Abstract
Potato is one of the most important food crops in the world. Many plant transcription factors (TFs) have been demonstrated to be essential for improvement of plant stress tolerance traits. However, very few TFs were used for improving potato stress tolerance. In this study, we presented the characterization of a new potato StNAC2 gene. The StNAC2 protein contains five subdomains of NAC proteins and belongs to NAP subfamily. StNAC2 is constitutively expressed in potato leaves, stems, tubers, flowers and roots. Transcripts of StNAC2 were significantly induced by Phytophthora infestans, the causal agent pathogen of potato late blight. StNAC2 also could be induced by wounding, salt, drought as well as signal molecules such as salicylic acid and abscisic acid, suggesting that StNAC2 transcription factor involved in the signal transduction cascades in responses to abiotic and biotic stresses in potato. Overexpression of StNAC2 in transgenic potato significantly enhanced salt tolerance in vitro and drought tolerance in pot growing condition. Thus, the functional analysis of the new StNAC2 gene in this study will enrich knowledge for understanding the function of the NAC genes in potato stress tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osmotic stresses, such as drought or salinity, are major abiotic environmental stressors exacerbated by global climate change that limits plant growth and development, and thus causing important economic losses of agronomical yield. Potato (Solanum tuberosum L.) is the world’s fourth most important food crop (FAO 2008). Compared to other crops, the potato is generally considered to be drought sensitive (van Loon 1981). Drought stress influences the development and growth of potato leaves, stem, shoots, roots and tubers (Ojala et al. 1990). Potato production in many areas of the world is increasingly effected by limited water supplies. The identification of drought tolerance traits and genes in potato would facilitate breeding for yield stability under water-limiting conditions (Evers et al. 2010).
Plants have developed a diversity of mechanisms to deal with different abiotic stresses, such as drought, salt, heat and cold (Vinocur and Altman 2005). Recent advances in understanding the genetic control of drought tolerance offered new opportunities to develop crops that are less damaged by limited water supplies through genetic manipulation (Hussain et al. 2011; Cominelli and Tonelli 2010; Umezawa et al. 2006). Among them, transcription factors (TFs) play essential roles in plant stress responses by regulating their target genes through binding to the cognate cis-acting elements (Umezawa et al. 2006; Yamaguchi-Shinozaki and Shinozaki 2006). Many plant TFs, have been implicated in abiotic stress tolerance, belong to different families including AP2/EREBP (Dietz et al. 2010), DREB/CBF (Agarwal et al. 2006), bZIP (Kobayashi et al. 2008), MYB/MYC (Dubos et al. 2010), NAC (Puranik et al. 2012), C2H2-ZFP (Huang et al. 2007) and WRKY (Chen et al. 2012).
Efforts have been undertaken to generate drought and salinity tolerant potato by manipulating the expression of stress-responsive genes. Those genes include TPS and TPP genes from yeast or bacteria (Kondrák et al. 2011; Goddijn et al. 1997; Yeo et al. 2000); GDP from Pleurotus sajor-caju (Jeong et al. 2001); coda from Arthrobacter globiformis (Ahmad et al. 2008); oxo from barley (Turhan 2005); BADH from spinach (Zhang et al. 2012); GalUR from the strawberry (Hemavathi et al. 2012); AtNDPK2; DREB1A and P5CS from Arabidopsis (Tang et al. 2007; Behnam et al. 2006; Hmida-Sayari et al. 2005); StMYB1R-1, SOD and APX from potato (Shin et al. 2011; Tang et al. 2006). Most of those genes come from other species rather than potato. Although a larger number of plants TFs have shown center role in manipulating plant stress tolerance, very few were used for improving potato stress tolerance.
NAC proteins are plant-specific TFs which have been shown to play important roles in abiotic and biotic stress responses (Puranik et al. 2012). The NAC domain was identified based on the conserved sequences from Petunia NAM and Arabidopsis ATAF1/2 and CUC2 proteins (Aida et al. 1997). Many NAC candidate genes have been used to improve stress tolerance ability including drought, cold, salt and dehydration in wide-range plants species covering Arabidopsis thaliana (Park et al. 2011; Liu et al. 2011a, b), Oryza sativa (Song et al. 2011; Jeong et al. 2010; Hu et al. 2008), Triticum aestivum (Tang et al. 2012; Xue et al. 2011), Nicotiana tabacum (Liu et al. 2011a, b) and Glycine max (Hao et al. 2011) and Gossypium hirsutum (Meng et al. 2009).
