Abstract
Phytopathogenic fungi would induce a variety of plant diseases, resulting in a severe reduction of agricultural output. However, the current plant disease control is mainly dependent on the environmentally and healthily hazardous chemical fungicides. Thus, the present work aimed to isolate an effective antagonistic microorganism against various soilborne phytopathogenic fungi. By dual culture with Rhizoctonia solani, a novel Streptomyces specie, Streptomyces sp. N2, was screened out from a total of 167 isolated actinomycetes, which displayed a strong inhibitory effect on R. solani (26.85 ± 1.35 mm of inhibition zone diameter). By means of macroporous resin and silica gel column chromatography coupled with preparative HPLC, an antifungal metabolite (3-methyl-3,5-amino-4-vinyl-2-pyrone, C6H7O2N) was isolated and purified from Streptomyces sp. N2. The bioassay results showed that the purified antifungal metabolite could not only possess a broad-spectrum inhibitory effect on a range of plant pathogenic fungi in vitro (e.g., R. solani, Pyricularia grisea, Fusarium oxysporum f. sp. niveum, F. oxysporum f. sp. vasinfectum, Penicillium italicum, and Colletotrichum gloeosporioides), but also had a significantly effective in vivo biocontrol efficacy on grape fruits anthracnose caused by C. gloeosporioides. Microscopic observation indicated that the antifungal metabolite from Streptomyces sp. N2 would exert its antimicrobial activity by disorganizing the cytoplasmic organelles of phytopathogenic fungi. The above results suggested that Streptomyces sp. N2 was one of promising fungicide for biocontrol of fungal plant diseases, especially due to its broad-spectrum and effective antagonist on various plant pathogens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rhizosphere is a playground and battlefield for the tripartite interactions between soilborne pathogens, beneficial microorganisms, and the plant [1]. Once the soilborne pathogens, especially the fungal pathogens, can effectively escape the rhizosphere battle zone, they would reach and further infect the host tissue, resulting in a series of plant diseases [2, 3]. It is estimated that the plant diseases induced by soilborne pathogens would cause 10 ∼ 16 % reduction of the global agricultural productivity [4].
Facing such a severe threat on global food security caused by plant diseases, it is extremely important to search and develop a highly effective fungicide against the soilborne plant pathogens. At present, chemical fungicides have been extensively used worldwide in current agriculture. However, the excessive use of chemical fungicides has led to many attendant problems, such as environmental pollution, deteriorating human health, development of pathogen resistance to fungicides, and phytotoxicity [5]. Due to being more environment-friendly, biological control is recognized as an alternative and promising approach to control the soilborne plant pathogens. To date, many antagonistic microbes have been developed as the biocontrol agents, like Bacillus spp., Streptomyces spp., Pseudomonas spp., Trichoderma spp., and nonpathogenic Fusarium spp. [6, 7]. For these antagonistic microbes, the mechanisms of pathogens suppression include production of antibiotics, cell wall-degrading enzymes, and parasitism (direct) and induction of host resistance (indirect) [8].
China is a large agricultural country with a great diversity of crops. Like other countries in the world, the outbreak of plant diseases is becoming an increasingly serious problem that affects the agricultural output. However, the control of plant diseases in China is mainly dependent on the chemical fungicides. Thus, the present work aimed to isolate an effective antagonistic microorganism, and a novel actinomycete strain, Streptomyces sp. N2, was screened out from the soil of Xishan National Forest Park of Kunming in Yunnan Province, which exerted a broad and efficient resistance to multiple plant pathogenic fungi.
Materials and Methods
Isolation and Screening of the Antifungal Streptomyces spp.
Soil samples were collected from China’s Anhui Province, Guangdong Province, Hubei Province, Jiangxi Province, and Yunnan Province. The suspensions of the 18 soil samples were prepared by mixing 10 g soil with 90 ml distilled water, and 0.1 ml aliquots of serially diluted samples were plated on Gause’s No.1 medium (soluble starch 20 g, KNO3 1 g, K2HPO4 0.5 g, MgSO4 0.5 g, NaCl 0.5 g, FeSO4·7H2O 0.01 g, agar 20 g, distilled water 1000 ml, pH 7.2 ∼ 7.4), respectively. After 7 days of incubation at 30 °C, the Streptomyces colonies on the agar plates were picked on the basis of their morphological characteristics and further purified on Gause’s No.1 agar.
