Abstract
The plant-specific NAC (for NAM, ATAF1, 2 and CUC2) transcription factors (TFs) have been implicated in different cellular processes involved in stress responses such as cold, high salinity or drought as well as abscisic acid (ABA) signalling. However, the roles of the chrysanthemum NAC TF genes in plant stress responses are still unclear. A full-length cDNA designated DgNAC1, containing a highly conserved N-terminal DNA-binding NAC domain, has been isolated from chrysanthemum by RACE (rapid amplification of cDNA ends). It encodes a protein of 284 amino acids residues (=~32.9 kDa) and theoretical pI of 7.13. The transcript of DgNAC1 was enriched in roots and flowers than in stems and leaves of the adult chrysanthemum plants. The gene expression was strongly induced by ABA, NaCl, drought and cold treatment in the seedlings. Subcellular localization revealed that DgNAC1:GFP fusion protein was preferentially distributed to nucleus. To assess whether DgNAC1 is a practically useful target gene for improving the stress tolerance of chrysanthemum, we ectopically over-expressed the full-length DgNAC1 cDNA in tobacco and found that the 35S:DgNAC1 transgenic tobacco exhibited a markedly increased tolerance to salt. Despite this increased salt stress tolerance, the transgenic tobacco showed no detectable phenotype defects under normal growth conditions. These results proposed that DgNAC1 is appropriate for application in genetic engineering strategies aimed at improving salt stress tolerance in chrysanthemum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
High salt is the major environmental stress inhibiting plant growth and development and limiting the geographical distribution and utilization of plants. Plants respond to survive for salt stress via a series of molecular, physiological, and cellular processes culminating in salt stress tolerance. In these responses and adaptations, many salt-inducible genes are induced (Bartels and Sunkar 2005; Chinnusamy et al. 2004; Knight and Knight 2001, Shinozaki and Yamaguchi-Shinozaki 2007). Among them, transcription factors (TFs) are regarded as master switch which regulate stress-responsive genes and function in establishing stress tolerances (Nakashima et al. 2009; Vinocur and Altman 2005; Zhu 2002). Among these TFs, some members of NAC, AP2/EREBP, bZIP, MYB, MYC, WRKY, Cys2/His2-type zinc finger proteins families paly crucial roles in regulating defense responses to biotic and environmental stress (Agarwal et al. 2006; Chinnusamy et al. 2006; Umezawa et al. 2006).
The NAC domain-containing proteins, which contain a highly conserved N-terminal DNA-binding NAC domain and a variable transcriptional regulation C-terminal domain, constitute one of the largest families of plant-specific TFs. They are involved in a wide range of processes such as abiotic and biotic stress responses, plant development, and senescence (Aida et al. 1997; Duval et al. 2002; Olsen et al. 2005; Ooka et al. 2003; Zheng et al. 2009). A number of stress-responsive NAC domain-containing proteins have been identified in various plant species. Several studies showed that overexpression of some NAC genes altered the expression of many stress-related genes in transgenic plants and enhanced tolerance to salt, cold, and/or dehydration stresses (Hu et al. 2006, 2008; Nakashima et al. 2007; Ohnishi et al. 2005; Tran et al. 2004). However, the roles of the NAC genes in plant stress responses are still not well known.
Chrysanthemum is one of the most famous ornamental species in the world. However, high soil salinity is a serious threat to chrysanthemum growth and production in the cutting-chrysanthemum industry. To improve salt tolerance in chrysanthemum, the isolation and functional characterization of a novel salt-responsive NAC TF gene, DgNAC1 from chrysanthemum is reported here. In addition to salt stress, DgNAC1 was induced by ABA, drought, and cold treatments. The 35S:DgNAC1 transgenic tobacco improved tolerance to continuous salt stress from stress assays.
Materials and methods
Plant materials and stress treatments
Chrysanthemum (Dendronthema grandiform) cv. Jinba seedlings growing in greenhouse were exposed to air on filter paper for dehydration, or subjected to 4°C for cold stress. For salt and ABA treatments, seedlings were placed in 250 mM NaCl and 0.1 mM ABA, respectively (Tong et al. 2009). All excised leaf samples of control and treated plants were taken for treatment for up to 24 h, frozen immediately in liquid N2, and stored at −80°C for RNA extraction.