Efforts to explore new genes in potato and utilize them to strengthen stress tolerance are of importance to potato production. NAC genes have emerged as important players in plant stress response. Up to now, only one characterized potato NAC gene StNAC which responds to Phytophthora infestans infection and wounding was reported by Collinge and Boller (2001). By analyzing the microarray data, we found that an EST similar to NAC gene was highly induced by biotic stimulus (Tian et al. 2006). However, the biological function of this NAC transcription factor (designated as StNAC2) in potato is currently unknown. In this study, we have focused on functional characterization of the potato StNAC2 gene using transgenic approach to evaluate its impact on drought and salt stress adaptation in potato. Transgenic potato plants overexpressing StNAC2 showed enhanced salt tolerance in vitro and drought tolerance in pot growing condition. Our results indicate that StNAC2 plays an important role in biotic and abiotic stress and may serve as a potential candidate gene for improving potato stress tolerance.
Materials and methods
Plant materials
E-potato-3 (Solanum tuberosum L.), a potato cultivar of China, was used as transgenic material. Plantlets were propagated in vitro on MS medium supplemented with 3 % sucrose and 0.8 % agar in transparent plastic boxes in a culture room with the 16 h light and 8 h dark growing period at 22 ± 2 °C.
Cloning of StNAC2 gene
Total RNA was isolated from E-potato-3 leaves using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. cDNA synthesis was performed using a SMART RACE cDNA amplification kit with SMART IV™ Oligonucleotide and CDSIII RACE Primers (Clontech, Palo Alto, CA, USA). Two gene-specific primers, GSP5 (5′-CCTAAAACTCCCTTTTCTCTCA-3′) and GSP3 (5′-TTAGCTCTGATCTTCCTCCTG-5′), were designed for rapid amplification of 3′ and 5′ cDNA ends according to the EST fragment sequence (GenBank accession No. CO267919) (Tian et al. 2006). RACE was performed according to protocol supplied by SMART RACE cDNA amplification kit (Clontech, Palo Alto, CA, USA) under the following PCR conditions: an initial 2 min at 94 °C and a final extension step of 72 °C for 5 min, each cycle consisted of 30 s at 94 °C, followed by 1 min at 60 °C and 1 min at 72 °C. The PCR reaction was performed for 32 cycles using Pyrobest™ DNA polymerase (Takara BIO INC, Japan). Nucleotide sequences were compared using bioinformatics software from NCBI (http://www.ncbi.nlm.nih.gov) and predicted amino acid sequences were aligned using the GeneDoc (http://www.psc.edu/biomed/genedoc). Phylogenetic analyses based on the resulting alignments were then constructed using the neighbor-joining method with the MEGA4 programme (http://www.megasoftware.net/mega4).
Analysis of gene expression
To investigate the organ-specific expression of the StNAC2, total RNAs were extracted from various tissues including leaves, stems, roots, flowers and tubers from 7-week-old greenhouse-grown potato plants. Total RNAs were isolated from samples using RNeasy plant mini kit (Mycomebio, Beijing, China) combined with a DNase digestion procedure (Takara, Dalian, China) to ensure DNA-free RNA preparations. DNase-treated RNA samples were used for reverse transcription (RT) of the first-strand cDNA using the ReverTra Ace Kit (Toyobo, Osaka, Japan) according to the manufacturer’s instructions. A ten-fold dilution of the reverse transcript cDNA was used as template for each RT-PCR reaction. The StNAC2 specific forward primer 1: 5′-TTATCAGTGCCAGTAGCCTTC-3′ and StNAC2 specific reverse primer 2: 5′-TTAGCTCTGATCTTCCTCCTG-3′ were used to perform RT-PCR amplification (expected length is 277 bp). The constitutively expressed β-tubulin gene was used as internal standard for each RT-PCR. β-tubulin forward primer: 5′-TTGGACAGTCTGGTGCTGGGAATA-3′, β-tubulin reverse primer: 5′-TGGCCAGGGAATCTCAAACAGCAAG-3′ (the amplified length is 481 bp). PCR was performed in a total volume of 20 μl containing: 1.5 mM MgCl2, 0.25 mM dNTPs, 0.5 mM of each forward and reveres primers, 2 μl reaction buffers, 1 U Taq DNA polymerase (Takara, Dalian, China) and 1 μl template. The cycling parameters of PCR amplification were as follows: the reaction was performed for 35 cycles with an initial denaturation step at 94 °C for 3 min and a final extension at 72 °C for 5 min, each cycle consisted of 30 s at 94 °C followed by 30 s at 55 °C and 1 min at 72 °C. The products were separated by electrophoresis on 1 % agarose gels.
To determine whether StNAC2 expression is induced by abiotic stresses and/or exogenous plant regulators, we collected cDNA microarray data from our unpublished data to investigate StNAC2 expression profile under various treatments. The details of cDNA microarray procedures were described in previous reports by Wang et al. (2005).