All the obtained isolates of Streptomyces spp. were tested and screened in vitro antagonistic activity against Rhizoctonia solani by using a dual culture assay, as described by Boukaew et al. [9]. Circular piece of agar (5 mm diameter) was excised from the 7-day-old Streptomyces spp. colony, which was placed on the center of potato dextrose agar (PDA) medium (leaching solution of potato 200 g, sucrose 20 g, agar 20 g, distilled water 1000 ml). Four mycelial plugs of 2-day-old R. solani were placed on the PDA plate, 5 cm away from the Streptomyces spp. After incubation at 28 °C for 2 days, the strongest antifungal Streptomyces sp. was screened out according to its inhibition zones of dual culture plates.
Morphological Characteristics and Identification of Isolate N2
The morphological characteristics of isolate N2 was observed by using a scanning electron microscope (JSM, 6360LV, JEOL, Tokyo, Japan): isolate N2 was grown on Gause’s No.1 agar at 28 °C for 7 days. The prepared samples were mounted on stubs, air-dried in a desiccator and splutter-coated with gold, and then and viewed on the scanning electron microscope at an accelerating voltage of 20 kV.
The identification of isolate N2 was according to its 16S rDNA gene sequencing and phylogenetic analysis: total genomic DNA was extracted using the Ezup DNA extraction kit (Sangon Biotech, Shanghai, China). The 16S rDNA was amplified using the primers, 27F (AGTTTGATCMTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT). The PCR products were gel purified with Gel purification Kit (Sangon Biotech, Shanghai, China). The obtained 16S rDNA sequence was analyzed by the NCBI database using BLAST algorithms at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST). A phylogenetic tree was constructed using the Phylogeny.fr software available at: http://www.phylogeny.fr/.
Isolation and Purification of the Antifungal Metabolite Produced by Streptomyces sp. N2
The fermentation of Streptomyces sp. N2 was carried out in a 250-ml Erlenmeyer flask containing 40 ml of fermentation medium (sucrose, 40 g; corn starch, 20 g; corn steep liquor, 20 g; (NH)2SO4, 2 g; KH2PO4, 1 g; MgSO4, 1 g; MnSO4·H2O, 0.01 g; ZnSO4·7H2O, 0.01 g; distilled water, 1000 ml; pH 7.2 ∼ 7.4), which was inoculated with a 2-cm2 cell plug from the fresh slant, and cultivated at 28 °C on a rotary shaker at 200 rpm for 6 days.
The fermentation broth was centrifuged at 5000 rpm for 20 min, and then the mycelium was extracted with n-butyl alcohol for 24 h at room temperature. Then the extract was filtered, and the filtrate was evaporated with a rotary evaporator at 45 °C. The obtained aqueous concentration was successively subjected to column chromatography on SIPI-21 macroporous resin and silica gel, using the gradient mixtures of methanol-water (0:100, 20:80, 40:60, 80:20, and 100:0, v/v) and chloroform-methanol (100:0, 80:20, 70:30, 60:40, and 50:50, v/v) as elutions, respectively. By using thin layer chromatography (TLC) together with the oxford plate assay system, the fractions that had the same retention factor (Rf) value and antifungal activity were pooled and evaporated to dryness in vacuo. Final purification of the antifungal factions was performed via a preparative HPLC system (Shimadzu, LC-6AD): Shim-pack Prep-ODS column (20 mm × 250 mm, 10 μm); flow rate, 5 ml/min; mobile phase, 65:35 (v/v) of methanol-water; detection wavelength, 254 nm.
In Vitro Antagonism of the Antifungal Metabolite Against Plant Fungal Pathogens
The 5-mm-diameter inoculum plugs of R. solani, Pyricularia grisea, Fusarium oxysporum f. sp. niveum, F. oxysporum f. sp. vasinfectum, Penicillium italicum, and Colletotrichum gloeosporioides were cut from the margin of 7-day-old colonies on PDA plates, respectively. Each plug was inoculated on the central of PDA pour plate mixed with 5 μg/ml antifungal metabolite isolated and purified from Streptomyces sp. N2 broth, and then incubated at 28 °C. The growth inhibitory effects on plant fungal pathogens were represented as inhibition percentage, which was calculated according to the following equation: Inhibition (%) = [(Growth diameter in untreated control − Growth diameter in treatment) × 100] / Growth diameter in untreated control [10].
In Vivo Biocontrol Capability of the Antifungal Metabolite
Grape fruits were used to test the in vivo biocontrol efficacy of the antifungal metabolite produced by Streptomyces sp. N2. Firstly, the grapes were surface disinfected in 70 % ethanol for 2 min, and followed by three rinses in sterile water. Next, each grape was cut into an approximately 8 mm length and 5 mm depth of triangular wound. The wound was artificially inoculated with a 5-mm-diameter inoculum plug of C. gloeosporioides, and then treated with 100 μl of the antifungal metabolite (10 μg/ml). After 5 days cultivation at 28 °C, the degree and size of the canker lesions were observed and recorded.