Isolation of the DgNAC1 gene
For 3′-RACE, one primer was designed GSP1 (5′-ITGGATAATGCAIGAGTAICGIT-3′) corresponding to conversed regions of the amino acid WIMHEYR. Primers for 5′ RACE were: GSP2, 5′-GTATTGATAGTGGCGGATGTGACGA-3′, and GSP3, 5′-CATATGTGTTGATTGTACATCCGAG-3′. The RACE reactions were performed according to the the manufacturer’s protocol (Invitrogen RACE cDNA amplification kit). A single full-length cDNA sequence by combining the 5′-RACE fragment and 3′-RACE fragment was obtained. Finally, a pair of primers (F1, 5′-TCCAAAGAGCGCAGACTAGCCACTGAGC-3′, and F2, 5′-AATAGTCAATAAGCCCACTTGCTTA-3′) was then designed from the putative 5′- and 3′-untranslated region (UTR) of the full-length cDNA sequence. The resultant DNA fragments and RACE products were purified by agarose gel and cloned into pMD18-T Vector (Takara) and sequenced (Invitrogen, Beijing).
RNA isolation and semi-quantitative RT-PCR assay
Total RNA from various chrysanthemum tissues was extracted by Trizol. The first strand cDNA was synthesized with 1 μg total RNA and 1 μl Superscript II enzyme (Invitrogen, USA) according to the manufacturer’s protocol. As a control, 18s rRNA gene was amplified from chrysanthemum various tissues. The primers used for detecting DgNAC1 gene expression were: forward 5′-GCAGATAAGCCAATTGGAAAGCCGA-3′, and reverse 5′-ATGGAGAGAGTTGAAAGTCATTGTA-3′. The PCR was performed as follows: pre-denaturation at 94°C for 5 min, followed by 34 cycles of 30 s at 94°C, 30 s at 59°C, 50 s at 72°C for DgNAC1, 31 cycles for 18s rRNA and a final extension of 8 min at 72°C. The amplified products were resolved in a 1.2% agarose gel and then detected by agarose gel electrophoresis.
Total RNA extracted from the leaves of the tobacco and the first strand cDNA synthesized were the same as above.
All RT-PCR experiments were repeated at least three times.
Subcellular localization
The DgNAC1 ORF were cloned into the SacI and EcoRI sites of the pSAT6-GFP-N1 vector. This vector contains a modified redshifted GFP at EcoRI-NcoI sites. The DgNAC1-GFP construct was transformed into onion epidermal cells by particle bombardment as described earlier (Wang and Fang 2002). The transient expression of the DgNAC1-GFP fusion protein was observed by using Confocal Microscopy.
Overexpression of DgNAC1 in tobacco plants
To overexpress DgNAC1 in tobacco (Nicotiana tabacum cv. Xanthi), the DgNAC1 cDNA was cloned in pBI121 (Clotech) by replacing the gus gene. The DgNAC1 gene driven under the cauliflower mosaic virus (CaMV) 35S promoter was introduced into tobacco plants by Agrobacterium-mediated GV3101 transformation (An et al. 1988).
Analysis of salt tolerance of transgenic tobacco plants
The T2 generation plants of lines OE-3, OE-12 and OE-20 were used in the subsequent experiments. Salt treatment I: About 100 seeds of wild type and DgNAC1-OX transgenic T2 lines were plated on 150 mM NaCl culture medium plate. The plate were placed in a growth chamber under a 16 h light/8 h dark cycle at 25°C. Germination rate was scored after 6 days. Germination was assayed as describer by Xiong et al. (2001). Salt treatment II: Wild type and DgNAC1-OX transgenic T2 lines were germinated on MS culture media for 14 days, after which, seedings were transferred to plastic pots filled with soil vermiculite at the ratio of 1:1. Seedings of WT and T2 transgenic tobacco plants were placed in a growth chamber under a 16 h light/8 h dark cycle at 25°C. Salt treatment was applied as follows. After 23 days the plant were watered with 400 mM NaCl for 7 days, after which, photos were taken (Saad et al. 2010). The tobacco plants were then re-watered regularly as a recovery process. The survival rate was scored after 6 days. Experiments on salt tolerance was conducted three times and the standard error (±SE) were measured relatively.