Vector construction and transformation of potato
For construction of over expression vector, the StNAC2 coding region was PCR amplified from cDNA with the primers NAC2EXF: 5′-CCGGATCCCCTAAAACTCCCTTTTCTCTCA-3′ and NAC2EXR: 5′-CCGAGCTCGTTGTCATAAACTTCTGATGTCACC-3′ which add BamHI and SacI restriction sites separately. The PCR fragments were cloned into a pMD-18 cloning vector (Takara, Dalian, China) to obtain pMD-18-StNAC2. After confirmation by sequencing, double digested StNAC2 coding region was inserted into BamHI and SacI sites of the pBI121 vector by replacing the GUS gene. The StNAC2 expression vector under the control of the CaMV 35S promoter was introduced into A. tumefaciens strain LB4404 by the freeze–thaw method and then to transform potato microtuber slices according to the protocol reported by Si et al. (2003).
Confirmation of transgenic potato plants
Regenerated shoots from transformed microtuber slices were cultured on rooting medium with 200 mg/l kanamycin and 250 mg/l carbenicillin. Genomic DNA was isolated from the leaves of well rooted and wild-type potato plants. Isolated DNA was subjected to PCR analysis to amplify the nptII gene using nptII specific primers. The PCR conditions were the same as those described in gene expression part.
Southern blot analysis was conducted to confirm the stable integration of transgenes into potato genomic DNA. Hybridization probes were prepared from a 520 bp nptII gene fragment which was labeled using the DIG DNA labeling kit (Roche Molecular Biochemicals, Germany). DNA isolation and digestion, pre-hybridization, hybridization, washing conditions and hybridization signal detection were same as the procedures described by Tian et al. (2010). Pst I enzyme was used to digest genomic DNA.
Real-time RT-PCR was used to detect transgene expression levels. In vitro cultured young leaves collected from transgenic and control plants were used to isolate total RNA for analyses of the StNAC2 expression level. The qStNAC2 specific forward primer: 5′-CATGCATGAATATCGTTTGAGTG-3′ and qStNAC2 specific reverse primer 2: 5′-CAACTTTCATCATCTCTATAGTT-3′ were used to perform real-time RT-PCR amplification (expected length is 140 bp) on the Bio-RAD CFX Connect optics module PCR System using SYBR Premix Ex Taq™ (TaKaRa, Dalian, China). The constitutively expressed ef-1α was used as internal reference gene for calculating relative transcript levels. Forward primer: 5′-GGAAACGGATATGCTCCA-3′, reverse primer: 5′-CTTACCTGAACGCCTGTCA-3′ (the amplified length is 101 bp). The PCR thermal cycle conditions were as following: denature at 95 °C for 3 min and 40 cycles for 95 °C, 10 s, 60 °C, 20 s, 72 °C, 15 s. The relative quantification value was calculated by the 2−∆∆CT method.
Assessment of salt tolerance of transgenic potato plants in vitro
The shoot tips of transgenic and control plants were cultured in plastic boxes containing 20 ml MS medium supplemented with 150 mM NaCl and kept in a culture room with the 16 h light and 8 h dark growing period at 22 ± 2 °C. After 4 weeks of the salt treatment, the plantlets were harvested and the heights, fresh weight, root numbers, root length of plantlets were measured. Each treatment contained 25 plantlets which were cultured in 5 boxes.
Evaluation of transgenic plants for drought stress tolerance
Tubers of transgenic and control potato plants were grown in plastic pots (Φ 20 cm) containing 2.5 kg compost soil mix in a naturally illuminated greenhouse for 5 weeks. Then, the selected 30 uniform plants of each line were moved from greenhouse to a well-sheltered environment. One repeat contains 10 plants per genotype. Same volume of water was irrigated in each pot in each time without outflow. Water was withheld from the plants being drought stressed. Drought tolerance was evaluated by relative wilt percentage of plant leaves 3, 5, 8 days after water deficit stress. Finally, the plants were allowed to recover by rewatering. Three rounds of water withholding were conducted.
Statistical analysis
All data were analyzed by analysis of variance using SAS statistics program. Statistical differences are referred to as significant when P < 0.05 or 0.01.
Results
StNAC2 gene cloning and structural features
A 1,257 bp full-length StNAC2 cDNA was obtained by assembling the overlapping 3′-RACE and 5′-RACE sequences. The ORF comprises 849 nucleotides and encodes a predicted protein of 282 amino acids (Fig. 1a). The ORF is flanked by 81 bp 5′-UTR and a 3′-UTR which is 327 nucleotides long including the poly (A) tail. The StNAC2 sequence has been deposited in GenBank under accession No. EF091874. NCBI Blastp results showed that the predicted protein contains no apical meristem (NAM) domain in the N-terminus from 9 to 135 amino acids, indicating it is a NAC family protein (Fig. 1a).
