may differ to a great extent in affecting the response to anti-EGFR therapy. In fact, a recent meta-analysis reported that mutations of PIK3CA exon 20 are associated with lower response rates and overall survival, thereby suggesting it to be a potential molecular marker for resistance to anti-EGFRs in KRAS wild-type metastatic CRC; however, no such role for exon 9 mutation has been seen yet [76]. One of the largest European study reported that among all PIK3CA mutations, E542K, E545K, and Q546K mutations at exon 9 accounted for 15.6, 26.8, and 4.2 %, respectively, while the H1047R and H1047L mutations at exon 20 accounted for 20.5 and 3.8 % of all the mutations [45]. However, in our cohort, E545K was the most recurrent mutation followed by T544I and Q546R, none of the cases showed E542K and Q546K in exon 9. Similarly, in exon 20, apart from H1047R, we also detected G1049S and D1056N.
Several studies in the past have reported the coexistence as well as a significant association of mutations of PIK3CA and KRAS in CRC tumors [17, 35, 45]. Notably, we found that PIK3CA mutation was present in 10.4 % patients with KRAS mutations, when compared with only 4.5 % patients with KRAS wild type. In fact, PIK3CA exon 9 mutations were found to be significantly associated with KRAS mutations, indicating that PIK3CA and KRAS gene mutation represents partially overlapping subgroups in colon carcinomas. In addition, we also found one case with KRAS G12A mutation and polymorphism (H1047H) of PIK3CA exon 20 in colorectal carcinomas. Furthermore, the PIK3CA mutation of exon 9 (codons 542, 545, 546) and exon 20 (codon 1047) has been previously reported to be oncogenic in nature by various cell line studies [6, 77–82]. No significant association between PIK3CA mutations and clinical characteristics was observed in the present study, which tallies with earlier studies [16, 26]; nevertheless, a recent study from Rosty et al. [58] reported a significant association with proximally located tumors and mucinous type of differentiation.
In conclusion, to our knowledge, this is the first comprehensive study to evaluate PIK3CA mutation in Indian CRC patients. The fact that these genetic markers play a crucial role in predicting drug response, their inclusion in the genetic evaluation of CRC patients may play a pivotal role in tailoring the therapy, and will certainly provide the necessary advance for true personalized medicine.
References
Jemal A, Bray F, Center MM, Ferlay J, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Mohandas K. Colorectal cancer in India: controversies, enigmas and primary prevention. Indian J Gastroenterol. 2011;30:3–6.
Ferlay J, Bray F, Pisani P, Parkin DM, editors. GLOBOCAN 2002: cancer incidence, mortality and prevalence worldwide. CancerBase No. 5, Version 2.0. Lyon: IARC Press; 2004.
Arnold D, Schmoll HJ. Colorectal cancer: (neo-) adjuvant treatments in colorectal cancer. Ann Oncol. 2005;16:133–40.
Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–62.
Samuels Y, Diaz LA Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–73.
Oikonomou E, Pintzas A. Cancer genetics of sporadic colorectal cancer: BRAF and PI3KCA mutations, their impact on signaling and novel targeted therapies. Anticancer Res. 2006;26:1077–84.
Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res. 2006;12:5268–72.
McCubrey JA, Steelman LS, Abrams SL, et al. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul. 2006;46:249–79.
Calistri D, Rengucci C, Seymour I, et al. Mutation analysis of p53, K-ras, and BRAF genes in colorectal cancer progression. J Cell Physiol. 2005;204:484–8.
Fransén K, Klintenäs M, Osterström A, et al. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis. 2004;25:527–33.
Wilson PM, Labonte MJ, Lenz HJ. Molecular markers in the treatment of metastatic colorectal cancer. Cancer J. 2010;16(3):262–72.
Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26(35):5705–12.