Results
Isolation of the DgNAC1 gene from chrysanthemum
Based on the conserved regions of Arabidopsis ATAF1, Oryza sativa OsNAC6, and soybean GmNAC2, degenerate primers to conduct the 3′-RACE were proposed to obtain a 678 bp fragment from leaves of chrysanthemum. The full-length cDNAs was obtained by 5′-RACE, and were designated as DgNAC1 (Genbank accession No. HQ317452). Sequence analysis showed that the DgNAC1 cDNA was 1137 bp in length, including a complete open reading frame of 855 bp flanking with a 5′-UTR of 117 bp and a 3′-UTR of 165 bp (Fig. 1). The predicted protein of DgNAC1 is composed of 284 amino acids with a calculated molecular mass of 32.9 kDa and its theoretical isoelectric point was 7.13.
The predicted amino acid sequence of DgNAC1 was compared to other NAC-domain contain proteins from rice, soybean, Arabidopsis, and Brassica napus by DNAMAN (Version 6.0) (Fig. 2). The result indicates that DgNAC1 included a highly conserved NAC DNA-binding domain, which consists of five consensus subdomains (A: 5–25, B: 34–51, C: 60–96, D: 103–130, E: 144–158) in the N-terminal region and a highly variable C-terminal transcriptional regulation domain. The conserved NAC domains were then retrieved for construction of a neighbor-joining phylogenetic by MEGA 4.1 (Fig. 3). Phylogenetic analysis demonstrated that the majority of 46 isolated proteins belong to eleven different subgroups of ATAF1, ATAF2, AtNAC3, OsNAC3, NAP, NAM, ANAC042, VND, NAM, TIP, and TERN. DgNAC1 was clustered into ATAF1 subgroup, and more closely related to the OsNAC6.
Comparison of the deduced amino acid sequence of DgNAC1 and other NAC domain proteins. The comparison was conducted by DNAMAN (version 6.0). Amino acid residues conserved in all seven sequences were shaded in black, and those conserved in six sequences were boxed in light grey. The five sub-domains (A–E) were underlined. Oryza sativa (OsNAC6, BAB89800); Arabidopsis thaliana (ATAF1, X74755.1; ATAF2, X74756.1); Brassica napus (BnNac5-1, AAP35050; BnNac5-11, AAP35053); Glycine max (GmNAC2, AAY46122)
Phylogenetic tree analysis of DgNAC1 and other plant NAC domain-containing proteins. The tree was constructed by neighbor-joining method with MEGA program (ver 4.1). Branch numbers represent as percentage of bootstrap values in 1000 sampling replicates and scale indicates branch lengths. The accession numbers as follows: AhNAC1 (ACD39411), GmNAC2 (AAY46122), SlNAM1 (GU256056), ATAF1 (X74755.1), BnNac5-11 (AAP35053), BnNac18 (AAP35054), OsNAC6 (BAB89800), BnNac1-1 (AAP35048), BnNac5-1 (AAP35050), BnNac5-7 (AAP35051), ATAF2 (X74756.1), BnNac5-8 (AAP35052), ANAC072 (AAM14367.1), GmNAC4 (AAY46124), GmNAC3 (AAY46123), ANAC019 (NP-175697), ANAC055 (NP-188169), AtNAC3 (AAP42729), OsNAC3 (BAA89797), GRAB1 (CAA09371), OsNAC19 (AAT02360), OsNAC4 (BAA89798), NAP (NP_564966), GmNAC1 (AAY46121), AtNAM (AAD17313), NAM-D2 (ABI94356), NAM-B1 (ABI94353), NAM-A1 (ABI94352), ANAC042 (NP_18188.1), ANAC009 (NP_174009), ANAC094 (NP_198798), VND2 (NP_195339.1), VND1 (NP_79397.1), VND4 (NP_172690), AtNAC1 (AAF21437), CUC3 (AAP82630), AtNAC2 (AAO41710), GmNAC5 (AAY46125), CUC1 (BAB20598), NAM (CAA63012), CUC2 (BAA19529), NTL9 (NP_001119122.1), TIP (AAM47025), TERN (BAA78417), GmNAC6 (AAY46126)
Expression analysis of DgNAC1
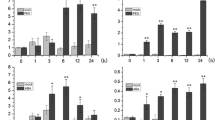
The spatial-specific expression of DgNAC1 in different tissues at the adult stage was determined by RT-PCR. The result shows that DgNAC1 mRNA is more abundant in roots and flowers than in stems and leaves (Fig. 4a).