Amino acid sequences alignment of StNAC2 with four NAC proteins and phylogenetic tree of 25 plant NAC proteins. a The NAM domain predicted by Blastp. b Amino acid sequence comparison of five plant NAC proteins. The NAM domain is shown by lines above the amino acid sequences. Identical amino acids are shaded in black, and similar amino acids are shaded in gray. Sequences were aligned using Clustal X and GeneDoc. c Phylogenetic analyses were conducted in MEGA4 using the neighbor-joining method. The numbers beside each node represent bootstrap values (≥50 %) based on 1,000 replications. The scale bar indicates the relative amount of change along branches. The five sub-subfamilies distinguished within the NAP, OsNAC3, ATAF, AtNAC3, and NAM subfamilies are shown in gray rectangles. The GenBank accession numbers for amino acid sequences are as follows: A. thaliana ATNAC2 (NP_188170), AtNAM (AAD17314), CUC1(BAB20598), CUC2 (BAA19529), CUC3 (AAP82630), NAP (CAA10955), ANAC047 (NP_187057), ATAF1 (NP_171677), ANAC055 (AAM61076), ATAF2 (CAC35884), AtNAC2 (BAB20600), AtNAC3 (BAB20599); Oryza sativa ANAC019 (AAT02360), SNAC1 (ABD52007), OsNAC1 (BAC53810), OsNAC2 (BAC53811), OsNAC3 (BAA89797), OsNAC4 (BAA89798), OsNAC6 (BAA89800); Petunia hybrida NAM (CAA63102); from soybean: GmNAC3 (AAX85980); Citrus sinensis CsNAC (ABQ96643); Solanum lycopersicum SlNAC (AAR88435); Solanum tuberosum StNAC2 (ABK96797.1) and StNAC (CAC42087); Capsicum annuum CaNAC1 (AAW48094)
Phylogenetic analysis of several NAC proteins
Based on an amino acids alignment of five plant NAC proteins (Fig. 1b), higher sequence similarities were found in the N-terminus rather than in the C-terminus. Amino acid sequence comparison of five plant NAC proteins showed that StNAC2 shares 38.4, 38.1 and 37.9 % similarity with StNAC (CAC42087), SlNAC (AAR88435) and CaNAC1 (AAW48094), respectively. StNAC2 shares higher 54.6 % similarity with the Arabidopsis NAP (CAA10955) rather than another potato NAC protein StNAC.
NAC proteins could be classified into several subfamilies on the basis of similarities in NAC domains (Puranik et al. 2012). We reconstructed a phylogenetic tree from the amino acid sequences of 25 NAC proteins. Phylogenetic analysis showed that the 25 NAC proteins belong to five NAC subfamilies. StNAC2 was clustered into the NAP subfamily, while another StNAC from potato clustered into ATAF group. It is clear that StNAC and StNAC2 belong to different group of NAC family (Fig. 1c), indicating that StNAC2 is a new member of potato NAC proteins.
Expression of StNAC2 in different organs and response to biotic and abiotic stimuli
RT-PCR was conducted to determine StNAC2 expression levels in various potato tissues at the flowering stage. The results showed that StNAC2 was constitutively expressed in leaves, stems, tubers, flowers and roots (Fig. 2a). The expression levels in leaves and stems were stronger than in tubers, flowers and roots.
Expression of StNAC2 in different organs and response to biotic and abiotic stresses and signal molecules. a Expression of StNAC2 in leaves stems, roots, tubers and flowers in potato. RT-PCR was performed with specific primers of StNAC2. b Expression of StNAC2 in response to biotic and abiotic stresses and signal molecules. For P. infestans treatments, the third and fourth fully expanded leaves of 6-week-old plants were detached and inoculated with zoospores of P. infestans as described previously (Tian et al. 2006). For signal molecules treatments, fully expanded leaves from 6-week-old greenhouse-grown potato plants were detached and sprayed with 10 mM salicylic acid (SA), 100 μM abscisic acid (ABA). Leaves sprayed with distilled water were used as a control. For wounding treatment, detached leaves were incised with a blade several times. Samples were collected and frozen in liquid nitrogen and stored at −70 °C before total RNA isolation at the indicated times. Microarray results from our unpublished data, the experimental design is same as described previously by Wang et al. (2005). Error bars show SD from three biological repeats
The induction patterns of StNAC2 response to biotic and abiotic stimuli are shown in Fig. 2b. The results indicated that StNAC2 expression was significantly induced by P. infestans and peaked at 8 h after inoculation. After mechanical wounding, accumulation of StNAC2 transcripts was increased after 8 h and kept until 36 h. Expression profiling showed that StNAC2 was up-regulated by ABA at an increasing rate from 2 to 12 h and reached peak at 12 h. After that, slight decline was observed at the 24-h time point and then increased slightly. Induction patterns of SA showed a peak at 2–8 h and then declined. ETH and MeJA induced StNAC2 expression at a low level with little variation in abundance during the time course (data not shown). The results demonstrated that StNAC2 is responsible for biotic and abiotic stresses.
