Abstract
The conversion of biomass into ethanol using fast, cheap, and efficient methodologies to disintegrate and hydrolyse the lignocellulosic biomass is the major challenge of the production of the second-generation ethanol. This revision describes the most relevant advances on the conversion process of lignocellulose materials into ethanol, development of new xylose-fermenting strains of Saccharomyces cerevisiae using classical and modern genetic tools and strategies, elucidation of the expression of some complex industrial phenotypes, tolerance mechanisms of S. cerevisiae to lignocellulosic inhibitors, monitoring and strategies to improve fermentation processes. In the last decade, numerous engineered pentose-fermenting yeasts have been developed using molecular biology tools. The increase in the tolerance of S. cerevisiae to inhibitors is still an important issue to be exploited. As the industrial systems of ethanol production operate under non-sterile conditions, microbial subpopulations are generated, depending on the operational conditions and the levels of contaminants. Among the most critical requirements for production of the second-generation ethanol is the reduction in the levels of toxic by-products of the lignocellulosic hydrolysates and the production of low-cost and efficient cellulosic enzymes. A number of procedures have been established for the conversion of lignocellulosic materials into ethanol, but none of them are completely satisfactory when process time, costs, and efficiency are considered.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Non-food or second-generation ethanol has great potential as a source of bioenergy due to the abundance of plant biomass on earth. Success in utilising this plant biomass will have a positive impact on the environment with political benefits for the society as a whole. It is well known that the main components of plant biomass are hemicellulose, cellulose and lignin [1].
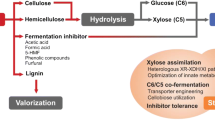
Technological trends, challenges and future perspectives related to production of the second-generation bioethanol have been exhaustively evaluated [2]. Recent trends favour the integration of three main key steps in the conversion of lignocellulose into ethanol [3, 4], and these steps comprise pre-treatment, hydrolysis and fermentation as illustrated in Fig. 1. The pre-treatments usually involve chemicals (acids, bases combined to oxidising agents and organic solvents) and physical methods, such as temperature and pressure to separate the lignin or the fractions from the lignocellulose containing both the lignin and hemicellulose [3, 5, 6]. The hydrolytic step can be chemically or enzymatically catalysed. The fermentation step is usually carried out using batch processes, but there are other processes, such as the integrated processes, which includes simultaneous saccharification and to fermentation [5].
Simplified diagram of the ethanol production process from lignocellulose indicating the steps where chemicals or industrial celluloses can be added to the process. The pre-treatment process produces a hemicellulose (soluble fraction) and water-insoluble fraction composed of cellulose and lignin, while these two fractions require further hydrolyses. In the integrated prepossesses, combinations between fermentation and hydrolysis strategies take place as follows [5]: simultaneous saccharification and fermentation processes (SSF); separate hydrolysis and fermentation processes (SHF); simultaneous saccharification and co-fermentation (SSCF) and consolidated biological processes or CBP processes
The main goal of the pre-treatments is to make cellulose accessible to hydrolysis [7], and critical improvements to reduce the effective-cost of the cellulosic ethanol are required [8, 9]. Although a variety of chemical, physical and biological approaches have been applied to the pre-treatment of the lignocellulose, only those involving chemicals offering higher yields and better release of the hemicellulose and the cellulose from the lignocellulose at a low cost are to be implemented [7]. Treatments with sodium hydroxide efficiently remove the lignin from the lignocellulosic material but with an increased degradation of the cellulose before hydrolysis step [9]. The following would be expected from a properly improved pre-treatment process [7]: (1) preservation of the hemicellulose fraction; (2) increases in the sugar formation during the hydrolysis step; (3) reduced crystallinity and increases in the porosity of both the cellulose and the lignocellulose and (4) decreases in the levels of inhibitors.
The hydrolytic step of the lignocellulose shows some limitations. For instance, the acid hydrolysis has disadvantages such as the generation of higher amounts of inhibitors and equipment corrosion [10]. Sulphuric acid at moderated concentrations (20% by weight for 2 h heating time) was able to give the highest yield of dextrose for the production of ethanol [11]. The direct use of the concentrated acid hydrolysis is carried out at low temperatures for longer periods, while diluted acid hydrolysis requires higher temperatures for shorter reaction periods [7, 10, 11]. The direct acid hydrolysis of sugarcane bagasse in a two-step process seems to be a promising strategy [12, 13].
In order to reduce the effective cost of bioethanol from lignocellulose, when enzymes are used, the following has been suggested [1, 14]: (1) the use of a less aggressive pre-retreatment processes; (2) stable and low-cost enzymes showing lower capacity of being irreversibly adsorbed by lignin; (3) use of enzymes which are less inhibited by both glucose or short-chain sugars formed during hydrolyses and (4) preference for an enzymatic hydrolysis able to be carried out at higher rates. Additionally, improvements in all areas of enzyme production have been made by looking for low-cost medium, selecting strains of fungi or bacteria and developing enzyme recycling strategies [15].
Saccharomyces cerevisiae is widely employed for the commercial production of bioethanol from sugars, such as the sucrose from sugar cane and the glucose or maltose from starch. However, there are crucial limitations related to the fermentation of lignocellulosic materials by S. cerevisiae. Natural strains of this yeast are unable to efficiently utilise xylose, which is the second major sugar [33 kg xylose and 60 kg glucose/ton of sugarcane] of the lignocellulosic hydrolysates [16]. On the other hand, glucose can be easily converted into ethanol by S. cerevisiae. A number of publications dedicated to the genetic improvement of S. cerevisiae strains are described in the present revision. Successful research involving genetic improvements has demonstrated that S. cerevisiae is able to co-ferment pentose and glucose as also discussed in this revision. On the other hand, non-Saccharomyces yeasts such as Pachysolen tannophilus, which is able to convert most of the xylose from hydrolysates into ethanol, giving yields as high as 0.32 g g−1 to 0.47 g g−1 [17]. The present revision also describes the following: (1) the tolerance and natural defense of xylose-fermenting yeasts against inhibitors; (2) monitoring of long-term ethanol production and formation of adapted yeast strain, which are dependent on operational conditions and contaminants (bacteria, wild yeast, variants induced by process conditions) and (3) the optimisation of strategies to improve the global process of ethanol production.
Ethanol Production by Xylose-Fermenting Microorganisms
S. cerevisiae has been habitually used to produce the ethanol from sugars and the major fermentable sugar from cellulose and hemicellulose are glucose and xylose. However, natural strains of xylose-fermenting yeasts (P. stipitis, P. tannophilus, Candida shehatae and Kluyveromyces marxianus), and strains of bacteria (Clostridium sp, Bacillus macerans, Clostridium saccharolyticum and Thermoanaerobacter ethanolicus) are candidates for use as biocatalysts to convert xylose or cellulose into ethanol [18]. C. shehatae, P. tannophilus and P. stipitis are the most exhaustively investigated organisms and most efficient xylose fermenters [18]. Species of fungi and bacteria are able to utilise cellulose within a temperature range of 28 °C to 74 °C, depending on the selected organism [19].
The disadvantages of using bacteria in large-scale fermentation are the low ethanol yields, by-product formation, intolerance to high ethanol concentrations and growth at narrow and neutral pHs varying from 6.0 to 8.0 [20]. The most promising bacteria to be improved for industrial applications are Escherichia coli, Klebsiella oxytoca and Zymomonas mobilis. These three bacteria have been engineered for ethanol production from xylose [21]. However, a genetically modified strain of S. cerevisiae was able to consume higher amounts of xylose from corn stove hydrolysates than the engineered strains of E. coli and Z. mobilis [22].
Some fungal strains such as Monilia sp., Neocallimastix sp., Trichoderma reesei and Fusarium oxysporum show the ability to directly convert cellulose/hemicellulose into ethanol/acetic acid in single-step fermentations [23]. The major disadvantage related to bioconversion of sugars from hydrolysates to ethanol by filamentous fungi is the slower fermentation rates than those obtained with yeasts and the secretion of a variety of by-products, including acids. A high amount of acetic acid (12.8 g L−1, acetate form) was produced from glucose by a strain of F. oxysporum under oxygen-limited conditions [24]. This fungi has been used for simultaneous saccharification and fermentation of the cellulose (SSF process), but the rate of ethanol production is low and a significant amount of acetic acid was formed.
