Abstract
The paper discusses the effect of microwave assisted aqueous extraction conditions on the recovery and quantification of bioactive compounds of Basella alba leaves. The experiment was carried out using the Box–Behnken design of response surface methodology (RSM) with microwave power (100–300 W), treatment time (5–15 min), and feed/solvent ratio (0.025–0.05 w/v) as independent variables. The response variables, total phenolic content (TPC), total flavonoid content (TFC), and antioxidant activity (AA) were predicted individually using both RSM and artificial neural network-genetic algorithm (ANN-GA). Experimental values of TPC, TFC and AA (% of DPPH scavenging assay) ranged from 2.64 to 5.46 mg GAE g−1, 7.38 to 15.71 mg QE g−1 and 0.12 to 0.32 mg GAEAC g−1 respectively. The predicted values of TPC, TFC, and AA for the optimized conditions extracted using RSM are 6.21 mg GAE g−1, 14.29 mg QE g−1, and 0.25 mg GAEAC g−1, respectively, whereas using ANN-GA were 6.23 mg GAE g−1, 11.2 mg QE g−1, and 0.24 mg GAEAC g−1, respectively. When compared to RSM, ANN-GA demonstrated a greater value of R2 and lower values of other statistical parameters. Additionally, the predicted value of ANN-GA was more closely aligned with the experimental value. Therefore, ANN-GA can be considered the best model for the optimization and modeling of aqueous MAE of bioactive components from Basella alba leaves.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malabar spinach is a climbing plant from a monotypic genus in the Basellaceae family. It is cultivated as a leafy vegetable or pot herb in the regions of India and Indonesia. It is commonly known as Poi saag, Ceylon spinach, Indian spinach, climbing spinach, and vine spinach [1,2,3]. The plant is also found in Bangladesh, China, Philippines, Africa, South America, and Thailand. Basella alba and Basella rubra were the two most commonly utilized varieties of this species. B. alba has a green stem, whereas B. rubra has a reddish-purple stem. The plant is a rich source of minerals, vitamins, dietary fiber, phenolic compounds, and antioxidants [4,5,6,7]. B. alba has demonstrated androgenic activity, antioxidant activity, antiulcer activity, cytotoxic and antibacterial activity, central nervous system depressant activity, anti-inflammatory activity, wound healing properties, nephroprotective, and antioxidative properties [8, 9].
Even though the basella species are consumed as a leafy vegetable and used medicinally, proper scientific studies on its phytochemical composition and novel extraction methods have not been explored. Identification of suitable extraction conditions for the recovery of bioactive components is essential to maximize their health-improving benefits. The extraction techniques used for the recovery of bioactive components are broadly categorized into conventional and non-conventional extraction methods. Maceration, Soxhlet extraction, and hydro-distillation are the commonly used conventional methods. The novel/non-conventional methods include ultrasound-assisted extraction (UAE), enzyme-assisted extraction (EAE), pulsed electric field (PEF) extraction, pressurized liquid extraction, microwave-assisted extraction (MAE), and supercritical fluid extraction [10, 11]. The main drawbacks of conventional methods are the long treatment time, need for high-purity solvents, low solvent recovery rate (which necessitates the use of a large quantity of solvent), and higher cost of extraction [12, 13].
MAE is gaining popularity due to the several benefits it provides, including enhanced extraction yields, reduced extraction time, and solvent usage. Physical parameters like solubility, dielectric constant (ε′), and dissipation factor must be considered to maximize the yield of MAE. Solvents with high dielectric constants (such as water) can absorb more microwave (MW) energy; hence, the polarity of the solvent is critical in MAE. Polar solvents are often considered to be superior to non-polar solvents [14]. MAE has recently been studied for extracting bioactive compounds from cashew apples [15], Allium sativum [16], fruit seeds [17], and pineapple peel [18, 19], and found that method is superior over other technologies in terms of higher yield and quality of the extract. Investigation into the MAE of B. alba has not yet been reported so far. Hence a research attempt has been made to study the effect of MAE conditions on the recovery of bioactive components from B. alba leaves and its optimization using RSM and ANN-GA techniques. In this study, a Box–Behnken design (BBD) was applied to evaluate the effect of MAE conditions on the recovery of TPC, TFC, and AA. These findings will aid in the assessment of bioactive potential of B. alba and its utilization, particularly in the food and pharmaceutical sector.