Effect of the Antifungal Metabolite on R. solani Mycelial Morphology
R. solani was inoculated on the PDA plate containing 5 μg/ml of the antifungal metabolite. After 5 days cultivation at 28 °C, a small tuft of R. solani mycelia was picked up and washed with 75 % (v/v) ethanol. The air-dry mycelium was put on a clean microscope slide and stained with a drop of Lacto-phenol cotton blue (phenol 10 g, cotton blue 0.5 g, glycerol 20 ml, lactic acid 10 ml, deionized water 10 ml). Then the stained mycelia was visualized by an optical microscope (Olympus CX31, Japan) equipped with camera.
Results
Isolation and Screening of Antifungal Actinomycetes
A total of 167 actinomycetes were isolated from the collected soil samples. According to the further screening with a dual culture assay, it was found that 12 isolates (serial number: N2, D8, E7, O3, I1, K3, JI, G8, P1, A10, F7, G1) exhibited the antagonistic activity against R. solani, and Fig. 1 showed the inhibition zone diameters of R. solani under the action of the above isolates.
From Fig. 1, it was found that strain N2 (isolated form Xishan National Forest Park of Kunming in Yunnan Province, China) was the most effective antagonist against R. solani among the 12 isolates, which inhibition zone diameter reached 26.85 ± 1.35 mm. Therefore, isolate N2 was chosen as a potential biocontrol actinomycete for further study.
Characterization and Identification of Isolate N2
The morphological characteristics of isolate N2 was visualized by using scanning electron microscope. From Fig. 2, it was observed that isolate N2 formed an extensively branched substrate mycelium, aerial hyphae which carried elliptic, smooth-surfaced spores in rectiflexibiles spore chains. A phylogenetic tree of isolate N2 was constructed using 16S rDNA gene sequence (1450 bp), as shown in Fig. 3. With an alignment covering at least 90 % of the gene length at greater than 90 % nucleotide identity using BLAST against nr/nt database at the NCBI website, twenty-two 16S rDNA sequences were found from the strains of genus Streptomyces, such as Streptomyces prasinopilosus, Streptomyces graminearus, Streptomyces phaeogriseichrom, Streptomyces griseofuscus, and Streptomyces costaricanus. However, it was difficult to differentiate between N2 isolate and the species of genus Streptomyces, which indicated that the isolate N2 was a novel Streptomyces specie. Therefore, isolate N2 was named as Streptomyces sp. N2.
Production of the Antifungal Metabolite from Streptomyces sp. N2
After the sequential column chromatography on macroporous resin and silica gel, the collected antifungal fraction was further purified using preparative HPLC, and Fig. 4 showed its HPLC spectrum. It was observed that the collected antifungal fraction had three sub-fractions (named metabolite I, II, and III, respectively), which retention time was 29.9, 31.1, and 38.4 min, respectively. Among the three sub-fractions, metabolite I was the main component, occupying 54.47 % of the total peak area ratio. Bioactivity determination showed that metabolite I exhibited the strongest antifungal activity. By further spectroscopic analyses, it was found that the antifungal metabolite I was 3-methyl-3,5-amino-4-vinyl-2-pyrone (molecular formula, C6H7O2N), which was validated as a novel structural biosubstance.
In Vitro and In Vivo Biocontrol Efficacy of the Antifungal Metabolite I
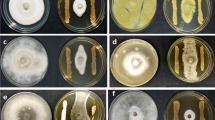
The antifungal metabolite I was prepared to assess the biocontrol efficacy of Streptomyces sp. N2. Firstly, the in vitro antifungal activity of metabolite I against phytopathogenic fungi was determined. It was observed visually that the antifungal metabolite I from Streptomyces sp. N2 displayed a broad-spectrum inhibitory effect on a range of plant pathogenic fungi, such as R. solani, Pyricularia grisea, F. oxysporum f. sp. niveum, F. oxysporum f. sp. vasinfectum, Penicillium italicum, and C. gloeosporioides, as shown in Fig. 5. By further calculating the growth diameters under metabolite I treatment and the untreated control, the inhibition percentage of the above phytopathogenic fungi was 78.07, 71.59, 81.48, 59.12, 88.60, and 48.65 %, respectively, as shown in Table 1.