Yeast S. cerevisiae has proved to be more robust than bacteria and other yeasts with respect to both the tolerance to ethanol and inhibitors present in hydrolysates and higher efficiencies of sugar conversion into ethanol [25]. However, this does not exclude the possibility of using bacteria for biofuel production (e.g. biobutanol). In addition, yeasts are more resistant to viral infections and high ethanol concentrations than bacteria. Viral infections can cause serious problems to bacterial fermentations in large biotechnological companies [26, 27].
Entry of Pentose into Cells of the S. cerevisiae
Xylose and arabinose are the two most abundant sugars in the lignocellulose hydrolysates obtained from agricultural and agro-industrial materials [4]. In S. cerevisiae, most of the glucose is transported to the yeast cells by a low-affinity transporter, while the high-affinity transporters are expressed under low glucose concentration [28]. Kinetic studies revealed that xylose can be transported by both high- and low-affinity glucose transporters. Affinity of the transporters for both glucose and xylose is very low under high glucose concentration, but increased affinities were observed under low glucose concentration [28]. Even though not metabolised by S. cerevisiae, the xylose and arabinose are taken up by the cells via the hexose transporters (Hxt), while xylose is usually consumed only after the glucose depletion from the medium [29]. The genes encoding for the expression of the different transporters that allow the accumulation of intracellular xylose were enumerated based on their increasing efficiencies as follows [29]: HXT7 > HXT5 > GAL2 > WT > HXT1 > HXT4 >>> RE700A. Heterologous transporters from different origins (Candida intermedia, P. stipitis, Arabidopsis thaliana) have been inserted into cells of S. cerevisiae, resulting in recombinant yeasts that exhibit an improved pentose transport at a lower exogenous glucose concentration as seen in native yeast strains [30].
Knowledge of the molecular aspects of hexose and pentose transporters in yeasts has increased considerably [31]. Nevertheless, a number of questions related to the pentose transport remain to be explored. It has been expected that, in near-term, physiological studies will be able to explored the apparent redundancy of sugar transporters identified within the gene transport families (genes showing similar properties) of S. cerevisiae. The identification of functional regions in FMS transporters (facilitator super family of proteins) is another open issue for further studies. Several lineages of sugar transporters consist of entirely uncharacterised proteins, whose biological functions remain to be elucidated [31].
Metabolic Pathways for Pentose Utilisation
As illustrated in Fig. 2, there are two different pathways available in nature for the conversion of pentose into ethanol [32, 33]: (1) oxidoreductase-based pathways (type I pathways) found in most fungi and (2) isomerase-based pathways (the type II pathway) found in most bacteria. The genes encoding for important enzymes of the pentose pyrophosphate pathway are XR (xylose reductase), XDH (NAD+-dependent xylitol dehydrogenase) and XK (xylulose kinase). However, these genes are expressed at such a low level that the xylose utilisation by S. cerevisiae is not allowed, while XK is the rate-limiting step of the pentose phosphate pathway [34].
Outline of the xylose metabolism pathway in fungi and bacteria as previously reported [32]
The yeast S. cerevisiae has shown crucial limitations in relation to the pentose fermentation. This is due to the following: (1) inability of S. cerevisiae to utilise pentose [35]: (2) low levels of expression of the xylose-fermenting genes that codify for xylose reductase (XR), xylitol dehydrogenase (XDH) and xylulose kinase (XK) [32]; (3) low expression of XK limits the rate of the xylose fermentation [34] and (4) imbalance of the redox cofactors (NAD+/NADH and NADP+/NADPH ratios) that reduces rates and ethanol yields from pentose [36]. The application of engineering approaches to traditional pathways of pentose utilisation allows the cells of the S. cerevisiae to catabolise xylose and arabinose [37, 38]. By using DNA technologies, the genes linked to the pentose metabolism from bacteria and other fungi have been inserted into the genome of S. cerevisiae for the utilisation of sugars from hemicellulose hydrolysates [37, 38].
Genetic Tools and Strategies to Improve Phenotypes of Yeast Strains
In fermentation processes, evolution can takes place in the yeast population generating diversity [39]. In the industrial processes of ethanol production, cells are reused 400 to 600 times for 200 to 300 days every year [40]. Yeast diversity can occur at laboratory scale [41] or during long-term evolutionary processes such as those occurring during the operation of the Brazilian industrial process of ethanol production with cell reuse. The screening and selection of yeast phenotypes during this process has given superior industrial strains for bioethanol production [42].
Advanced improvements in the optimisation of xylose-fermenting yeasts require novel metabolic engineering tools and strategies. Several genetic tools, such as protein engineering, metabolic engineering, rational metabolic engineering and inverse metabolic engineering are available to improve the ethanol production by yeast strains, as elegantly described [43, 44]. Protein engineering consists of the design and construction of new proteins or enzymes with novel or desired functions by modifying amino acid sequences of proteins using recombinant DNA technologies. An example of protein engineering is the production of both the dehydrogenase (LDH) and the formate dehydrogenase (FDH), which participates in important process of the food and pharmaceutical industries where they are related to each other in terms of cellular level of NADH and generation of chiral acids [45]. In essence, rational metabolic engineering requires powerful theoretical methods, as the pathway analysis, which involves the topology or cellular locations of the metabolic pathways [46]. Inverse metabolic engineering means the elucidation of a metabolic engineering strategy as follows [47]: First, identify and construct a desired phenotype; second, determine the genetic or particular environmental factors conferring a particular phenotype and third, transfer the selected phenotypes to another strain of yeast by direct genetic or environmental manipulation.
The association between the tools of metabolic engineering and synthetic biology (creation of biological functions not found in nature) [48] provides powerful approaches for the engineering of complex industrial phenotypes. The new xylose-fermenting strains of S. cerevisiae obtained using these approaches are able to efficiently express genes from other xylose-fermenting microorganism [49]. These tools are genome shuffling, global transcriptome machine engineering (gTME, a tool for global perturbations in the transcriptome) and directed evolutionary engineering that allows simultaneous modifications of multiple genes and the combination of useful alleles from many parental strains into a single cell [50, 51]. In evolutionary engineering, interactive and repetitive cycles of genetic variations, screening and selection process are followed by data analysis in order to select the best clones evolved during the continuous evolution in large microbial populations [52].
It has been observed that most cellular phenotypes are affected by many genes [51]. Complex phenotypes can be studied using DNA shuffling that combines the advantage of multi-parental crossing with the recombination of entire genomes, which are normally associated to the conventional breeding and also provide information on complex phenotypes. The whole transcriptional control process changes the transcription with improvements in the expression of target genes [50]. The rapid phenotypic improvements in cells of the yeast population are observed due to accelerated evolutionary processes. Improvements in complex phenotypes such as tolerances to temperature and ethanol yields have been described as a result of the use of genome shuffling and evolutionary processes [53–55]. In addition, the gTME approaches offer the advantage of identifying simultaneously genetic changes at different positions of the entire genome without the previous need for DNA sequencing [50].
Despite its importance, the process of evolutionary adaptation occurring in a large microbial population is still poorly understood [52, 53]. Genetic variations take place during adaptation, and they can be due to spontaneous mutation or adaptive mutations (beneficial mutations) that allow the fitness of the cells to the environmental conditions [56]. It has been difficult to identify the precise genetic changes underlying adaptation at a genome-wide scale. In recent years, a series of high-throughput DNA sequencing technologies capable of producing giant DNA sequence information in a single experiment have been developed [53]. Such technologies allow the genome of parental and evolved strains to be rapidly determined. By analysing a large number of clones, the sequencing libraries of the DNA from evolved yeast strains have been obtained at laboratory scale in nutrient-limited chemostat cultures [53].