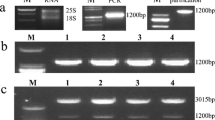
Confirmation of transformates by PCR and Southern blotting
After Agrobacterium-mediated transformation using CaMV 35S-StNAC2 expression cassettes, twelve independent kanamycin-resistant putatively transformed potato lines were generated and further confirmed by PCR analysis using nptII specific primers and Southern blotting. Partial results are shown in Fig. 3a, b. The insertion number of transgene is about 1–2 copies in three selected transgenic overexpression lines OE2, OE3 and OE4 (Fig. 3b). Real-time RT-PCR revealed that transgenic lines OE2, OE3 and OE4 displayed constitutively elevated expression of StNAC2 and were used for further investigation (Fig. 3c).
Molecular characterization of transgenic potato lines. a PCR analyses of genomic DNA to detect the presence of nptII gene in putative transgenic plants. The 450 bp fragment of the nptII gene was amplified by the nptII specific primers. Lane M: molecular marker (DL2000), Lane P: plasmid (positive control), Lane CK: negative control and Lane OE1-OE4: four selected putative overexpressing transgenic lines. b Southern blot analysis of Pst I digested genomic DNA isolated from three different transformants and control plants and hybridized with the DIG labeled nptII probe. Lanes P represent plasmid (positive control), Lanes CK and OE: DNA samples isolated from control and three transgenic potato plants. c qRT-PCR analysis of relative transcript levels of StNAC2 in four transgenic lines under normal conditions. Transcript levels of control (CK) were used as reference, which was set at a relative expression level of ‘1’. Error bars indicate the standard error; the experiments were repeated three times alone with at least five independent repetitions of the biological experiments. Asterisks above each column indicate a significant difference (P < 0.01) between control and transgenic lines
Transgenic plants confers salt tolerance in vitro
We analyzed the salt tolerance of transgenic potato in vitro. As shown in Fig. 4, control and transgenic potato lines (OE2, OE3 and OE4) grew normally on MS media without NaCl. When they grew on media supplemented with 150 mM NaCl, the growth of both was inhibited, however, growth performance of transgenic potato lines is better than control plants (Fig. 4a). We further tested the gene expression level under the salt stress. Compared to in vitro plants grown on MS media, StNAC2 gene expression levels were up-regulated both in control and transgenic potato lines, but gene expression level is still significantly high in transgenic potato lines than in control plants (Fig. 4b). As reflected in quantitative estimation (Table 1), the root numbers, height of plants of stressed transgenic potato lines were significantly higher compared with controls. Root length and fresh weight of transgenic potato lines are still higher than control, even if difference is not significant in all three transgenic plants. The results indicated that the transgenic potato plants conferred tolerance to salt stress in vitro.
Overexpression of StNAC2 in potato plants confers salt tolerance in vitro. a Appearance of transgenic and control plants after salt stress. Plants were grown on MS media supplemented with or without 150 mM NaCl for 4 weeks. OE2, OE3 and OE4 represent overexpressing transgenic lines. b qRT-PCR analysis of relative transcript levels of StNAC2 in transgenic lines under salt stress. Bar represents the standard error of three biological repeats. Asterisks above each column indicate a significant difference (P < 0.01) between control and transgenic lines
Overexpression of StNAC2 in transgenic potato enhances tolerance to drought stress
To determine whether the transgenic lines confer tolerant to drought conditions, control and transgenic plants (OE2, OE3 and OE4) were grown in the same pots and growing conditions. Under normal growth conditions, three transgenic lines showed no obvious abnormal morphological phenotype compared with control. Five weeks later, water was withheld for up to 8 days and the wilt percentages of leaves were measured. Control plants started to show wilted symptoms after 3 days post-treatment (Fig. 5a). Three overexpression lines were slightly wilted, but the wilting percentage of leaves is less than that of control (Fig. 5d). With the development of water deficiency, the degree of wilting of leaves on both control and transgenic plants became severe. But wilting symptom in transgenic lines is less severe compared with control. After 8 days of drought stress, control plants were wilted around 100 % percentage, while the transgenic plants showed about 90 % wilting percentage (Fig. 5d). After plants were totally wilted, the pots were watered fully again, and then the second round water withholding was performed. The transgenic plants recovered faster than control. Similar to the first round of water withholding, the transgenic lines show less wilting symptom than the control plants. After three rounds of water withholding the transgenic lines survived but display chlorotic leaves, while control plants are almost dead (Fig. 5b). Expression pattern of StNAC2 under drought stress condition was tested by real-time RT-PCR on potato leaves. As shown in Fig. 5c, StNAC2 expression was gradually increased after water withholding both in the control and transgenic lines. But the transcripts increased significantly in three transgenic plants. The most elevation follows 5 days water deficiency. After that, transcripts of StNAC2 are still keeping in higher level in transgenic plants. The high level expression StNAC2 is consistent with low wilting percentage. These results revealed that overexpression of StNAC2 in transgenic potato enhances tolerance to drought stress.