Expression patterns of DgNAC1 in different organs and in response to various treatments. a Expression patterns of DgNAC1 in roots (R), stem (S), leaves (L), and flowers (F) under normal conditions. b–f Expression patterns of DgNAC1 in the seeding leaves under H2O, 0.1 mM ABA, 250 mM NaCl, and exposed to cold (4°C) and drought (on filter paper) conditions, respectively. a–f Ethidium bromide staining of PCR products using DgNAC1-specific primers with (top) and without (middle) prior reverse transcription, and the RT-PCR products with 18s rRNA-specific primers (bottom). Total RNA was isolated at 0, 1, 5, 12, 24 h after each treatment. Each experiment point was repeated at least three times. Representative results were shown here
To investigate the expression patterns of DgNAC1 gene under stress such as high salinity, drought, low temperature and exposure to ABA, the analysis with RT-PCR was performed, respectively. The expression of DgNAC1 kept at low affected level in normal conditions (Fig. 4b). By ABA treatment, the expression level of DgNAC1 increased rapidly and reached the maximum in 5 h and maintained the stable high level for 24 h (Fig. 4c). The concentration of DgNAC1 mRNA increased rapidly and peaked within 5 h by NaCl treatment (Fig. 4d). The expression of DgNAC1 increased slowly and peaked within 12 h in response to cold treatment (Fig. 4e). The expression of DgNAC1 obviously increased after 1 h dehydration and maintained the stable high level for 24 h (Fig. 4f). RT-PCR analysis revealed that the expression of this gene could be induced by drought, salt, cold and ABA.
Localization of DgNAC1 in the nucleus
To examine subcellular localization of DgNAC1 protein, the DgNAC1-GFP fusion protein was introduced into onion epidermal cells by particle bombardment. As shown in Fig. 5, confocal microscopic examination showed that the DgNAC1-GFP fusion protein was targeted into the nucleus, whereas the control GFP alone was distributed in both the entire cytoplasm. These results suggested that the DgNAC1 protein is a nuclear localization protein.
Subcellular localization of DgNAC1. Transient expression in onion epidermal cells of 35S-GFP and 35S-DgNAC1-GFP translational product were visualized by fluorescence microscopy. The transient vector harboring 35S-GFP and 35S-DgNAC1-GFP cassettes were transformed into onion epidermal cells by particle bombardment. The photos were taken in the bright light (a, d), in the dark for GFP images (b, e) and DAPI-stained images (c, f) after incubation for 20 h
Overexpression of DgNAC1 confers tolerance to salt stress in transgenic tobacco
Overexpression of DgNAC1 in tobacco plants under the control of the CaMV 35S promoter was generated to investigate the role of DgNAC1 for salt stresses in plant. Among 34 lines of transformants, five independent transgenic lines (OE-3, OE-7, OE-12, OE-14, and OE-20) were confirmed by using RT-PCR analysis (Fig. 6a). The 35S:DgNAC1 transgenic tobacco showed no detectable phenotype defects (such as root depth and volume, and plant height and flowers) (data not shown) under normal conditions.