Methodology
Raw materials
Fresh Basella alba leaves were purchased from the local market in Kharagpur, West Bengal, India for the experiments. Cleaned leaves were blotted for surface water removal and dried in a laboratory model freeze dryer (IIC—INSTIND, Kolkata) at a temperature of − 35 °C, vacuum pressure of 150 mbar, and condenser temperature of 20 °C for 24 h. The dried leaves are then ground to flour with a particle size of less than 425 µm using a mesh screen (BS 36). The dried leaves were powdered and stored for further analysis.
Chemicals and reagents
Chemicals and reagents of analytical quality from Himedia Laboratories (Mumbai, India) and Merck Specialties Private Limited (Mumbai, India) were used for the research. The chemicals and reagents include 2,2-diphenyl-1-picrylhydrazyl (DPPH), Folin–Ciocalteu (FC) reagent, sodium hydroxide, sodium carbonate, sodium nitrite, hydrochloric acid, aluminum chloride, ethanol, and methanol.
Microwave-assisted extraction
The extraction of bioactive compounds from Basella leaf powder was performed using a combined extraction system (NuWav-Promaster, Nutech Analytical Technologies Pvt. Ltd., India) with a platinum temperature sensor. The experiments were carried out at varying power levels of 100–300 W and treatment time of 5–15 min. The solvent used was water. The feed-to-solvent ratio varied from 0.025 to 0.05. After extraction, the samples were centrifuged at 10,000 rpm for 10 min, followed by filtration through Whatman No. 4 filter paper. The extract thus obtained was collected in 10 mL plastic vials and stored in refrigerated conditions until further analysis.
Experimental design
Based on previous studies [13], three-level BBD of RSM with five central points was used for the experiments. The independent variables selected for the study are MW power, extraction time, and feed/solvent ratio. The levels for the independent parameters in the study are presented in Table 1 with their coded forms. The experiment contained 17 runs and the treatment combinations are presented in Table 2. The extract was prepared according to the design, and the dependent variables of the study, such as TPC, TFC, and AA, were determined using the protocols specified in the subsequent sessions. The influence of independent variables on the responses such was then modeled mathematically using RSM and ANN-GA technique.
Total phenolic content
TPC analysis was performed as per the method described by Kumar and Srinivasa Rao [20] with minor modifications. About 0.2 mL liquid extract was taken (diluted by 2.8 mL) and was mixed with 0.5 mL of Folin–Ciocalteu reagent. The mixed solution was incubated for 5 min and then mixed with 2 mL of Na2CO3 (7%, w/v) solution. The samples were kept in the dark for 30 min. The absorbance was then read at 760 nm using a UV-V spectrophotometer (UV Plus, Mortras Scientific, India). Calculated TPC was expressed as mg of Gallic acid equivalent (GAE) per gram of sample.
Total flavonoid content
The determination of TFC was done according to Kumar and Srinivasa Rao [20] in a modified form. About 1 mL of liquid extract was taken and mixed with 5 mL of distilled water. From this, 2 mL of extract was taken followed by 0.15 mL of NaNO2, and added with 2 mL of AlCl3 solution. After 3 min, 1 M NaOH was added and the mixture was incubated for 2 h. The spectrophotometric readings were measured at wavelength 510 nm which were further expressed as quercetin equivalent per gram of sample.
Antioxidant activity
For AA, the methodology was adopted from Rout et al. [13] with slight modifications was used. About 0.1 mL liquid extract was taken and mixed with 3.9 mL DPPH solutions. After keeping the sample for 30 min in the dark, the spectrophotometric readings were taken at a wavelength of 517 nm. Pure methanol was used for zeroing. Gallic acid was used as the reference, and the results were reported as mg of gallic acid equivalent antioxidant capacity (mg GAEAC g−1 sample).