Co-fermentation of Substrates Containing Pentose
The lignocellulosic hydrolysates contain mainly glucose, xylose and other sugars such as arabinose, mannose and galactose [4], and new yeasts able to co-ferment the mixture of three sugars (glucose, xylose, arabinose) are preferred strain [9]. As S. cerevisiae has limitations concerning pentose fermentation, the co-utilisation of hexose and pentose has been obtained by inserting pathways from different organism into the cells of S. cerevisiae. The fermentation of arabinose was first described by combining the expression of the genes encoding for the enzymes of the non-oxidative pentose pathway of Lactobacillus plantarum using an evolutionary engineering strategy. As a result of this study, a high ethanol yield (0.43 g g−1) from arabinose was obtained without formation of xylitol and arabitol [57]. In another study [58], a bacterial arabinose isomerase was combined to different xylose utilisation pathways as follows: The xylose reductase/xylitol dehydrogenase and xylose isomerase pathways and this combination resulted in both higher pentose uptake and overall ethanol production than those obtained with the combination of the xylose isomerase and the bacterial arabinose pathway [58]. At last, a l-arabinose and d-xylose co-fermenting strains of S. cerevisiae, expressing pathways of entirely fungal origin produced ethanol and biomass, showed a high arabitol yield (0.48 g g−1) and a low xylitol yield (0.07 g g−1) during the aerobic fermentation of xylose [38]. Application of evolutionary engineering to recombinant xylose-fermenting yeast resulted in two types of clones showing different phenotypes. The largest clonal subpopulation was able to grow anaerobically on xylose for 460 generations showing simultaneous xylose and glucose utilisation in batch culture, but an impaired growth on glucose was observed. Surprisingly, clones of the smaller subpopulation were incapable of anaerobic growth on xylose [59]. On the other hand, the application of gTME to a engineered strain of S. cerevisiae improved xylose utilisation and ethanol tolerance [60], as shown by the utilisation ratio of 90.8% glucose, and 97.3% xylose in 50 g/l glucose–xylose mixed culture (proportion 1:1, w/w). Despite the progress in optimising pentose utilisation by yeasts, limitations related to simultaneous exogenous sugar metabolism still persists [61]. Optimisation of the interdependent process such as transport and metabolism, using novel combinatory techniques and global engineering, are needed [61].
Improving Cofactor Balance in Pentose Utilisation
The utilisation of pentose by yeasts requires a great demand for NADH, NADP+ or both, and this causes depletion in the energy pool of the cells and leads to metabolic limitations that reduce ethanol yields [57]. The performance of the entire metabolism of a cell depends on the cellular proportion between the reducing and oxidising forms of the nicotinamide cofactors. Since xylose fermentation depends on the activity of a reductase and a dehydrogenase (xylulose reductase (XR) enzyme and xylitol dehydrogenase (XDH) enzyme), the insertion of a combined xylose reductase/xylitol dehydrogenase pathway and a bacterial arabinose isomerase pathway into S. cerevisiae cells resulted in both higher pentose sugar uptake and overall ethanol production [32, 62]. Enzymes catalysing the first steps in the metabolic pathway of xylose metabolism show different coenzyme specificities for nicotinamide cofactors, a strain of S. cerevisiae transformed with genes encoding XR and XDH from P. stipitis and XL3 (encoding xylokinase) was able to convert xylose into ethanol followed by an unfavourable excretion of xylitol [63]. This unfavourable excretion of xylitol is due to intracellular redox imbalance caused by the different coenzyme specificity of both XR and XDH [64]. Another improved strain of S. cerevisiae transformed with XR and XDH form P. stipitis was able to convert xylose to ethanol with an unfavourable excretion of xylitol due to the intracellular co-factor imbalance generated by different coenzyme specificities between NADH-preferring XR and NAD+-dependent XDH enzymes [65]. A cofactor–redox-engineered strain of xylose-fermenting showed increased ethanol yields and reduced xylitol and glycerol formation associated to an improved ethanol yield from xylose under anaerobic conditions. Improved ethanol and xylitol yields were due to the over-expression of the GPD1 gene, encoding for NADP+-dependent d-glyceraldehyde dehydrogenase from Kluyveromyces lactis, associated with the deletion the gene ZWF1 that encodes for glucose-6-phosphate dehydrogenase in this cofactor–redox-engineered strain [36]. In a recent publication, a recombinant strain was constructed based on the coenzyme preference shown by the xylose reductase from Candida tenuis, which was inserted into the cells of S. cerevisiae. The coenzyme preference of the reductase from C. tenuis changed from NADPH (strain BP000) to NADH (strain BP10001) when expressed in S. cerevisiae with increases in the xylose consumption and decreases in xylitol formation under conditions of low-glucose concentrations (<4 g L−1), showing a fivefold enhanced xylose uptake [62].
Tolerance and Yeast Defense Mechanisms Against Lignocellulose Inhibitors
Lignocellulosic hydrolysates contain a variety of toxic compounds. These compounds are predominantly weak organic acids, furan aldehydes and phenolic compounds, and they exhibit negative effects on yeast growth, metabolism and ethanol yields [66]. One of the major obstacles for the development of large-scale ethanol production from lignocellulose is the negative effect of the organic compounds on the growth of S. cerevisiae, the ethanol yield and the productivity [66].
Genetic engineering tools have been used to elucidate the cellular mechanisms able to minimise the effects of the toxic products from the hydrolysates and develop strategies to overcome inhibition. Complex and advanced strategic approaches, based on the use of genetic engineering tools, are available to relieve the inhibition and increase the innate tolerance of the yeast cells to inhibitors as revised [67]. These tools allow the over-expression of genes encoding for enzymes against lignocellulosic inhibitors and alterations in co-factor balance. Adaptation is a strategy that proved to be very useful in displaying evolutionary processes in a cell population due to stimulations of changes in the cell metabolome such as such levels of intracellular metabolites, activity of most of the glycolytic enzymes and levels of storage materials of the yeast cells [68]. At increasing concentrations of furfural and 5-hydroxymethyl-2-furaldehyde (HMF) [69, 70], the observed adaptation is due to induction of the expressions of genes codifying for enzymes able to metabolise the inhibitors [71]. The increased tolerance (~300 generations) of evolved strains to inhibitors shows three main characteristics [69]: (1) growth at higher hydrolysate concentrations; (2) significantly reduced lag phases and (3) a shorter process time. As shown by the maintenance of viability and morphological alterations typical to apoptosis, adaptation to acetic acid stress (80 mM in acetic acid at pH 3.0 for 30 min) protects cells of S. cerevisiae against acetic acid-induced cell death [72].
Effects of Acetic Acid and Medium pH on Fermentation
Acetic acid is a by-product of the alcoholic fermentation that can cause fermentation arrest and to reduce ethanol productivity. This acid is the major weak acid of the lignocellulose hydrolysates [66], and it is also produced by contaminant acetic acid bacteria [73]. Lactic acid bacteria and their metabolic products can severely interfere with the yeast growth [73].
Acetic acid is a minor by-product formed by cells of S. cerevisiae during fermentation, and its formation is a process dependent on the medium pH. When acetic acid is added to the medium, yeast cells show decreases in the metabolic flux through the tricarboxylic acid cycle at pH 6.9, and these decreases are followed by a reduced glucose uptake and increased acetate formation [74]. It is believed that the regulation process of acetate formation is not completely elucidated, whereas the medium pH appears to regulate acetic acid formation [74]. At a pH of 5.0, the external acetic acid increases the glucose consumption rates, whereas xylose consumption rates are not significantly affected [75]. At a pH 3.5, addition of acetic acid to the medium had a strong negative effect on xylose consumption rate after the glucose depletion from the medium [75]. In a genetically engineered strain, 42A (LNH-ST) of S. cerevisiae, the sugar consumption rates and both the ethanol and biomass formation decreased when the acetate concentration increased during the co-fermentation of a mixture of glucose and xylose, and this inhibitory effect on the xylose consumption was severely affected [76]. The authors concluded that the inhibitory effect of acetic acid against yeast cells could be reduced by increasing the medium pH, while the proportion between levels of the dissociated form and the non-dissociated form of acetic acid confirmed the inhibitory effect of the of the non-ionised form of the acetic acid molecule [76].