Drought tolerance assays of StNAC2-overexpressing transgenic potato. A water withholding assay was performed with 5-week-old plants for three rounds (8 × 3 days) up to 24 days. a Plants performance after 3 days at first round water withholding. b Performance of plants recovered after the third round water withholding. c qRT-PCR analysis of relative transcript levels of StNAC2 in three transgenic lines under drought stress condition. d Wilt percentage of transgenic potato plants after 3, 5 and 8 days when water was withheld at first round. In (a) and (b), OE2, OE3 and OE4 represent three StNAC2-overexpressing transgenic lines. CK is non-transgenic control potato. In (c) error bars indicate the standard error; Asterisks above each column indicate a significant difference (P < 0.01) between control and transgenic lines. In (d), each bar represents the standard error of three independent experiments (n = 10). Asterisks above each column indicate a significant difference (P < 0.05) between control and transgenic lines
Discussion
Recently, 110 NAC genes were identified in potato and several NAC genes were highly induced by abiotic stress (Singh et al. 2013). Up to date, only one potato NAC gene StNAC was characterized in detail. StNAC was rapidly and strongly induced by wounding and P. infestans (Collinge and Boller 2001). In this study, we have cloned and characterized another potato StNAC2 gene. StNAC2 is a NAP family gene rather than ATAF family to which StNAC belongs. In this regard, StNAC2 is a new NAC gene cloned from potato in the present research. Since the identification of first NAC domain gene NAM from Petunia hybrid which involved in shoot apical meristem development (Souer et al. 1996), many NAC proteins have been reported to be involved in processes of plant development, for example, lateral root formation (Xie et al. 2000), cell expansion of specific flower organs (Sablowski and Meyerowitz 1998) and secondary wall thickening in woody tissues (Mitsuda et al. 2007). Besides their roles in plant development, NAC domain genes are also involved in various plant stress response including water, salt, cold, wounding, insect feeding and pathogen infection (Collinge and Boller 2001; Hegedus et al. 2003; Nogueira et al. 2005; Oh et al. 2005; Lin et al. 2007). In this study, we found that StNAC2 expression was significantly induced by potato late blight agent pathogen P. infestans. Besides, StNAC2 could be induced by wounding, salt and drought as well as signal molecules including ABA and SA (Figs. 2, 4, 5). Recent studies suggest a crosstalk between plant responses to pathogens and abiotic stresses (Abuqamar et al. 2009; Mauch-Mani and Flors 2009). Thus, StNAC2 may play important role in the signal transduction network responses to abiotic and biotic stresses in potato. ABA signal pathway is well documented involving in abiotic stresses such as drought, low temperature and osmotic stress (Danquah et al. 2014). Expression analysis revealed StNAC2 transcription was induced by ABA treatment (Fig. 3b), so we presume that StNAC2 might play roles in potato responses to abiotic stress, probably through ABA-dependent pathway.
Over expression of many NAC genes has been shown to improve drought, salt and cold tolerance in transgenic plants (Puranik et al. 2012), which promotes us to investigate function of StNAC2 in potato drought and salt stresses. Our results showed that overexpression of StNAC2 in transgenic potato significantly enhanced salt tolerance in vitro and drought tolerance in pot growing condition. One of the problems is the constitutive overexpression of stress-related genes including NAC genes that often cause abnormal development or yield penalty (Priyanka et al. 2010; Kasuga et al. 1999; Nakashima et al. 2007). We did not find obvious developmental differences between the transgenic plants and the wild-type plants under normal growth condition. However, we still cannot rule out the possibility that StNAC2 plays a role in potato development.
Although the molecular bases of the improved stress tolerance of the StNAC2 transgenic potato have not been completely resolved in this study, our data suggest that the biological role of StNAC2 is mainly associated with plant adaptation to abiotic and biotic stresses. Overexpression of this gene could enhance drought and salt tolerance in potato, making it a potential candidate for engineering stress tolerant potato. The growth and productivity of potato are often threatened by environmental factors, such as drought, salt, and cold. Further study of the role of the StNAC2 gene in responding to different stresses may contribute to a deeper understanding of the cross-reactions of potato plants to multiple abiotic stresses. Especially it is worth to test the ability of the late blight disease resistance on overexpression plants, since StNAC2 could be significantly induced by P. infestans. Whether the overexpression of StNAC2 could achieve the ultimate goal of improved yield in potato in drought- and salt-prone environments awaits field trials in the future.
Author contribution
Qinfen Xu: Gene cloning, transformation. Evaluation of salt tolerance in in vitro transgenic plants. Qin He: Gene expression analysis, qRT-PCR. Southern blotting. Shuai Li: Evaluation of drought tolerance of transgenic plants. Zhendong Tian: Design experiments. Phylogenetic analysis. Writing MS.