RSM modeling and optimization
Process parameter combinations were modeled and optimized using RSM. The empirical second-order polynomial regression model equation (Eq. 1) was used in the study to analyze the response variables and predict the optimal values.
where \({y}_{j}\) are the different responses, \({\beta }_{0}\), \({\beta }_{i}\), \({\beta }_{ii}\) and \({\beta }_{ij}\) are the regression coefficients for the mean, linear, quadratic, and interaction terms, respectively, and \({x}_{i}\) & \({x}_{j}\) are the independent variables in coded values ranging from − 1 to + 1.
The analysis was carried out using commercial software (Design Expert 11, Stat-Ease Inc., Minneapolis, USA). To validate the model, the best MAE conditions were evaluated experimentally in triplicate. The optimization of the various process variables was done with the objective of maximizing the TPC, TFC, and AA. Each response was converted into a dimensionless desirability function (di) with a scale of 0–1. The overall desirability function (G), which was formed by adding the individual desirability values of each response, represented the maximization of each response.
where di is the desirability of the response variable and n is the number of responses in the study. Under optimal conditions predicted by the model, the validity of the presently developed model was tested. The average results of the experiments were compared to the expected values of the generated model, and the efficacy of the established model was evaluated.
ANN modeling and GA optimization
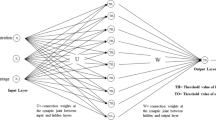
ANN modeling was carried out using a neural network fitting tool of MATLAB (version R2022a, The Math Works, Inc., USA). ANN model consists of input layer, hidden layer, and output layer. The input layer comprised of three neurons [microwave power (W), time (min), and feed-to-solvent ratio (w/v)]. The trial-and-error method was used to determine the number of neurons in the hidden layer and it was selected based on the highest R2 and lowest MSE values. For the study, the number of neurons thus achieved was 10. The output layer contains a single neuron that represents the responses of the extraction process, such as TPC (Y1 mg GAE g−1), TFC (Y2 mg QE g−1), and AA (Y3 mg GAEAC g−1). The input and output layers of the proposed ANN model are shown in Fig. 1. For each response, the training was done individually. The input and output data points were coded for the ANN model development [15]. The transfer function for the hidden layer was a hyperbolic sigmoid function (tansig), and the transfer function for the output layer was a linear function (purelin). The data were trained using the Levenberg–Marquardt back-propagation (trainlm) algorithm, with 15% of the total data used for validating, 15% for testing, and 70% for training. The network is trained until it has the highest R2 and lowest MSE. After training, the predicted output was calculated using the weights and bias values represented by the following equation.
where xi and yi are the input and predicted coded output parameters; WOH and UIH are the weights between the hidden and output layers and the input and hidden layers; TO and TH are the bias values of the output and hidden layer neurons, respectively.
Genetic algorithm (GA) optimization was carried out using the Global optimization toolbox of MATLAB (version R2022a, The Math Works, Inc., USA). The population type (double vector), size (200), and crossover fraction (0.8) were the primary parameters chosen for GA optimization. In addition, the creation function, fitness scaling function, selection function, crossover function, and mutation function were selected as feasible population, rank, roulette function, scattered, and adaptive feasibility, respectively [21]. The fitness function (f) was generated to maximize all output responses and is shown below.
where Y1, Y2, and Y3 are the ANN-predicted actual values of responses, and the negative sign shows the maximization of the specified function (f) in GA.
Statistical analysis
The evaluation of the RSM and ANN models was conducted by employing various statistical metrics, including the coefficient of determination (R2), average absolute deviation (AAD), mean square error (MSE), normal mean square error (NMSE), mean percentage error (MPE), root mean square error (RMSE), and normal root mean square error (NRMSE). These parameters were calculated using the respective equations associated with each evaluation criterion. The model exhibiting the lowest values for the Average Absolute Deviation (AAD), Mean Squared Error (MSE), normalized Mean Squared Error (NMSE), Root Mean Squared Error (RMSE), Normalized Root Mean Squared Error (NRSME), and the highest value for the coefficient of determination (R2) is deemed to be the most effective model for accurately representing the responses.
where xp is the predicted data, xa is the experimental data, xm is the mean experimental data, and n is the number of experiments.