Bokemeyer C, Kohne C, Rougier P, et al. Cetuximab with chemotherapy (CT) as first-line treatment for metastatic colorectal cancer (mCRC): analysis of the CRYSTAL and OPUS studies according to KRAS and BRAF mutation status. J Clin Oncol. 2010;28:15s.
Samuels Y, Wang ZH, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554.
Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–7.
Janku F, Lee JJ, Tsimberidou AM, et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011;6:e22769. doi:10.1371/journal.pone.0022769.
Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–8.
Douillard J, Siena S, Cassidy J et al. Randomized phase 3 study of panitumumab with FOLFOX4 compared to FOLFOX4 alone as 1st-line treatment (tx) for metastatic colorectal cancer (mCRC): the PRIME trial. Eur J Cancer. 2009;7S(3):6. Abstract LBA10.
Bagadi SB, Sanghvi M, Nair SB, et al. Combined mutational analysis of KRAS, NRAS and BRAF genes in Indian patients with colorectal carcinoma. Int J Biol Markers. 2012;27:27–33.
Patil H, Korde R, Kapat A. KRAS gene mutations in correlation with clinicopathological features of colorectal carcinomas in Indian patient cohort. Med Oncol. 2013;30:617.
Sinha R, Hussain S, Mehrotra R, et al. Kras gene mutation and RASSF1A, FHIT and MGMT gene promoter hypermethylation: indicators of tumor staging and metastasis in adenocarcinomatous sporadic colorectal cancer in Indian population. PLoS One. 2013;. doi:10.1371/journal.pone.0060142.
Malhotra P, Anwar M, Nanda N, et al. Alterations in K-ras, APC and p53-multiple genetic pathway in colorectal cancer among Indians. Tumour Biol. 2013;34:1901–11.
Kalikaki A, Koutsopoulos A, Trypaki M, et al. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–9.
Sapio MR, Posca D, Troncone G, et al. Detection of BRAF mutation in thyroid papillary carcinomas by mutant allele-specific PCR amplification (MASA). Eur J Endocrinol. 2006;154:341–8.
Mao C, Zhou J, Yang Z, et al. KRAS, BRAF and PIK3CA mutations and the loss of PTEN expression in Chinese patients with colorectal cancer. PLoS One. 2012;7:e36653. doi:10.1371/journal.pone.0036653.
Barbi S, Cataldo I, De Manzoni G, et al. The analysis of PIK3CA mutations in gastric carcinoma and metanalysis of literature suggest that exon-selectivity is a signature of cancer type. J Exp Clin Cancer Res. 2010;29:32. doi:10.1186/1756-9966-29-32.
Bachman KE, Argani P, Samuels Y, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–5.
World Health Organisation Cancer Incidence in Five Continents. Lyon: The World Health Organisation and The International Agency for Research on Cancer; 2002.
Mandel JS, Bond JH, Church TR, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N Engl J Med. 1993;328:1365–71.
Wingo PA, Tong T, Bolden S. Cancer Statistics, 1995. CA Cancer J Clin. 1995;45:8–30.
Liévre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5.
Freeman DJ, Juan T, Reiner M, et al. Association of K-ras mutational status and clinical outcomes in patients with metastatic colorectal cancer receiving panitumumab alone. Clin Colorectal Cancer. 2008;7:184–90.
Perkins G, Lièvre A, Ramacci C, et al. Additional value of EGFR downstream signaling phosphoprotein expression to KRAS status for response to anti-EGFR antibodies in colorectal cancer. Int J Cancer. 2010;127:1321–31.
Velho S, Oliveira C, Ferreira A, et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41:1649–54.
Barault L, Veyrie N, Jooste V, et al. Mutations in the RASMAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122:2255–9.
Cappuzzo F, Varella-Garcia M, Finocchiaro G, et al. Primary resistance to cetuximab therapy in EGFR FISH-positive colorectal cancer patients. Br J Cancer. 2008;99:83–9.