Determinants of Tolerance to Acetic Acid
A great number of genes (490 to 650 genes) seem required for tolerance to acetic acid and other processes as follows [77]: transcription, internal pH, homeostasis, carbohydrate metabolism, cell wall assembly, biogenesis of the mitochondria, ribosome and vacuoles, in sensing signaling and uptake of various nutrients in particular iron, potassium, glucose and amino acids. The genes involved in these processes are novel candidates for genetic engineering to obtain more robust yeast strains against acetic acid toxicity.
Aquaporins or water channels (encoded by gene FPS1) are membrane pore proteins that selectively conduct water molecules in and out of the cell, while preventing the passage of ions and other solutes. Some aquaglyceroporins also transport other small uncharged solutes, such as glycerol, CO2, ammonia and urea [78]. When the gene FPS1 is disrupted, the S. cerevisiae cells acquire an elevated tolerance to acetic acid as shown by the increases in growth and fermentation [79]. In addition, there are other genes (gene network) that seem to provide additional and strong protective effects toward the acetic acid stress when expressed, and they are as follows [80]: (1) SAP30 encoding for the histone deaceltylase complex; (2) HRK1 encoding for the protein kinase involved in the regulation of membrane transporter activity and (3) HAA1 encoding for the transcriptional factor haa1p involved in the reduction of intracellular concentration of acetate. An evidence of this protective effect is the increase tolerance to the acetic acid resulting from, the deletion of HRK1 (protein kinase gene) involved in the reduction of acetic acid in yeast-stressed cells [79]. Using genome-wide approaches and more detailed gene-by-gene analysis [80], the involvement of several transcriptional regulatory networks, under the control of known transcriptional factors (Msn2p/Msn4p, War1p, Haap1p, Rim101p and Pder1p/Pdr3p) were identified based on the genomic responses, mainly related to the growth of S. cerevisiae in the presence of weak acids at different conditions [81]. A strain of xylose-fermenting yeast (strain RWB2018) was able to grow on media containing 5–6 g/L xylose at pH ≤ 4.0 after continuous sequential-batch cultivations under anaerobic conditions (~400 generation) at increasing concentrations of acetic acid added to the medium. This continuous batch-strategy resulted in an adapted yeast culture, where the specific xylose-consumption rate showed a 75% increase [82].
Resistance to Formic Acid
Although poorly utilised as electron donor for glycerol formation, formate is the most harmful inhibitor of the S. cerevisiae cells. It induces apoptosis-like yeast cell death in the budding yeast cells of S. cerevisiae via Yca1p, a metacaspase that induces Yca1p-independent apoptosis-like cell death process in S. cerevisiae [83]. Yeast cell death caused by formic acid is associated with the reactive oxygen species (ROS) burst, while ROS is accumulated more rapidly into the mitochondria the YAC1 disruptants (metacaspase gene) than in wild-type strains [83]. Metacaspases are cysteine-dependent proteases found in protozoa, fungi and plants that show multifunctional proteases essential for the normal physiology of non-metazoan organisms [84]. The metacaspase Yca1 (also termed Mca1) is a regulator of oxidative stress-induced and senescence-associated to death of yeast cells [85].
In anaerobic cultures of S. cerevisiae, the cells oxidise the excess of NADH formed in biosynthesis via glycerol production. The relatively low tolerance of the yeast to formic acid is dependent on the expression of a mitochondrial NAD+-dependent formate dehydrogenase gene that is expressed at very low levels [86]. In order to increase the availability of intracellular NADH in vivo, a metabolic and genetic engineering approach was used to obtain novel recombinant strains of S. cerevisiae in which FDH1 gene (encoding for formate dehydrogenase) and/or GPD2 gene (encoding glycerol-3-phosphate dehydrogenase) were over-expressed. This strain consumed up to 70% of the formate added to the feed in an anaerobic and glucose-limited chemostat culture with increasing glycerol yields [87]. Co-consumption of formate and glucose was obtained in an aerobic glucose-limited chemostat culture using a recombinant strains of S. cerevisiae (strain CEN. PK113-7D) expressing another structural gene of formate dehydrogenase (FAHD2). In this culture, significant increases in the tolerance of S. cerevisiae (CEN. PK113-7D) to acetic acid and formic acid were observed when small amounts of formic acid were added to the feed, due to the formate utilisation via glycerol formation in order to oxidise NADH in excess [87].
Furan Aldehydes Derived from Pentose
One of the major causes of the high toxicity of furfural and 5-hydroxymethyl-2-furfuraldehyde towards S. cerevisiae cells is that these two inhibitors are able to elevate the levels of ROS generated by the mitochondria [69, 88]. The highly oxidative radicals (ROS) cause damages to DNA, proteins and cell membranes, and induce apoptosis [88, 89]. Depending on the conditions of the hydrolytic process, significant amounts of furan aldehydes are formed during the lignocellulose hydrolysis.
In order to detoxify hydrolysates containing 5-hydromethyl-2-furfuraldehyde, the main strategies are as follows: (1) overexpression of dehydrogenases and reductases of the pentose pathways aiming the conversion of corresponding aldehydes into alcohols [90] and (2) adaptation of the yeast cells to furfural and 5-hydromethyl-2-furfuraldehyde [67]. In addition, aldehyde dehydrogenase 6 plays an important role in direct oxidation of furan derivatives and supply of NADPH to the cofactor-depending reactions [91]. Overexpression of dehydrogenases and reductases increases the specific growth rate and ethanol productivity [92]. At a lower furfural concentration (15 mM), the overexpression of the NADH-dependent oxidoreductase is the main defense mechanism against the furfural inhibition, due to the effective action of the NADPH-dependent oxidoreductase on the furanaldehyde [92]. On the other hand, the use of an adaptation strategy based on evolution leads to yeast strains able to grow at concentrations of hydrolysates that effectively kill the parental strains followed by a significant improvement in the ethanol production, mainly due to a prolonged lag phase induced by furfural that allows the yeast cells to remain viable in the toxic environment [69].
Proteomic analysis has revealed that genes involved in glucose fermentation (glycolysis) and the tricarboxylic acid cycle are upregulated in cells treated with furfural, whereas proteins involved in glycerol biosynthesis are downregulated [93]. In addition, the levels of alcohol dehydrogenase (ADH genes) seem relate the tolerance of the ethanologenic yeasts towards furfural. Furfural affects the redox balance of the cellular metabolism by downregulating levels of genes involved in the glycerol biosynthesis. As a consequence of the effects of furfural on the redox balance, NADH/NAD+ ratio decrease, while ATP/ADP ratio increase [93]. Furfural downregulates genes involved in both the transcriptional and translation controls, while acetic acid downregulates genes encoding for mitochondrial ribosomal proteins and genes involved in the carbohydrate metabolism. For this reason, mitochondria play important roles in the resistance to both acetic acid and furfural [93]. Nevertheless, the molecular mechanism involved in tolerance and adaptation of S. cerevisiae to inhibitors of the lignocellulosic hydrolysates are still unclear [93].
Organic Compounds Derived from Lignin
The inhibitory organic compounds present in lignocellulosic hydrolysates are monomeric phenolic compounds containing a number of substituted groups (aliphatic, hydroxyl, carboxylic, aldehyde and the acyl groups of the vanillin structure), and the structure of these substituted groups confers differentiated inhibitory capacities to the inhibitors [94]. In addition, lignocellulose contains lignin, which is a complex molecule composed of phenyl propane units that exhibit wide variety of substituted groups linked to the aromatic rings of the compounds derived from lignin and they affect the growth and fermentation of strains of S. cerevisiae [94]. Nevertheless, S. cerevisiae also exhibits a few defense mechanisms towards the aromatic compounds derived from lignin hydrolysis. Even so, the inhibitory effects of the aromatic compounds are greatly dependent on their chemical structure as described above [94]. Low-molecular-mass phenolic compounds show significant inhibitory effects on S. cerevisiae and lower stability, while their effects can be suppressed by the action of polymerases that catalyse the polymerisation of the low molecular phenolic compounds [94]. Resistance to lignocellulose-derived inhibitors can result from the over-expression of the following genes [95]: (1) ATR1 codifying for multidrug resistance and stress responses and also contributing to a more efficient ethanol production from hydrolysates; (2) YAP1 encoding for protein acting as transcription factor that display hyper-resistance to coniferyl aldehyde and (3) FLR1 encoding for a multidrug transporter of the major facilitator superfamily of transporters, which confer enhanced resistance to coniferyl aldehyde and 5-hydroxy-2-methylfuraldehyde.