Results and discussion
RSM modeling
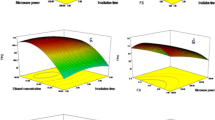
The independent variables chosen for this study were power, treatment time, and feed-to-solvent ratio. These variables were investigated to determine their impact on the levels of three bioactive compounds TPC, TFC, and AA in the leaves of B. alba using MAE. The predicted and experimental values of all the dependent parameters were recorded and are presented in Table 2. The effect of process parameters on response variables is shown in Fig. 2. The multiple linear regression equations developed were evaluated using statistical parameters like R2, predicted R2, and adjusted R2. All regression models showed higher R2 value above 0.9 and the lack of fit was insignificant.
The equations presented below represents the second-order polynomial regression models showing the relationship between dependent and independent variables of the study.
where A, B, and C are the coded form of the independent variables, microwave power, treatment time, and feed-to-solvent ratio, respectively.
Effect of process variables on total phenolic content
The variation in TPC during different treatment combinations of the MAE process is presented in Table 2. The maximum value of TPC (5.46 mg GAE g−1) was observed at 200 W for 5 min and the minimum value (2.64 mg GAE g−1) at 200 W for 10 min. Initially TPC value gradually decreased with the power (up to 200 W) and as the power level increased further to 300 W, it showed an increasing trend. It is also evidenced in RSM plots that TPC gradually decreased initially to 4.5–5 mg GAE g−1 value up to 200 W after which it increased to 3.5–4 mg GAE g−1 up to 300 W. In a study on Pistacia lentiscus leaves, Dahmoune et al. [22] also reported an increase in phenols recovery initially due to increased heat and mass transfer phenomena up to a certain power level (300 W), and subsequently they also observed degradation of phenolics due to thermal effects at increasing densities. In another study, Song et al. [23] reported that the phenolics contents increased when treated at higher power for a short time in Ipomoea batatas leaves. As the ratio of feed to solvent is decreased, an increase in the TPC was observed. In addition, the TPC values tend to increase over time, indicating a positive correlation between prolonged exposure time and TPC levels.
Effect of process variables on total flavonoid content
The maximum value of TFC (9.65 mg QE g−1) was obtained at 300 W for 10 min and the minimum (7.37 mg QE g−1) was obtained at 100 W for 10 min. As evident from the RSM plots, the TFC value decreased with an increase in power level and treatment duration. The observed phenomenon of a decrease in TFC is due to flavonoid decomposition at higher temperatures [24]. Similar results have been observed by Pan et al. [25] and Jokić et al. [26]. At higher temperatures, glycosidic linkages in flavonoid molecules hydrolyze, generating unstable intermediate compounds like aglycones, semiquinoids, etc. that easily oxidize to generate brown, high molecular weight compounds [27, 28].
Effect of process variables on antioxidant activity
The antioxidant activity (AA) is calculated in terms of gallic acid equivalent (GAE). The AA of MAE samples were higher than the control samples. The highest value of AA (0.12 mg GAEAC g−1) was observed for 200 W treated samples for 5 min. AA increased with power and time and decreased with the feed-to-solvent ratio as it is mentioned in Fig. 2. Similar results were reported in the MAE of Buddleia officinalis where the antioxidant value is gradually increasing with the power and time [25].
RSM optimization
RSM was used to maximize the bioactive recovery of TPC, TFC, and AA form B. alba leaves. The regression coefficients as well as other statistical parameters like F-values, coefficient of determination (R2), adjusted R2, and coefficient of variation (CV) are summarized in Table 3. Besides, additional statistical parameters AAD, MSE, NMSE, RSME, NRSME, and MPE were shown in Table 4. High R2 confirmed shows good accuracy of the model. The high degree of correlation is confirmed between the predicted and the experimental values as the adjusted R2 values are nearly identical to the R2 values. Additionally, the CV in every instance was less than 5%, confirming the model’s great reproducibility and good precision. According to Abdullah et al. [21], for a model to be considered preferable, its R2 value must be greater than 0.80. From Table 3, the R2 and adjusted R2 values of TPC, TFC, and AA were 0.99 mg GAE g−1, 0.89 mg QE g−1, and 0.92 mg GAEAC g−1, demonstrating the acceptable fit of the developed model. Furthermore, the lowest values of the other statistical parameters (Table 4) confirm the model’s acceptability. The lack of fit of all three responses was equally non-significant.