The 35S:DgNAC1 transgenic tobacco (lines OE-3, OE-12 and OE-20) improved salt tolerance. a The expression level of DgNAC1 in wild type and DgNAC1-OX transgenic T0 lines. Ethidium bromide staining of PCR products using DgNAC1-specific primers with (top) and without (middle) prior reverse transcription, and the RT-PCR products with 18s rRNA-specific primers (bottom). b About 100 seeds of wild type and DgNAC1-OX transgenic T2 lines were plated on 150 mM NaCl culture medium plate. Germination rate was scored after 6 days. c The seedlings in wild type and DgNAC1-OX transgenic T2 lines were watered with 400 mM NaCl solution for 7 days. d Survival rates of seeding in wild type and DgNAC1-OX transgenic T2 lines after 6 days recovery. About 100 seedings were used for each treatment. Datas are indicated as a percentage relative to controls. All treatments are repeated at least three times and represent results showed here
The rate of seed germination of transgenic lines (OE-3, OE-12, and OE-22) and WT on 150 mM NaCl culture medium plate for 7 days was scored. As shown in Fig. 6b, the transgenic seeds showed higher germination rate (70–86%) than wild-type seeds (26%). When the seedlings were watered with 400 mM NaCl solution for 7 days, the WT were more wilted and yellower than transgenic lines (Fig. 6c). The survival rate (86–94%) of transgenic seedings was higher than that (35%) of wild-type seeding after 6 days recovery (Fig. 6d).
Discussion
Some NAC TF genes usually play critical roles in response to multiple environmental stresses in plants (Nakashima et al. 2007; Tran et al. 2004; Zheng et al. 2009). A NAC gene, termed DgNAC1, was isolated from chrysanthemum and characterized in our study. Sequence analysis showed that it contains a highly conserved NAC DNA-binding domain in the N-terminal region and a highly variable C-terminal transcriptional regulation domain. The DgNAC1 was structurally similar to OsNAC6 which was isolated from Oryza sativa under high-salt, drought and cold stress (Ohnishi et al. 2005). Phylogenetic analysis demonstrated that DgNAC1 was clustered into ATAF1 subgroup and more closely related to the OsNAC6. These results indicate that DgNAC1 is a novel member of the NAC TF genes family.
The mRNA expression analysis showed that DgNAC1 substantially induced by the treatment of NaCl, drought, cold, and ABA, may be involved in the abiotic-stress response via the ABA-dependent pathway. The expression patterns of DgNAC1 were similar to OsNAC6 and ATAF1 during several different stresses (Lu et al. 2007; Ohnishi et al. 2005). The 35S:DgNAC1 transgenic tobacco exhibited a markedly increased tolerance to salt. In Oryza sativa, overexpression of OsNAC6 also enhanced the tolerance to salt stress in transgenic lines (Nakashima et al. 2007). The overexpression of another stress-responsive NAC gene ATAF1 in Arabidopsis, a member of ATAF1 subfamily, has been reported to enhance the tolerance to drought (Wu et al. 2009). Interestingly, ATAF1 plays a complex role in abiotic stresses responses, which results from the ataf1-1 mutant line with high tolerance to drought stress in Arabidopsis (Lu et al. 2007; Wu et al. 2009). Further experiment needs to identify whether the DgNAC1 is also functional in a manner similar to OsNAC6 and ATAF1 in abiotic stresses responses.
Growth supression may be helpful for enhancing defense responses to abiotic stresses in plants, because it is more accommodable to stresses (Mittler et al. 2001; Vinocur and Altman 2005). Overexpression of different NAC TF genes, such as ATAF1 or OsNAC6, caused growth retardation and enhanced stresses tolerance in transgenic lines (Nakashima et al. 2007; Wu et al. 2009). However, overexpression of SNAC1 or SNAC2 in rice significantly increased stress resistance while plant morphology and yield of transgenic plants were not affected under normal growth conditions (Hu et al. 2006, 2008). Moreover, the 35S:DgNAC1 transgenic tobacco showed indetectable phenotype defects such as abnormal morphology and growth retardation under normal growth conditions. Further studies need to determine the relation of the growth to stresses tolerance in different NAC TF genes.