Vanillin or hydroxymethoxylbenzaldehyde is a major phenolic compound, which seems to be a stronger inhibitor of bioethanol fermentation than other inhibitors because it acts at low concentrations on the fermentation of the lignocellulosic hydrolysates by yeasts and bacteria [96]. Tolerant strains of S. cerevisiae can grow in a media containing 0 or 15 mM vanillin [97]. Tolerance to vanillin is linked to the expression of genes involved in biosynthesis of ergosterol, which are as follows [97, 98]: ERG28 encoding for a membrane protein that facilitate the interactions between the Erg26p dehydrogenase and the Erg27p 3-ketoreductase; HMG1, encoding for mitochondrial NADH-cytochrome b5 reductase; ERG5 encoding for the sterol C-22 desaturase that is a cytochrome P450 oxidase and ERG7 encoding for 2,3-epoxysqualene-lanosterol cyclase or lanosterol synthase. In recent publication, an enhanced resistance of S. cerevisiae to vanillin was obtained by the overexpression of lacA, a laccase gene from Trametes sp. AH28-2 [99].
A laccase-producing strain of S. cerevisiae was obtained, and this transformant showed the ability to ferment hydrolysates in the presence of coniferyl aldehyde [100]. Coniferyl aldehyde can be converted into coniferyl alcohol in S. cerevisiae cells due to the enzymatic reaction that catalyses the reduction of the aldehyde [100].
Yeast Tolerance to Ethanol and Other Industrial Stresses
The following factors are involved in ethanol tolerance in S. cerevisiae cells [101]: plasma membrane composition, trehalose levels, heat-shock proteins, heat-shock factors, stress response elements and others. Genome-wide analysis and statistical analysis of S. cerevisiae during growth of strains under ethanol stress allow the identification of deletion genes (~4,828 genes) involved in the tolerance of these yeasts to various environmental stresses [102]. The identified genes are sensitive to ethanol and other stresses, such as tolerance to methanol and 1-propanol, and heating, H2O2 and increasing NaCl concentrations. Among these genes are also those related to organelles and functions as follows [102]:(1) VAM genes involved in vacuolar assembly and morphology, (2) genes related to H+ ATPase functions (V-ATPase that protect cells against ethanol stress); (3) biogenesis and organisation of cellular components; (4) protein fate; (5) cellular transport, (6) signal transduction and (7) cell cycle, DNA processing and wall integrity (dependent on the genes OCH1 and MNN11 encoding for mannosyl transferases). As genes involved in mitochondrial functions seem involved in the tolerance to several stresses, the identification of these genes will help to elucidate the molecular mechanisms of the stress responses [102].
Genome shuffling was applied to the industrial strain SM-3 of S. cerevisiae, and both the thermal (maximal growth temperature at 55 °C) and ethanol tolerances (tolerating 25%, v/v ethanol at 45 °C to 48 °C) were greatly improved as shown by growth assays on solid media [54]. Another interesting report, a novel gene ETP1/YHLO10c was identified as required for the adaptation of S. cerevisiae to ethanol, since it activates the ENA1 gene that encodes for Etp1p protein involved in the yeast growth activity on ethanol as sole carbon source. The exact molecular mechanism of action of Etp 1p is not currently known [103].
Monitoring a Long-Term Ethanol Production Process and Generation of Adapted Yeast Strains
A lower cost-effective ethanol production process depends on a rapid and high yielding-conversion of carbohydrates to ethanol and biomass, which in turn is dependent on the survival and performance of yeast cells under industrial conditions. S. cerevisiae is exposed at industrial scale to a number of stresses, including high ethanol concentrations, high osmotic pressures, oxidative stresses, temperature fluctuations and genomic variability during ethanol production process [41].
Strains of S. cerevisiae have been selected from Brazilian distilleries with the aim to obtain more adapted and robust strains to be used as staters of long-term industrial processes. Based on genetic analysis, the genome of a S. cerevisiae cell does not remain stable during consecutive generations [39], while the prevalence and dominance of a starter strains in the yeast population changes with the process conditions as shown by the differences in karyotypes of the isolates obtained from the fermentation process. Even so, three isolates (PE-2, CAT-1 and BG-1) behaved as the best starters for the bioethanol production [42]. This three isolates selected during long-term industrial processes are genetically adapted strains that resulted from chromosomal changes. Although little attention has been given to the integration among yeasts in mixed cultures, yeast strains compete with each other, leading to changes in product yields (ethanol, glycerol, succinic acid, acetic acid and others) and growth [104].
Depending on the process conditions, the chromosomal changes observed for the isolates obtained from the industrial fermentation process indicate the possibility of occuring the following genetic events: (1) hybridisation [39]; (2) mitotic recombination [105]; (3) transposition of DNA-segments (transposons) or Ty elements [106] and (4) gene duplication [107, 108]. Most of random mutations (nucleotide changes in the DNA sequence) can be harmful or neutral, while adaptive mutations are essential to the fitness of a strain to its environment. It seems that the frequency of the adaptive mutations can be as low as one in 1011 cell divisions [56]. Newer genetic models indicate that those beneficial mutations are more abundant than previously suspected and sufficiently common to coexist and compete for fixation within cell populations [109].
An increased severity (acidity, inhibitors) is expected during the fermentation lignocellulose hydrolysates. As lignocellulosic hydrolysates are supposed to be fermented at industrial scale under non-sterile conditions in a near future, the development and use of monitoring procedures will be essential. Procedures for the selection of strain resistant to lignocellulosic hydrolysates have to be developed.
Strategies for Improvement of the Bioprocesses of Ethanol Production from Biomass
One of the major bottlenecks in the production of second-generation bioethanol is the high cost of the cellulolytic enzymes [110]. The need for lower amounts of enzymes could be achieved by the production of new enzymes and improvements in the hydrolytic processes. An effective strategy for reducing effective-cost of the lignocellulosic ethanol would be the selection of new fungal strains and the use of lignocellulosic substrates showing improved accessibility to enzymes. The new fungi should be able to secrete enzymes at high levels during short fermentation periods. In addition, the novel cellulase preparations should be able to efficiently disrupt the crystalline polysaccharide structures of the cellulose at higher rates and temperatures below 50 °C in order to be applied to SSF processes using lignocellulosic materials [111]. Developments in supported metal catalysis (e.g. particles associate to Pt, TiO2, ZrO2) seems an attractive alternatives for cellulose hydrolysis [112]. Other improvements are those related to detoxification (degradation of toxic by-products) of the hydrolysates, and this can be achieved by using high cell density fermentations, ion- exchange columns, absorbent resins [113], granulated activated carbon [114] and evaporation of hydrolysates for eliminating volatile inhibitors such as acetic acid and furfural [115]. Improved sugar yields and hydrolysis of softwoods can be obtained when laccases (p-diphenol: dioxygen oxidoreductase, EC 1.10.3.2) are produced by fungi associated to cellulases [110]. Laccases are fungal enzymes (e.g. enzyme from Phanerochaete chrysosporium) that oxidise a broad range of substrates such as polyphenols, substituted phenols and diamines [116].
Recently, several integrated processes have been optimised to improve ethanol production from lignocellulose materials. Process configurations for ethanol production from lignocellulose involve a variety of integrative approaches that are summarised as follows [5]: (1) simultaneous saccharification and fermentation processes (SSF); (2) separate hydrolysis and fermentation processes (SHF); (3) simultaneous saccharification and co-fermentation (SSCF) and (4) consolidated biological processes (CBP processes). In the CBP process, cellulase production, hydrolysis and fermentation take place in a single step [23]. Hydrolysates from wheat straw gave higher yields of ethanol in a simultaneous saccharification and fermentation process (SSF) than SHF process operated under the same conditions [117]. Among the research targets of particular interest to the second-generation ethanol are the improvements in process design, development of processes to obtain co-fermentation of pentose and hexose and the establishment of techniques to overcome bacterial contaminations [118]. Although the recycling of fermenting cells and enzyme is difficult, repeated fed-batch process systems using cell recycling have been increasingly applied to ethanol production due to the greater volumetric ethanol productivity and the possibility of fermenting of sugar mixtures containing glucose and pentose [119]. The ethanol production from lignocellulose is negatively affected by the low levels of final ethanol. Fermentation at high sugar concentrations is essential to maximise final ethanol and minimise ethanol production costs [5].