ANN modeling
Model fitting
The TPC, TFC, and AA were estimated using the help of the Levenberg–Marquardt (LM) algorithm within an ANN framework. The selection of the optimal model was based on the criteria of minimizing the MSE and maximizing R2 value. The selected model for TPC, TFC, and AA, which was obtained at epochs 5, 4, and 8, respectively, demonstrates a final architecture. This architecture consists of an input layer with three neurons, a hidden layer with 10 neurons, and an output layer with one neuron for each response. The visual representation of this architecture can be observed in Fig. 1. To achieve the most rigorous validation, the 17 datasets were partitioned into three distinct sets during the experimental runs. In the context of TPC, the training dataset consisted of experimental runs 3, 4, 6, 7, 9, 10, 11, 12, 13, 14, and 17. The R2 values of the training, validation, and testing were 0.9865, 0.9715, and 0.9929, and the MSE value of these runs were 0.0302, 0.0796, and 0.1421, respectively. Similarly, for TFC, the experimental runs 1, 2, 3, 4, 6, 7, 8, 11, 13, 14, 16, and 17 were used for training 11 observations for validation, and 3 observations for testing, respectively. The corresponding R2 values for training, validation, and testing were 0.9983, 0.9808, and 0.9431, and MSE values were 0.0018, 0.0433, and 2.7121, respectively. In the same manner, the experimental runs 2, 3, 4, 6, 7, 8, 11, 12, 14, 15, and 17 were used for training 11 observations for validation 3 observations for testing, respectively used for the modeling of the antioxidant activity. The corresponding R2 for training, validation, and testing were 0.9913, 0.9725, and 0.8682, and MSE values were 0.001, 0.001, and 0.0079, respectively. The confirmation of ability of the developed artificial neural network (ANN) model to predict unknown data is evidenced by the observed decrease in mean squared error (MSE) and increase in R-squared (R2) values. Figure 3 shows the performance and error histogram of the model that has been developed. The optimization of the Artificial Neural Network (ANN) was conducted through the assessment of the output with the input data. The utilization of weights was employed to minimize the error function during the optimization procedure. The weights and bias values of the final model are presented in Eqs. 15–26. The post-training performance and error histogram of the responses are presented in Fig. 3.
where U denotes bias values, W denotes the values of the weights, TH is the threshold value between the input and hidden layer, and TO is the threshold value between the hidden and output layer. Suffixes a, b, and c to represents the TPC, TFC, and AA, respectively.
GA optimization
The optimization process involved the use of a genetic algorithm (GA) to maximize the extraction efficiency of TPC, TFC, and AA. The independent parameters, namely MW power, treatment time, and feed-to-solvent ratio were subjected to optimization through the GA methodology. The optimization process was pursued until achieving significantly reduced values of the MSE and RSME between the mean and individual fitness values. The optimization cycle was repeatedly pursued following the introduction of mutation. If the desired solution was not achieved, the entire population was once again employed for reproduction, crossover, and mutation in subsequent cycles. By the application of GA, the optimum conditions for the MAE of bioactive components from B. alba were obtained in 142 iterations and are given as follows: MW power of 100 W, treatment time of 8.162 min, and feed-to-solvent ratio of 0.039. The predicted values of the response variables generated using the developed ANN model were presented in Table 5. The obtained optimal process parameters were evaluated experimentally and measured responses were found to be, 6.01 mg GAE g−1 TPC, 11.06 mg QE g−1 TFC, 0.23 mg GAEAC g−1 AA. These experimental values aligned well with the predicted values, demonstrating the developed ANN-GA model’s good prediction and optimisation abilities.
Conclusion
In this study, the microwave-assisted aqueous extraction of bioactive components from B. alba was investigated. The effect of MAE conditions on the response variables (TPC, TFC and AA) was analyzed, predicted, and optimized by RSM and ANN-GA technique. The predicted values of TPC, TFC, and AA extracted using RSM are 6.21 mg GAE g−1, 14.29 mg QE g−1, and 0.25 mg GAEAC g−1, respectively, whereas using ANN-GA were 6.23 mg GAE g−1, 11.2 mg QE g−1, and 0.24 mg GAEAC g−1, respectively. Both the generated ANN and RSM models were highly predictive. However, the ANN model outperformed the RSM model, with a higher R2 and lower AAD, MSE, NSME, RSME, NRSME, and MPE values. Thus, it can be inferred that the ANN-GA model is an efficient quantitative tool for optimizing the process parameters for the MAE of B. alba.