Moreover, the in vivo biocontrol efficacy of the antifungal metabolite I was tested in controlling decays of grape fruits caused by C. gloeosporioides. When the grapes were treated with the antifungal metabolite I from Streptomyces sp. N2, the degree and size of the canker lesions were significantly smaller than those untreated, as shown in Fig. 6. Compared to the untreated grapes with a 2.56 ± 0.07 cm2 of lesion area, the treated grapes had almost no symptoms of lesions, suggesting that the antifungal metabolite I had an effective biocontrol efficacy in grapes.
Interaction Mechanism of the Antifungal Metabolite I on Phytopathogenic Fungi
The biocontrol mechanism of the antifungal metabolite I on phytopathogenic fungi was investigated by using R. solani. Microscopic observations showed that the antifungal metabolite I from Streptomyces sp. N2 would cause significant morphological changes and structural alterations in R. solani mycelium, as shown in Fig. 7. The untreated mycelium had regular hyphae with a smooth surface and integrated cell organelles. However, after treated with the antifungal metabolite I, R. solani hypha became clearly bending, short swollen, and irregularly branching. Furthermore, the treated R. solani showed a significant cellular disorganization, in which the damaged and disintegrated cytoplasmic organelles were visually observed. According to the morphological changes and structural alterations in R. solani mycelium, it could be inferred that the interaction mechanism of the antifungal metabolite I on phytopathogenic fungi was via changing the structure of cell membranes and further disorganizing the cytoplasmic organelles.
Discussion
During the whole growth processes, plants are under continuous attack by a vast number of soilborne pathogens, especially the fungal pathogens. For example, the outbreak of the destructive rice blast disease and rice sheath blight disease is a serious and recurrent problem in the rice-growing regions of the world, which were caused by Magnaporthe grisea (anamorph. Pyricularia grisea) and R. solani, respectively [10, 11]. Fusarium wilt is known to be one of the most severe and prevalent soilborne diseases that result in serious economic loss [12], such as the fusarium wilt of watermelon caused by F. oxysporum f. sp. niveum [13] and the fusarium wilt of cotton caused by F. oxysporum f. sp. vasinfectum [14]. The predominant and destructive plant disease anthracnose caused by fungi in the genus Colletotrichum would affect the yield and quality of many plants, including vegetables, fruits, and trees [15]. Additionally, frequent occurrence of postharvest diseases would result in a significant loss to fruit, such as the green and blue mold decay of caused by Penicillium digitatum and Penicillium italicum in citrus fruit [16].
However, plant pathogens are difficult to control because their populations are variable in time, space, genotype, and even self-evolution [17]. The current plant disease control is mainly dependent on the environmentally and healthily hazardous chemical fungicides. Therefore, it is urgent to search and develop a highly effective biofungicide. It is well known that actinomycetes have the ability to produce a variety of bioactive secondary metabolites. According to statistics, approximately 60 % of agriculturally useful antibiotics are produced by different Streptomyces species [18]. For example, many antifungal metabolite compounds have been successfully explored for commercial use in the fungal diseases control for rice, vegetables, and fruits, such as Blasticidin S of Streptomyces griseochromogenes, Kasugamycin of Streptomyces kasugaensis and Streptomyces kasugaspinus, Polyoxins of Streptomyces cacaoi var. asoensis, and Validamycin A of Streptomyces hygroscopicus var. limoneus [19].
In the present work, a novel actinomycete, Streptomyces sp. N2, was successfully isolated and screened out from Xishan National Forest Park of Kunming in Yunnan Province, China. One antimicrobial metabolite (3-methyl-3,5-amino-4-vinyl-2-pyrone, C6H7O2N) was separated and purified from Streptomyces sp. N2, which exerted its antifungal activity by disorganizing the cytoplasmic organelles of phytopathogenic fungi. Interestingly, this antifungal component exhibited a broad-spectrum and effective antagonistic activity on a range of phytopathogenic fungi, suggesting that Streptomyces sp. N2 had a potential ability to be developed as an efficient biocontrol agent against various fungal plant diseases. However, it should be pointed out that further studies were needed to identify the chemical structure of the purified antifungal metabolite and assess its biocontrol efficacy in planta.
References
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., & Moënne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant and Soil, 321, 341–361.
Berendsen, R. L., Pieterse, C. M., & Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends in Plant Science, 17, 478–486.
Doornbos, R. F., van Loon, L. C., & Bakker, P. A. H. M. (2012). Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agronomy for Sustainable Development, 32, 227–243.
Chakraborty, S., & Newton, A. C. (2011). Climate change, plant diseases and food security: an overreview. Plant Pathology, 60, 2–14.