Another strategy for process improvement would consist in an all-encompassing monitoring of the fermentation variables (microbial population, levels lignocellulosic inhibitors and fermentation by-products) using computerised devices and data analysis via neural networks [120]. Such monitoring would allow the prediction of the process performance so that earlier corrections could be made to avoid sluggish fermentation or the process collapse. The levels of by-products can affect the yeast metabolism as shown by effects of acetaldehyde (a by-product) on the growth. Acetaldehyde inhibits yeast growth at concentrations above 0.3 g L−1, but low concentrations greatly reduce the lag phase and increase the specific growth rate in ethanol-containing medium [121].
Despite the high discriminatory capacity of the genetic molecular tools used to identify yeast strains, it would be very convenient for the monitoring of the industrial process to use simple and fast methods to detect changes in the traits (characteristics) of yeast strains able to interfere with the fermentative capacity, and tolerance to ethanol. As an example of an unwanted trait is the loss in capacity of assimilating sucrose at both laboratory- and industrial-scale fermentations [41, 42]. Loss in the capacity to assimilate sucrose has a great negative impact on long-term fermentations processes. Changes in the characteristics of an isolated strain of S. cerevisiae, such as the increase in the growth capacity on solid medium containing acetic acid concentration increasing up to 4 g/L [122] might reflect its adaptation to acidity and inhibitors during the fermentation of the lignocellulosic materials. Naturally occurring strains of S. cerevisiae are usually very sensitive to cyclohexymide [123]. However, a homozygous strain (e.g. cyh2R/cyh2R strain) of S. cerevisiae might be easily monitored during the bioethanol production by simple plating of samples on a medium containing high amounts of cycloheximide, where the adapted strains would be able to grow.
Conclusions and Prospects
A variety of genes linked to the pentose metabolism of bacteria and other fungi have been cloned and inserted into the genome of S. cerevisiae for the utilisation of pentose [37, 38]. The application of the modern genetic engineering tools has demonstrated that some yeast phenotypes are complex traits dependent on the expression of a variety of genes [42]. The elucidation of the genes involved in the expression of industrials phenotypes might be a great contribution to the control and improvements of the fermentation processes. Researches related to the production of cheap cellulase preparations and the discovery of new process designs should be more encouraged. The most critical factors involved in the cellulase production seem to be as follows: (1) the search for cellulosic enzymes showing lower optimal temperatures (30 °C to 37 °C); (2) enzymatic catalysts showing shorter reaction times and (3) minimised inhibition towards the by-products of the biomass hydrolysis. Shorter and less expensive pre-treatments and strategies to reduce the number of steps of the entire process are needed.
In order to have bioethanol from plant biomass available on the market at competitive prices compared with those obtained from sugarcane or corn starch, great improvements are still required. However, ethanol is not the only fuel that can be produced from lignocellulosic materials. Biobutanol has been attracting the attention of the researchers due to its lower corrosiveness, volatility, energy density and easy separation from the process. Strains of Clostridium acetobutyricum with improved tolerance to n-butanol have been obtained using evolutionary engineering and screening strategies [124]. On the other hand, strains of S. cerevisiae need to be engineered in order to have more efficient n-butanol pathways from other microorganism inserted into their cells [125]. A simple gas, such as the carbon monoxide derived from the gasification of biomass, can be converted into ethanol by some species of Clostridium [126]. Microalgae are also candidates for biofuel production such as ethanol, hydrogen and biodiesel [127, 128]. It is not currently clear which will be the ideal biofuel and/or the appropriate organism to use in the future.
In the last decade, numerous engineered microorganisms have been developed, but none of them have been successfully applied for commercial ethanol production. Due to the importance of chemical feedstock, the production of higher alcohols, alkenes or fatty-acid derivatives including biodiesel, and alkanes from renewable sources are becoming increasingly attractive [129]. For instance, Amyris, Inc. (NASDAQ: AMRS) built a plant in Brazil to produce biofene from sugarcane syrup using an engineered yeast [130]. Biofene is a source of a broad range of renewable products including squalene, base oil, finished lubricants and diesel.
References
Sun, Y., & Cheng, J. (2002). Bioresource Technology, 83, 1–11.
Mussatto, S. I., Dragone, G., Guimarães, P. M. R., Silva, J. P. A., Carneiro, L. M., Roberto, I. C., et al. (2010). Biotechnology Advances, 28, 817–830.
Hendriks, A. T. W. M., & Zeeman, G. (2009). Bioresource Technology, 100, 10–18.
Margeot, A., Hahn-Hagerdal, B., Edlund, M., Slade, R., & Monot, F. (2009). Current Opinion in Biotechnology, 20, 372–380.
Gírio, F. M., Fonseca, C., Carvalheiro, F., Duarte, L. C., Marques, S., & Bogel-Lukasik, R. (2010). Bioresource Technology, 101, 4775–4800.
Zhao, X., Cheng, K., & Liu, D. (2009). Applied Microbiology and Biotechnology, 82, 815–827.
Kumar, P., Barrett, D. M., Delwiche, M. J., & Stroeve, P. (2009). Industrial and Engineering Chemistry Research, 48, 3713–3729.
Yang, B., & Wyman, C. E. (2008). Biofuels, Bioproducts and Biorefining, 2, 26–40.
Hamelinck, C. N., van Hooijdonk, G., & Faaij, A. P. C. (2005). Biomass and Bioenergy, 28, 384–410.
Carvalheiro, F., Duarte, L. C., & Gírio, F. M. (2008). Journal of Scientific and Industrial Research, 67, 849–864.
Iranmahboob, J., Nadim, F., & Monemi, S. (2002). Biomass and Bioenergy, 22, 401–404.
Rossell, C. E. V., Lahr Filho, D., Hilst, A. G. P., & Leal, M. R. L. V. (2005). International Sugar Journal, 107, 192–195.
Taherzadeh, M., & Karimi, K. (2007). BioResources, 2, 472–499.
Gregg, D. J., & Saddler, J. N. (1996). Biotechnology and Bioengineering, 51, 375–383.
Gautam, S. P., Bundela, P. S., Pandey, A. K., Khan, J., Awasthi, M. K., & Sarsaiya, S. (2011). Biotechnology Research International. doi:10.4061/2011/810425.
Stambuk, B. U., Eleutherio, E. C. A., Florez-Pardo, L. M., Souto-Maior, A. M., & Bom, E. P. S. (2008). Journal of Scientific and Industrial Research, 67, 918–926.
Slininger, P. J., Bolen, P. L., & Kurztman, C. P. (1987). Enzyme and Microbial Technology, 9, 5–15.
Olsson, L., & Hahn-Hägerdal, B. (1996). Enzyme and Microbial Technology, 18, 312–331.
Lynd, L. R., Weimer, P. J., van Zyl, W. H., & Pretorius, I. S. (2002). Microbiology and Molecular Biology Reviews, 66, 506–577.
Bothast, R. J., Nichols, N. N., & Dien, B. S. (1999). Biotechnology Progress, 15, 867–875.
Dien, B. S., Cotta, M. A., & Jeffries, T. W. (2003). Applied Microbiology and Biotechnology, 63, 258–266.
Lau, M. W., Gunawan, C., Balan, V., & Dale, B. E. (2010). Biotechnology for Biofuels, 3, 11–15.
Xu, Q., Singh, A., & Himmel, M. E. (2009). Current Opinion in Biotechnology, 20, 364–371.
Panagiotou, G., Villas-Bôas, S. G., Christakopoulos, P., Nielsen, J., & Olsson, L. (2005). Journal of Biotechnology, 115, 425–434.