Data Availability
Not applicable.
References
A. Azad, W. Wan Azizi, Z. Babar, Z.K. Labu, S. Zabin, An overview on phytochemical, anti-inflammatory and anti-bacterial activity of Basella alba leaves extract. Middle-East J. Sci. Res. 14(5), 650–655 (2013)
A. Chaurasiya, R.K. Pal, P.K. Verma, A. Katiyar, N. Kumar, An updated review on Malabar spinach (Basella alba and Basella rubra) and their importance. J. Pharmacogn. Phytochem. 10(2), 1201–1207 (2021)
S.K. Roy, G. Gangopadhyay, K.K. Mukherjee, Is stem twining form of Basella alba L. a naturally occurring variant? Curr. Sci. 98, 1370–1375 (2010)
M. Ranganatha, N.N. Rao, P. Giridhar, Micropropagation and in vitro flowering in Basella spp. Res. Sq. (2022). https://doi.org/10.21203/rs.3.rs-2166622/v1
P. Karthika, M.S. Priyanka, T.P. Vijayakumar, Nutritional Significance of Basella alba Linn. An Underutilized Medicinal Green Leafy Vegetable (Thanuj International Publishers, Tamil Nadu, 2022), p.350
S.S. Kumar, P. Manoj, G. Nimisha, P. Giridhar, Phytoconstituents and stability of betalains in fruit extracts of Malabar spinach (Basella rubra L.). J. Food Sci. Technol. 53, 4014–4022 (2016)
M.A. Nur, M. Khan, M.A. Satter, M.M. Rahman, M.Z. Amin, Assessment of physicochemical properties, nutrient contents and colorant stability of the two varieties of Malabar spinach (Basella alba L.) fruits. Biocatal. Agric. Biotechnol. 51, 102746 (2023)
G. Kumar, J. Joshi, P.S. Rao, P. Manchikanti, Effect of thin layer drying conditions on the retention of bioactive components in Malabar spinach (Basella alba) leaves. Food Chem. Adv. 3, 100419 (2023). https://doi.org/10.1016/j.focha.2023.100419
G. Akinniyi, S. Adewale, O. Adewale, I. Adeyiola, Qualitative phytochemical screening and antibacterial activities of Basella alba and Basella rubra. J. Pharm. Bioresour. 16(2), 133–138 (2019)
T. Alvi, Z. Asif, M.K.I. Khan, Clean label extraction of bioactive compounds from food waste through microwave-assisted extraction technique—a review. Food Biosci. 46, 101580 (2022)
M.A. Shanker, A.C. Khanashyam, R. Pandiselvam, T.J. Joshi, P.E. Thomas, Y. Zhang, S. Rustagi, S. Bharti, R. Thirumdas, M. Kumar, A. Kothakota, Implications of cold plasma and plasma activated water on food texture-a review. Food Control (2023). https://doi.org/10.1016/j.foodcont.2023.109793
T.J. Joshi, S.M. Singh, P.S. Rao, Novel thermal and non-thermal millet processing technologies: advances and research trends. Eur. Food Res. Technol. 249(5), 1149–1160 (2023). https://doi.org/10.1007/s00217-023-04227-8
R.K. Rout, A. Kumar, P.S. Rao, A multivariate optimization of bioactive compounds extracted from oregano (Origanum vulgare) leaves using pulsed mode sonication. J. Food Meas. Charact. 15, 3111–3122 (2021)
L. Wang, C.L. Weller, Recent advances in extraction of nutraceuticals from plants. Trends Food Sci. Technol. 17(6), 300–312 (2006)
A. Patra, S. Abdullah, R.C. Pradhan, Microwave-assisted extraction of bioactive compounds from cashew apple (Anacardium occidenatale L.) bagasse: modeling and optimization of the process using response surface methodology. J. Food Meas. Charact. 15, 4781–4793 (2021)
E.O. Oke, O. Adeyi, B.I. Okolo, J.A. Adeyi, C.J. Ude, J.A. Okolie, J.A. Adeyanju, O.O. Ajala, K. Nwosu-Obieogu, Microwave-assisted extraction proof-of-concept for phenolic phytochemical recovery from Allium sativum L. (Amaryllidaceous): optimal process condition evaluation, scale-up computer-aided simulation and profitability risk analysis. Clean. Eng. Technol. 13, 100624 (2023)