Abbreviations
- TFs:
-
Transcription factors
- RACE:
-
Rapid amplification of cDNA end
- NAC proteins:
-
NAM/ATAF/CUC proteins
- SA:
-
Salicylic acid
- ABA:
-
Abscisic acid
References
Abuqamar S, Luo H, Laluk K, Mickelbart MV, Mengiste T (2009) Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J 58:347–360
Agarwal PK, Agarwal P, Reddy MK, Sopory SK (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25:1263–1274
Ahmad R, Kim MD, Back KH, Kim HS, Lee HS, Kwon SY, Murata N, Chung WI, Kwak SS (2008) Stress-induced expression of choline oxidase in potato plant chloroplasts confers enhanced tolerance to oxidative, salt, and drought stresses. Plant Cell Rep 27:687–698
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Behnam B, Kikuchi A, Celebi-Toprak F, Yamanaka S, Kasuga M, Yamaguchi-Shinozaki K, Watanabe KN (2006) The Arabidopsis DREB1A gene driven by the stress-inducible rd29A promoter increases salt-stress tolerance in proportion to its copy number in tetrasomic tetraploid potato (Solanum tuberosum). Plant Biotechnol 23:169–177
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819:120–128
Collinge M, Boller T (2001) Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol 46:521–529
Cominelli E, Tonelli C (2010) Transgenic crops coping with water scarcity. New Biotechnol 27:473–477
Danquah A, de Zelicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32:40–52
Dietz KJ, Vogel MO, Viehhauser A (2010) AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signaling. Protoplasma 245:3–14
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Evers D, Lefevre I, Legay S, Lamoureux D, Hausman JF, Rosales ROG, Marca LRT, Hoffmann L, Bonierbale M, Schafleitner R (2010) Identification of drought-responsive compounds in potato through a combined transcriptomic and targeted metabolite approach. J Exp Bot 61:2327–2343
FAO (2008) International year of potato. Food and Agriculture Organization of the United Nations, Rome. http://www.potato2008.org/en/index.html
Goddijn OJ, Verwoerd TC, Voogd E, Krutwagen RW, de Graaf PT, van Dun K, Poels J, Ponstein AS, Damm B, Pen J (1997) Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol 113:181–190
Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, Ma B, Zhang JS, Chen SY (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J 68:302–313
Hegedus YuDM, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397
Hemavathi, Upadhyaya CP, Akula N, Kim HS, Jeon JH, Ho OM, Chun SC, Kim DH, Park SW (2012) Biochemical analysis of enhanced tolerance in transgenic potato plants overexpressing d-galacturonic acid reductase gene in response to various abiotic stresses. Mol Breed 28:105–115
Hmida-Sayari A, Gargouri-Bouzid R, Bidani A, Jaoua L, Savoure A, Jaoua S (2005) Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers salt tolerance in transgenic potato plants. Plant Sci 169:746–752
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Huang J, Yang X, Wang MM, Tang HJ, Ding LY, Shen Y, Zhang HS (2007) A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochem Biophys Acta 1769:220–227
Hussain SS, Kayani MA, Amjad M (2011) Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol Prog 27:297–306
Jeong MJ, Park SC, Byun MO (2001) Improvement of salt tolerance in transgenic potato plants by glyceraldehyde-3 phosphate dehydrogenase gene transfer. Mol Cells 12:185–189
Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Choi DY, Kim M, Reuzeau C, Kim JK (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291
Kobayashi F, Maeta E, Terashima A, Kawaura K, Ogihara Y, Takumi S (2008) Development of abiotic stress tolerance via bZIP-type transcription factor LIP19 in common wheat. J Exp Bot 59:891–905
Kondrák M, Marincs F, Kalapos B, Juhász Z, Bánfalvi Z (2011) Transcriptome analysis of potato leaves expressing the trehalose-6-phosphate synthase 1 gene of yeast. PLoS One 6:e23466
Lin R, Zhao W, Meng X, Wang M, Peng Y (2007) Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci 172:120–130
Liu QL, Xu KD, Zhao LJ, Pan YZ, Jiang BB, Zhang HQ, Liu GL (2011a) Overexpression of a novel Chrysanthemum NAC transcription factor gene enhances salt tolerance in tobacco. Biotechnol Lett 33:2073–2082
Liu X, Hong L, Li XY, Yao Y, Hu B, Li L (2011b) Improved drought and salt tolerance in transgenic Arabidopsis overexpressing a NAC transcriptional factor from Arachis hypogaea. Biosci Biotechnol Biochem 75:443–450
Mauch-Mani B, Flors V (2009) The ATAF1 transcription factor: at the convergence point of ABA-dependent plant defense against biotic and abiotic stresses. Cell Res 19:1322–1323
Meng CM, Cai CP, Zhang TZ, Guo WZ (2009) Characterization of six novel NAC genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci 176:352–359
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress responsive gene expression in rice. Plant J 51:617–630