Olsson, L., & Hahn-Hägerdal, B. (1993). Process Biochemistry, 28, 249–257.
Jones, D. T., Shirley, M., Wu, X., & Keis, S. (2000). Journal of Molecular Microbiology and Biotechnology, 2, 21–26.
Los, M., Golec, P., Los, J. M., Weglewska-Jurkiewicz, A., Czyz, A., Wegrzyn, A., et al. (2007). BMC Biotechnology, 7, 13–18.
Lee, W.-J., Kim, M.-D., Ryu, Y.-W., Bisson, L. F., & Seo, J.-H. (2002). Applied Microbiology and Biotechnology, 60, 186–191.
Sedlak, M., & Ho, N. W. Y. (2004). Yeast, 21, 671–684.
Runquist, D., Hahn-Hägerdal, B., & Radstrom, P. (2010). Biotechnology for Biofuels, 3, 5–11.
Leandro, M. J., Fonseca, C., & Gonçalves, P. (2009). FEMS Yeast Research, 9, 511–525.
Matsushika, A., Inoue, H., Kodaki, T., & Sawayama, S. (2009). Applied Microbiology and Biotechnology, 84, 37–53.
Chandel, A. K., Chandrasekhar, G., Radhika, K., Ravinder, R., & Ravindra, P. (2011). Biotechnology and Molecular Biology Review, 6, 8–20.
Toivari, M. H., Aristidou, A., Rouhonen, L., & Penttilä, M. (2001). Metabolic Engineering, 3, 236–249.
Toivari, M. H., Salusjârvi, L., Rouhonen, L., & Penttilä, M. (2004). Applied and Environmental Microbiology, 70, 3681–3686.
Verho, R., Londesborough, J., Penttilã, M., & Richard, P. (2003). Environmental Microbiology, 69, 5892–5897.
Wisselink, H. W., Toirkens, M. J., Wu, Q., Pronk, J. T., & van Maris, A. J. A. (2009). Applied and Environmental Microbiology, 75, 907–914.
Bettiga, M., Bengtsson, O., Hahn-Hägerdal, B., & Gorwa-Grauslund, M. F. (2009). Microbial Cell Factories, 8, 40–51.
Mortimer, R. K. (2000). Genome Research, 10, 403–409.
Amorim, H. V., Lopes, M. L., Oliveira, J. V. C., Buckeridge, M. S., & Godman, G. H. (2011). Applied Microbiology and Biotechnology, 91, 1267–1275.
Souza, C. S., Thomaz, D., Cides, E. R., Oliveira, K. F., Tognoli, J. O., & Laluce, C. (2007). World Journal of Microbiology and Biotechnology, 23, 1667–1677.
Basso, L. C., Amorim, H. V., Oliveira, A. J., & Lopes, M. L. (2008). FEMS Yeast Research, 8, 1155–1163.
Nevoigt, E. (2008). Microbiology and Molecular Biology Reviews, 72, 379–412.
Patnaik, R. (2008). Biotechnology Progress, 24, 38–47.
Karagüler, N. G., Sessions, R. B., Binay, B., Ordu, E. B., & Clarke, A. R. (2007). Biochemical Society Transactions, 35, 1610–1615.
Schuster, S., Dandekar, T., & Fell, D. A. (1999). Trends in Biotechnology, 17, 53–60.
Bailey, J. E., Sburlati, A., Hatzimanikatis, V., Lee, K., Renner, W. A., & Tsai, P. S. (2002). Biotechnology and Bioengineering, 79, 568–579.
Lee, S. K., Chou, H., Ham, T. S., Lee, T. S., & Keasling, J. D. (2008). Current Opinion in Biotechnology, 19, 556–563.
Jeffries, T. W. (2006). Current Opinion in Biotechnology, 17, 320–326.
Gong, J., Zheng, H., Wu, Z., Chen, T., & Zhao, X. (2009). Biotechnology Advances, 27, 996–1005.
Alper, H., & Stephanopoulos, G. (2007). Metabolic Engineering, 9, 258–267.
Sauer, U. (2001). Advances in Biochemical Engineering. Biotechnology, 73, 130–166.
Araya, C. L., Payen, C., Dunhum, M. J., & Fields, S. (2010). BMC Genomics, 11, 88–98.
Shi, D., Wang, C., & Wang, K. (2009). Journal of Industrial Microbiology and Biotechnology, 36, 139–147.
Hou, L. (2009). Biotechnology Letters, 31, 671–677.
Zeyl, C. (2004). Research in Microbiology, 155, 217–223.
Wisselink, H. W., Toirkens, M. J., del Rosario Franco Berriel, M., Winkler, A. A., van Dijken, J. P., Pronk, J. T., et al. (2007). Applied and Environmental Microbiology, 73, 4881–4891.
Bettiga, M., Hahn-Hägerdal, B., & Gorwa-Grauslund, M. F. (2008). Biotechnology for Biofuels, 1, 16–23.
Sonderegger, M., & Sauer, U. (2003). Applied and Environmental Microbiology, 69, 1990–1998.
Liu, H., Yan, M., Lai, C., Xu, L., & Ouyang, P. (2010). Applied Biochemistry and Biotechnology, 160, 574–582.
Young, E., Lee, S. M., & Alper, H. (2010). Biotechnology for Biofuels, 3, 24–35.
Krahulec, S., Petschacher, B., Wallner, M., Longus, K., Klimacek, M., & Nidetzky, B. (2010). Microbial Cell Factories, 9, 16–29.
Verduyn, C., van Kleef, R., Frank, J., Schreuder, H., van Dijken, J., & Scheffers, A. (1985). Biochemical Journal, 226, 669–677.
Jeffries, T. W. (1985). Trends in Biotechnology, 3, 208–212.
Watanabe, S., Saleh, A. A., Pack, S. P., Annaluru, N., Kodaki, T., & Makino, K. (2007). Microbiology, 153, 3044–3054.
Almeida, J. R. M., Modig, T., Petersson, A., Hahn-Hägerdal, B., Lidén, G., & Gorwa-Grauslund, M. F. (2007). Journal of Chemical Technology and Biotechnology, 82, 340–349.
Parawira, W., & Tekere, M. (2011). Critical Reviews in Biotechnology, 31, 20–31.
Mashego, M. R., Jansen, M. L. A., Vinke, J. L., van Gulik, W. M., & Heijnen, J. J. (2005). FEMS Yeast Research, 5, 419–430.
Heer, D., & Sauer, U. (2008). Microbial Biotechnology, 1, 497–506.
Liu, Z. L., Slininger, P. J., & Gorsich, S. W. (2005). Applied Biochemistry and Biotechnology, 121–124, 451–460.
Almeida, J. R. M., Bertilsson, M., Gorwa-Grauslund, M. F., Gorsich, S., & Lidén, G. (2009). Applied Microbiology and Biotechnology, 82, 625–638.
Giannattasio, S., Guaragnella, N., Corte-Real, M., Passarela, S., & Marra, E. (2005). Gene, 354, 93–98.
Vanderbergh, P. A. (1993). FEMS Microbiology Reviews, 12, 221–237.
Heyland, J., Fu, J., & Blank, L. M. (2009). Microbiology, 155, 3827–3837.
Bellissimi, E., van Dijken, J. P., Pronk, J. T., & van Maris, A. J. A. (2009). FEMS Yeast Research, 9, 358–364.
Casey, E., Sedlak, M., Ho, N. W. Y., & Mosier, N. S. (2010). FEMS Yeast Research, 10, 385–393.
Mira, N. P., Palma, M., Guerreiro, J. F., & Sá-Correia, I. (2010). Microbial Cell Factories, 9, 79–91.
Araya-Secchi, R., Garate, J. A., Holmes, D. S., & Perez-Acle, T. (2011). BMC Genomics, 12(Suppl 4), S8.
Zhang, J. G., Liu, X. Y., He, X. P., Guo, X. N., Lu, Y., & Zhang, B. (2011). Biotechnology Letters, 33, 277–284.
Mira, N. P., Becker, J. D., & Sá-Correia, I. (2010). OMICS, 14, 587–601.