Simi L, Pratesi N, Vignoli M, et al. High-resolution melting analysis for rapid detection of KRAS, BRAF, and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol. 2008;130:247–53.
Velho S, Moutinho C, Cirnes L, et al. BRAF, KRAS and PIK3CA mutations in colorectal serrated polyps and cancer: primary or secondary genetic events in colorectal carcinogenesis? BMC Cancer. 2008;8:255.
Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20:84–90.
Ogino S, Nosho K, Kirkner GJ, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–84.
Souglakos J, Philips J, Wang R, et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101:465–72.
Baba Y, Nosho K, Shima K, et al. Phosphorylated AKT expression is associated with PIK3CA mutation, low stage, and favorable outcome in 717 colorectal cancers. Cancer. 2011;117:1399–408.
Baldus SE, Schaefer KL, Engers R, et al. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16:790–9.
De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62.
Lurkin I, Stoehr R, Hurst CD, et al. Two multiplex assays that simultaneously identify 22 possible mutation sites in the KRAS, BRAF, NRAS and PIK3CA genes. PLoS One. 2010;5:e8802. doi:10.1371/journal.pone.0008802.
Di Nicolantonio F, Arena S, Tabernero J, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010;120:2858–66.
Saridaki Z, Tzardi M, Papadaki C, et al. Impact of KRAS, BRAF, PIK3CA mutations, PTEN, AREG, EREG expression and skin rash in ≥2 line cetuximab-based therapy of colorectal cancer patients. PLoS One. 2011;6:e15980. doi:10.1371/journal.pone.0015980.
Wong NS, Fernando NH, Nixon AB, et al. A phase II study of capecitabine, oxaliplatin, bevacizumab and cetuximab in the treatment of metastatic colorectal cancer. Anticancer Res. 2011;31:255–61.
Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–606.
Palomba G, Colombino M, Contu A, et al. Prevalence of KRAS, BRAF, and PIK3CA somatic mutations in patients with colorectal carcinoma may vary in the same population: clues from Sardinia. J Transl Med. 2012;10:178.
Bozzao C, Varvara D, Piglionica M, et al. Survey of KRAS, BRAF and PIK3CA mutational status in 209 consecutive Italian colorectal cancer patients. Int J Biol Markers. 2012;27:e366–74.
Guedes JG, Veiga I, Rocha P, et al. High resolution melting analysis of KRAS, BRAF and PIK3CA in KRAS exon 2 wild-type metastatic colorectal cancer. BMC Cancer. 2013;13:169.
Neumann J, Wehweck L, Maatz S, et al. Alterations in the EGFR pathway coincide in colorectal cancer and impact on prognosis. Virchows Arch. 2013;463:509–23.
Derbel O, Wang Q, Desseigne F, et al. Impact of KRAS, BRAF and PI3KCA mutations in rectal carcinomas treated with neoadjuvant radiochemotherapy and surgery. BMC Cancer. 2013;13:200.
Smith CG, Fisher D, Claes B, et al. Somatic profiling of the epidermal growth factor receptor pathway in tumors from patients with advanced colorectal cancer treated with chemotherapy ± cetuximab. Clin Cancer Res. 2013;19:4104–13.
Yanus GA, Belyaeva AV, Ivantsov AO, et al. Pattern of clinically relevant mutations in consecutive series of Russian colorectal cancer patients. Med Oncol. 2013;30:686.
Rosty C, Young JP, Walsh MD, et al. PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival. PLoS One. 2013;8:e65479. doi:10.1371/journal.pone.0065479.
Berg M, Danielsen SA, Ahlquist T, et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One. 2010;5:e13978. doi:10.1371/journal.pone.0013978.
Liao W, Liao Y, Zhou JX, et al. Gene mutations in epidermal growth factor receptor signaling network and their association with survival in Chinese patients with metastatic colorectal cancers. Anat Rec. 2010;293:1506–11.
Kwon MJ, Lee SE, Kang SY, et al. Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers: comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathol Res Pract. 2011;207:762–8.