Nogueira FTS, Schlogl PS, Camargo SR, Fernandez JH, Rosa Junior VE de, Pompermayer P, Arruda P (2005) SsNAC23, a member of the NAC domain protein family, is associated with cold, herbivory and water stress in sugarcane. Plant Sci 169:93–106
Oh SK, Lee S, Yu SH, Choi D (2005) Expression of a novel NAC domain-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens. Planta 222:76–887
Ojala JC, Stark JC, Kleinkopf GE (1990) Influence of irrigation and nitrogen management on potato yield and quality. Am Potato J 67:29–43
Park J, Kim YS, Kim SG, Jung JH, Woo JC, Park CM (2011) Integration transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol 156:537–549
Priyanka B, Sekhar K, Reddy VD, Rao KV (2010) Expression of pigeonpea hybrid-proline-rich protein encoding gene (CcHyPRP) in yeast and Arabidopsis affords multiple abiotic stress tolerance. Plant Biotechnol J 8:76–87
Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381
Sablowski RW, Meyerowitz EM (1998) A homolog of no apical meristem is an immediate target of the floral homeotic genes APETAL3. Cell 92:93–103
Shin D, Moon SJ, Han S, Kim BG, Park SR, Lee SK, Yoon HJ, Lee HE, Kwon HB, Baek D, Yi BY, Byun MO (2011) Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought tolerance. Plant Physiol 155:421–432
Si HJ, Xie CH, Liu J (2003) An efficient protocol for Agrobacterium-mediated transformation with microtuber and the introduction of an antisense class I patatin gene into potato. Acta Agronomica Sinica 29:801–805
Singh AK, Sharma V, Pal AK, Acharya V, Ahuja PS (2013) Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res 20:403–423
Song SY, Chen Y, Chen J, Dai XY, Zhang WH (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234:331–345
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85:159–170
Tang L, Kwon SK, Kim SH, Kim JS, Choi JS, Cho KY, Sung CK, Kwak SS, Lee HS (2006) Enhanced tolerance of transgenic potato plants expressing both superoxide dismutase and ascorbate peroxidase in chloroplasts against oxidative stress and high temperature. Plant Cell Rep 25:1380–1386
Tang L, Kim MD, Yang KS, Kwon SK, Kim SH, Kim JS, Yun DJ, Kwak SS, Lee HS (2007) Enhanced tolerance of transgenic potato plants overexpressing nucleoside diphosphate kinase against multiple environmental stresses. Transgenic Res 17:705–715
Tang Y, Liu M, Gao S, Zhang Z, Zhao X, Zhao C, Zhang F, Chen X (2012) Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol Plant 144:210–224
Tian ZD, Liu J, Wang BL, Xie CH (2006) Screening and expression analysis of Phytophthora infestans induced genes in potato leaves with horizontal resistance. Plant Cell Rep 25:1094–1103
Tian ZD, Liu J, Zhang Y, Xie CH (2010) Novel potato C2H2-type zinc finger protein gene StZFP1, which responses to bio- and abiotic-stresses, play a role in salt tolerance. Plant Biol 12:689–697
Turhan H (2005) Salinity response of transgenic potato genotypes expressing the oxalate oxidase gene. Turk J Agric 29:187–195
Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol 17:113–122
van Loon CD (1981) The effect of water stress on potato growth, development, and yield. Am Potato J 58:51–69
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress, achievements and limitations. Curr Opin Biotechnol 16:123–132
Wang BL, Liu J, Tian ZD, Song BT, Xie CH (2005) Monitoring the expression profiles of genes associated with quantitative resistance to late blight in R-gene-free potato by cDNA microarray analysis. Plant Sci 169:1155–1167
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036
Xue GP, Way HM, Richardson T, Drenth J, Joyce PA, McIntyre CL (2011) Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4:697–712
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yeo ET, Kwon HB, Han SE, Lee JT, Ryu JC, Byu MO (2000) Genetic engineering of drought resistant potato plants by introduction of the trehalose-6-phosphate synthase (TPS1) gene from Saccharomyces cerevisiae. Mol Cells 10:263–268
Zhang N, Si HJ, Wen G, Du HH, Liu BL, Wang D (2012) Enhanced drought and salinity tolerance in transgenic potato plants with a BADH gene from spinach. Plant Biotechnol Rep 5:71–77
Acknowledgments
This work was partially supported by the National High Technology Project of China (2013AA102603), National Natural Science Foundation of China (31171603), Doctoral Fund of Ministry of Education of China (20110146110019) and the Fundamental Research Funds for the Central Universities (2011PY145).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Wang.
Rights and permissions
About this article
Cite this article
Xu, Q., He, Q., Li, S. et al. Molecular characterization of StNAC2 in potato and its overexpression confers drought and salt tolerance. Acta Physiol Plant 36, 1841–1851 (2014). https://doi.org/10.1007/s11738-014-1558-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-014-1558-0