Salt tolerance is an important trait in chrysanthemum breeding, which has led researchers to focus on the characterization of salt-induced genes and the development of more salt-tolerant transgenic chrysanthemum by using these genes, aimed at providing economically important chrysanthemum. The 35S:DgNAC1 transgenic tobacco of the enhanced tolerance of to high salt stress without the severe growth retardation and dwarfing was observed in our study. Therefore, DgNAC1 is a potentially excellent genetic resource for the improvement of salt tolerance in chrysanthemum. In addition, a genetic transformation protocol was established for chrysanthemum from leaf explants for the production of Agrobacterium tumefaciens-mediated transgenic chrysanthemum (Jiang et al. 2010). Thus, it is possible that we would transfer the DgNAC1 to chrysanthemum to investigate its ability to improve chrysanthemum tolerance to high salinity. Further investigation would be essential to obtain the 35S:DgNAC1 transgenic chrysanthemum and conclude the precise mechanism of DgNAC1 in salt tolerance.
References
Agarwal PK, Agarwal P, Reddy MK, Sopory SK (2006) Roles of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25:1263–1274
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
An G, Watson BD, Chang CC (1988) Transformation of tobacco, tomato, potato, and Arabidopsis thaliana using a binary Ti vector system. Plant Physiol 81:301–305
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J Exp Bot 55:225–236
Chinnusamy V, Zhu J, Zhu JK (2006) Gene regulation during cold acclimation in plants. Physiol Plant 126:52–61
Duval M, Hsieh TF, Kim SY, Thomas TL (2002) Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol Biol 50:237–248
Hu HH, Dai MQ, Yao JL, Xiao BZ, Li XH, Zhang QF, Xiong LZ (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Hu HH, You J, Fang YJ, Zhu XY, Qi ZY, Xiong LZ (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Jiang D, Liang JL, Chen XL, Hong B, Jia WS, Zhao LJ (2010) Transformation of Arabidopsis flowering gene FT to from cut chrysanthemum ‘Jinba’ by Agrobacterium mediate. Acta Horticulturae Sinica 37(3):441–448
Knight H, Knight MR (2001) Abiotic stress signaling pathways: specificity and cross tolerance. Trends Plant Sci 6:262–267
Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63:289–305
Mittler R, Merquiol E, Hallak-Herr E, Kaplan A, Cohen M (2001) Living under a ‘dormant’ canopy: a molecular acclimation mechanism of the desert plant Retama raetam. Plant J 25:407–416
Nakashima K, Tran LP, Nguyen DV, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149:88–95
Ohnishi T, Sugahara S, Yamada T, Kikuchi K, Yoshiba Y, Hirano HY, Tsutsumi N (2005) OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet Syst 80:135–139
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247
Saad RB, Zouari N, Ramdhan WB, Azaza J, Meynard D, Guiderdoni E, Hassairi A (2010) Improved drought and salt stress tolerance in transgenic tobacco overexpressing a novel A20/AN1 zinc-finger ‘‘AlSAP’’ gene isolated from the halophyte grass Aeluropus littoralis. Plant Mol Biol 72:171–190
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227
Tong Z, Hong B, Yang YJ, Li QH, Ma N, Ma C, Gao JP (2009) Overexpression of two chrysanthemum DgDREB1 group genes causing delayed flowering or dwarfism in Arabidopsis. Plant Mol Biol 71:115–129
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress inducible NAC transcription factors that bind to a drought responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Cur Opin Biotechnol 17:113–122
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Cur Opin Biotechnol 16:123–132
Wang GL, Fang HY (2002) Gene engineering in plant, 2nd edn. Press of Science, Beijing, pp 734–736
Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Yang C, Xie Q (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19:1279–1290
Xiong LM, Lee BH, Ishitani M, Lee HJ, Zhang CQ, Zhu JK (2001) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15:1971–1984
Zheng XN, Chen B, Lu GJ, Han B (2009) Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun 379:985–989
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgments
This research was supported by the Key Scientific Research Project of Education Department of Sichuan Province (07ZA082, 09ZA065 and 10ZA051). We thank Prof. Tao Wang (State key Laboratories of AgroBiotechnology, China Agricultural University) for providing the pSAT6-GFP-N1 vector.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Liu, QL., Xu, KD., Zhao, LJ. et al. Overexpression of a novel chrysanthemum NAC transcription factor gene enhances salt tolerance in tobacco. Biotechnol Lett 33, 2073–2082 (2011). https://doi.org/10.1007/s10529-011-0659-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-011-0659-8