A.A. Manoj, A. Fathima, B. Naushad, S. Sunilkumar, M.A. Shanker, S. Abdullah, Valorization of fruit seeds by polyphenol recovery using microwave-assisted aqueous extraction: modelling and optimization of process parameters. J. Food Meas. Charact. (2023). https://doi.org/10.1007/s11694-023-01955-z
R.A. Abraham, J. Joshi, S. Abdullah, A comprehensive review of pineapple processing and its by-product valorization in India. Food Chem. Adv. (2023). https://doi.org/10.1016/j.focha.2023.100416
S.P. Bansod, J.K. Parikh, P.K. Sarangi, Pineapple peel waste valorization for extraction of bio-active compounds and protein: Microwave assisted method and Box Behnken design optimization. Environ. Res. 221, 115237 (2023)
A. Kumar, P. Srinivasa Rao, Optimization of pulsed-mode ultrasound assisted extraction of bioactive compounds from pomegranate peel using response surface methodology. J. Food Meas. Charact. 14(6), 3493–3507 (2020)
S. Abdullah, R.C. Pradhan, D. Pradhan, S. Mishra, Modeling and optimization of pectinase-assisted low-temperature extraction of cashew apple juice using artificial neural network coupled with genetic algorithm. Food Chem. 339, 127862 (2021)
F. Dahmoune, G. Spigno, K. Moussi, H. Remini, A. Cherbal, K. Madani, Pistacia lentiscus leaves as a source of phenolic compounds: microwave-assisted extraction optimized and compared with ultrasound-assisted and conventional solvent extraction. Ind. Crops Prod. 61, 31–40 (2014)
J. Song, D. Li, C. Liu, Y. Zhang, Optimized microwave-assisted extraction of total phenolics (TP) from Ipomoea batatas leaves and its antioxidant activity. Innov. Food Sci. Emerg. Technol. 12(3), 282–287 (2011)
X.D. Le, M.C. Nguyen, D.H. Vu, M.Q. Pham, Q.L. Pham, Q.T. Nguyen, Q.T. Tran, Optimization of microwave-assisted extraction of total phenolic and total flavonoid contents from fruits of Docynia indica (Wall.) Decne. using response surface methodology. Processes 7(8), 485 (2019)
Y. Pan, C. He, H. Wang, X. Ji, K. Wang, P. Liu, Antioxidant activity of microwave-assisted extract of Buddleia officinalis and its major active component. Food Chem. 121(2), 497–502 (2010)
S. Jokić, M. Cvjetko, Đ Božić, S. Fabek, N. Toth, J. Vorkapić-Furač, I.R. Redovniković, Optimisation of microwave-assisted extraction of phenolic compounds from broccoli and its antioxidant activity. Int. J. Food Sci. Technol. 47(12), 2613–2619 (2012)
J. Kroll, H.M. Rawel, S. Rohn, Reactions of plant phenolics with food proteins and enzymes under special consideration of covalent bonds. Food Sci. Technol. Res. 9(3), 205–218 (2003)
I. Scibisz, S. Kalisz, M. Mitek, Thermal degradation of anthocyanins in blueberry fruit. Zywnosc Nauka Technologia Jakosc (Poland) (2010). https://doi.org/10.15193/zntj/2010/72/056-066
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shende, A.S., Jayasree Joshi, T. & Srinivasa Rao, P. Microwave-assisted aqueous extraction of bioactive components from Malabar spinach (Basella alba) leaves and its optimization using ANN-GA and RSM methodology. Food Measure 18, 287–298 (2024). https://doi.org/10.1007/s11694-023-02182-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11694-023-02182-2