Mira, N. P., Teixeira, M. C., & Sá-Correia, I. (2010). OMICS, 14, 525–540.
Wright, J., Bellisimi, E., Hulster, E., Wagner, A., Pronk, J., & van de Maris, A. J. A. (2011). FEMS Yeast Research, 11, 299–306.
Du, L., Su, Y., Sun, D., Zhu, W., Wang, J., Zhuang, X., et al. (2008). FEMS Yeast Research, 8, 531–539.
Tsiatsiani, L., van Breusegem, F., Gallois, P., Zavialov, A., Lam, E., & Bozhkov, P. V. (2011). Cell Death and Differentiation, 18, 1279–1288.
Madeo, F., Herker, E., Maldener, C., Wissing, S., Lächelt, S., Herlan, M., et al. (2002). Molecular Cell, 9, 911–917.
Overkamp, K. M., Kötter, P., van der Hoek, R., Schoondermark-Stolk, S., Luttik, M. A. H., et al. (2002). Yeast, 19, 509–520.
Geertman, J. M. A., van Dijken, J. P., & Pronk, J. T. (2006). FEMS Yeast Research, 6, 1193–1203.
Salmon, T. B., Evert, B. A., Song, B., & Doetsch, P. W. (2004). Nucleic Acids Research, 32, 3712–3723.
Perrone, G. G., Tan, S. X., & Dawes, I. W. (2008). Biochimica et Biophysica Acta, 1783, 1354–1368.
Carmona-Gutierrez, D., Eisenberg, T., Büttner, S., Meisinger, C., Kroemer, G., & Madeo, F. (2010). Cell Death and Differentiation, 17, 763–773.
Park, S., Koo, H. M., Park, Y. K., Park, S. M., Park, J. C., Lee, O., et al. (2011). Bioresource Technology, 102, 6033–6038.
Heer, D., Heine, D., & Sauer, U. (2009). Applied and Environmental Microbiology, 75, 7631–7638.
Lin, F. M., Qiao, B., & Yuan, Y. J. (2009). Applied and Environmental Microbiology, 75, 3765–3776.
Larsson, S., Quintana-Sáinz, A., Reimann, A., Nilvebrant, N. O., & Jönsson, L. J. (2000). Applied Biochemistry and Biotechnology, 84–86, 617–632.
Alriksson, B., Horváth, I. S., & Jönsson, L. J. (2010). Process Biochemistry, 45, 264–271.
Klinke, H. B., Thomsen, A. B., & Ahring, B. K. (2004). Applied Microbiology and Biotechnology, 66, 10–26.
Endo, A., Nakamura, T., & Shima, J. (2009). FEMS Microbiology Letters, 299, 95–99.
Endo, A., Nakamura, T., Ando, A., Tokuyasu, K., & Shima, J. (2008). Biotechnology for Biofuels, 1, 3–9.
Ji, L., Shen, Y., Xu, L., Peng, B., Xiao, Y., & Bao, X. (2011). Bioresource Technology, 102, 8105–8109.
Larsson, S., Cassland, P., & Jönsson, L. J. (2011). Applied and Environmental Microbiology, 67, 1163–1170.
Ding, J., Huang, X., Zhang, L., Zhao, N., Yang, D., et al. (2009). Applied Microbiology and Biotechnology, 85, 253–263.
Auesukaree, C., Damnernsawad, A., Kruatrachue, M., Pokethitiyook, P., Boonchird, C., Kaneko, Y., et al. (2009). Journal of Applied Genetics, 50, 301–310.
Snowdon, C., Schierholtz, R., Poliszczuk, P., Hughes, S., & van der Merwe, G. (2009). FEMS Yeast Research, 9, 372–380.
Favale, S., Pietromarch, P., & Ciolfi, G. (2007). Vitis, 46, 39–43.
Aguilera, A., Chávez, S., & Malagón, F. (2000). Yeast, 16, 731–754.
Gabriel, A., Dapprich, J., Kunkel, M., Gresham, D., Pratt, S. C., & Dunham, M. J. (2006). PLoS Genetics, 2, 2026–2038.
Ames, R. M., Rash, B. M., Hentges, K. E., Robertson, D. L., Delneri, D., & Lovell, S. C. (2010). Genome Biology and Evolution, 2, 591–601.
Fawcett, J. A., & Innan, H. (2011). Genes, 2, 191–209.
Sniegowski, P. D., & Gerrish, P. J. (2010). Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 365, 1255–1263.
Pallone, H., & Viikari, L. (2004). Biotechnology and Bioengineering, 86, 550–557.
Eijsink, V. G. H., Vaaje-Kolstad, G., Varum, K. M., & Horn, S. J. (2008). Trends in Biotechnology, 26, 228–235.
Kobayashi, H., Komanoya, T., Guha, S. K., Hara, K., & Fukuoka, A. (2011). Applied Catalysis A: General, 409–410, 13–20.
Fargues, C., Lewandowaki, R., & Lameloise, M. L. (2010). Industrial and Engineering Chemistry Research, 49, 9248–9257.
Sainio, T., Turku, I., & Heinonen, J. (2011). Bioresource Technology, 12, 6048–6057.
Dehkoda, A. (2008). Master Thesis, University College of Boras, Örnsköldsvik, Sweeden.
Jhadav, A., Vamsi, K. K., Khaimar, Y., Boraste, A., Gupta, N., et al. (2009). International Journal of Microbiology Research, 1, 9–12.
Tomás-Pejó, E., Oliva, J. M., Ballesteros, M., & Olsson, L. (2008). Biotechnology and Bioengineering, 100, 1122–1131.
Brethauer, S., & Wyman, C. E. (2010). Bioresource Technology, 101, 4862–4874.
Kim, S. R., Lee, K. S., Choi, J. H., Ha, S. J., Kweon, D. H., Seo, J. H., et al. (2010). Journal of Biotechnology, 150, 404–407.
Insa, G., Sablayrolles, J. M., & Douzal, V. (1995). Bioprocess and Biosystems Engineering, 13, 171–176.
Stanley, G. A., Douglas, N. G., Every, E. J., Tzanatos, T., & Pamment, N. B. (1993). Biotechnology Letters, 15, 1199–1204.
Zheng, D. Q., Wu, X. C., Tao, X. L., Wang, P. M., & Li, P. (2011). Bioresource Technology, 3, 3020–3027.
Pérez, F., Regodón, J. A., Valdés, M. E., de Miguel, C., & Ramírez, M. (2000). Food Microbiology, 17, 119–128.
Li, J., Zhao, J. B., Zhao, M., Yang, Y. L., Jiang, W. H., & Yang, S. (2010). Applied Microbiology, 50, 373–379.
Steen, E. J., Chan, R., Prasad, N., Myers, S., Petzold, C. J., Redding, A., et al. (2008). Microbial Cell Factories, 7, 36–43.
Wilkins, M. R., & Atiyeh, H. K. (2011). Current Opinion in Biotechnology, 22, 1–5.
Scott, S. A., Davey, M. P., Dennis, J. S., Horst, I., Howe, C. J., Lea-Smith, D. J., et al. (2010). Current Opinion in Biotechnology, 21, 277–286.
Brennan, L., & Owende, P. (2010). Renewable & Sustainable Energy Reviews, 14, 555–577.
Yan, Y., & Liao, J. C. (2009). Journal of Industrial Microbiology and Biotechnology, 36, 471–479.
Amyris: The latest on the U. C. Berkeley-spawned Agrofuel Firm 2011. Available from: www.berkeleydailyplanet.com. Accessed December 02, 2011.
Acknowledgements
The authors wish to express their gratitude to FAPESP for all the financial support given to their research on bioethanol production for so many years and particularly to the Bioenergy/FAPESP program, which has encouraged them to work on the production of bioethanol from sugarcane bagasse.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Laluce, C., Schenberg, A.C.G., Gallardo, J.C.M. et al. Advances and Developments in Strategies to Improve Strains of Saccharomyces cerevisiae and Processes to Obtain the Lignocellulosic Ethanol−A Review. Appl Biochem Biotechnol 166, 1908–1926 (2012). https://doi.org/10.1007/s12010-012-9619-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-012-9619-6