Kim, H. J., Lee, E. J., Park, S. H., Lee, H. S., & Chung, N. (2014). Biological control of anthracnose (Colletotrichum gloeosporioides) in pepper and cherry tomato by Streptomyces sp. A1022. The Journal of Agricultural Science, 6, 54–62.
Weller, D. M., Raaijmakers, J. M., Gardener, B. B. M., & Thomashow, L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens 1. Annual Review of Phytopathology, 40, 309–348.
Spadaro, D., & Gullino, M. L. (2005). Improving the efficacy of biocontrol agents against soilborne pathogens. Crop Protection, 24, 601–613.
Palaniyandi, S. A., Yang, S. H., Zhang, L., & Suh, J. W. (2013). Effects of actinobacteria on plant disease suppression and growth promotion. Applied Microbiology and Biotechnology, 97, 9621–9636.
Boukaew, S., Chuenchit, S., & Petcharat, V. (2011). Evaluation of Streptomyces spp. for biological control of Sclerotium root and stem rot and Ralstonia wilt of chili pepper. Biocontrol, 56, 365–374.
Xiong, Z. Q., Tu, X. R., Wei, S. J., Huang, L., Li, X. H., Lu, H., & Tu, G. Q. (2013). In vitro antifungal activity of antifungalmycin 702, a new polyene macrolide antibiotic, against the rice blast fungus Magnaporthe grisea. Biotechnology Letters, 35, 1475–1479.
Harikrishnan, H., Shanmugaiah, V., Balasubramanian, N., Sharma, M. P., & Kotchoni, S. O. (2014). Antagonistic potential of native strain Streptomyces aurantiogriseus VSMGT1014 against sheath blight of rice disease. World Journal of Microbiology and Biotechnology, 30, 3149–3161.
Summerell, B. A., Laurence, M. H., Liew, E. C., & Leslie, J. F. (2010). Biogeography and phylogeography of Fusarium: a review. Fungal Diversity, 44, 3–13.
Zhao, S., Liu, D. Y., Ling, N., Chen, F. D., Fang, W. M., & Shen, Q. R. (2014). Bio-organic fertilizer application significantly reduces the Fusarium oxysporum population and alters the composition of fungi communities of watermelon Fusarium wilt rhizosphere soil. Biology Fertility of Soils, 50, 765–774.
Li, C. H., Zhao, M. W., Tang, C. M., & Li, S. P. (2010). Population dynamics and identification of endophytic bacteria antagonistic toward plant-pathogenic fungi in cotton root. Microbial Ecology, 59, 344–356.
Araújo, L., Gonçalves, A. E., & Stadnik, M. J. (2014). Ulvan effect on conidial germination and appressoria formation of Colletotrichum gloeosporioides. Phytoparasitica, 42, 631–640.
Liu, P., Cheng, Y. J., Yang, M., Liu, Y. J., Chen, K., Long, C. A., & Deng, X. X. (2014). Mechanisms of action for 2-phenylethanol isolated from Kloeckera apiculata in control of Penicillium molds of citrus fruits. BMC Microbiology, 14, 242.
Strange, R. N., & Scott, P. R. (2005). Plant disease: a threat to global food security. Annual Review of Phytopathology, 43, 83–116.
Couillerot, O., Loqman, S., Toribio, A., Hubert, J., Gandner, L., Nuzillard, J. M., Ouhdoucha, Y., Clémentc, C., Barkac, E. A., & Renault, J. H. (2014). Purification of antibiotics from the biocontrol agent Streptomyces anulatus S37 by centrifugal partition chromatography. Journal of Chromatography B, 944, 30–34.
Kim, B. S., & Hwang, B. K. (2007). Microbial fungicides in the control of plant diseases. Journal of Phytopathology, 155, 641–653.
Acknowledgments
This work was financially supported by the Training Program for Young Scientists of Jiangxi Provincial Department of Science and Technology (20142BCB23025), International Scientific and Technological Cooperation Projects of Jiangxi Provincial Department of Science and Technology (20141BDH80033), and Jiangxi Undergraduate Training Programs for Innovation and Entrepreneurship (201410410008).
Compliance with Ethical Standards
ᅟ
Conflict of Interest
The authors declare that they have no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, B., Chen, W., Wu, Zm. et al. A Novel and Effective Streptomyces sp. N2 Against Various Phytopathogenic Fungi. Appl Biochem Biotechnol 177, 1338–1347 (2015). https://doi.org/10.1007/s12010-015-1818-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1818-5