Aoyagi H, Iida S, Uetake H, et al. Effect of classification based on combination of mutation and methylation in colorectal cancer prognosis. Oncol Rep. 2011;25:789–94.
Hsieh LL, Er TK, Chen CC, et al. Characteristics and prevalence of KRAS, BRAF, and PIK3CA mutations in colorectal cancer by high-resolution melting analysis in Taiwanese population. Clin Chim Acta. 2012;413:1605–11.
Ling Y, Ying JM, Qiu T, et al. Detection of KRAS, BRAF, PIK3CA and EGFR gene mutations in colorectal carcinoma. Zhonghua Bing Li Xue Za Zhi. 2012;41:590–4.
Bando H, Yoshino T, Shinozaki E, et al. Simultaneous identification of 36 mutations in KRAS codons 61 and 146, BRAF, NRAS, and PIK3CA in a single reaction by multiplex assay kit. BMC Cancer. 2013;13:405. doi:10.1186/1471-2407-13-405.
Yip WK, Choo CW, Leong VCS, et al. Molecular alterations of Ras-Raf-mitogen-activated protein kinase and phosphatidylinositol 3-kinase-Akt signaling pathways in colorectal cancers from a tertiary hospital at Kuala Lumpur, Malaysia. APMIS. 2013;121:954–66.
Nakanishi R, Harada J, Tuul M, et al. Prognostic relevance of KRAS and BRAF mutations in Japanese patients with colorectal cancer. Int J Clin Oncol. 2013;18:1042–8.
Soeda H, Shimodaira H, Watanabe M, et al. Clinical usefulness of KRAS, BRAF, and PIK3CA mutations as predictive markers of cetuximab efficacy in irinotecan- and oxaliplatin-refractory Japanese patients with metastatic colorectal cancer. Int J Clin Oncol. 2013;18:670–7.
Vaughn C, Zobell S, Furtado L, et al. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosom Cancer. 2011;50:307–12.
Neumann J, Zeindl-Eberhart E, Kirchner T, et al. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205:858–62.
Li HT, Lu YY, An YX, et al. KRAS, BRAF and PIK3CA mutations in human colorectal cancer: relationship with metastatic colorectal cancer. Oncol Rep. 2011;25:1691–7.
Di Fiore F, Sesboue R, Michel P, et al. Molecular determinants of anti-EGFR sensitivity and resistance in metastatic colorectal cancer. Br J Cancer. 2010;103:1765–72.
Tol J, Dijkstra JR, Klomp M, et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer. 2010;46:1997–2009.
Siena S, Sartore-Bianchi A, Di Nicolantonio F, et al. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308–24.
Naguib A, Cooke JC, Happerfield L, et al. Alterations in PTEN and PIK3CA in colorectal cancers in the EPIC Norfolk study: associations with clinicopathological and dietary factors. BMC Cancer. 2011;11:123.
Mao C, Yang ZY, Hu XF, et al. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2012;23:1518–25.
Ikenoue T, Kanai F, Hikiba Y, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–7.
Miled N, Yan Y, Hon WC, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–42.
Guo XN, Rajput A, Rose R, et al. Mutant PIK3CA-bearing colon cancer cells display increased metastasis in an orthotopic model. Cancer Res. 2007;67:5851–8.
Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci. 2007;104:5569–74.
Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci. 2008;105:2652–7.
Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci. 2005;102:802–7.
Greene FL, Page DL, Fleming ID, et al. AJCC cancer staging manual. 6th ed. New York: Springer; 2002.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bisht, S., Ahmad, F., Sawaimoon, S. et al. Molecular spectrum of KRAS, BRAF, and PIK3CA gene mutation: determination of frequency, distribution pattern in Indian colorectal carcinoma. Med Oncol 31, 124 (2014). https://doi.org/10.1007/s12032-014-0124-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-014-0124-3