Abstract
An efficient protocol was developed for Agrobacterium tumefaciens-mediated transformation of tomato (Solanum lycopersicum) cultivars using cotyledon explants. The transformation frequency was assessed in response to several different factors, including seed germination medium, seedling age, pre-culture duration, pre-culture and co-cultivation media, inoculation medium, medium pH, washing medium, and kanamycin concentration in initial selection medium. Cotyledons excised from 6-d-old seedlings germinated on half-strength Murashige and Skoog’s (MS) basal medium containing 8.9 μM benzyladenine (BA) produced the most suitable explant material. Six days of explant pre-culture and 5 min inoculation with Agrobacterium culture in MS medium, containing 8.9 μM BA, 9.3 μM kinetin, and 0.4 mg l−1 thiamine at pH 5.0, significantly improved the transformation frequency. The addition of a tobacco feeder cell layer, however, did not lead to any significant improvement in the transformation rate. Kanamycin at 20 mg l−1 in the selection medium for the initial 10 d resulted in the highest transformation frequency. Combining the best conditions for each parameter resulted in an overall transformation efficiency of 44.3 %. Gene transfer was confirmed through PCR and Southern blot analyses. Mendelian inheritance ratios were found in 71.5 % of the independent transgenic lines from self-fertilized T1 progeny. The optimized transformation procedure showed high transformation frequencies for all three tomato cultivars tested, namely, Kashi Vishesh (H-86), Hisar Anmol (H-24), and Kashi Amrit (DVRT-1), and is also expected to give reproducible results with other tomato cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent times, genetic transformation of plants by introducing a specific DNA fragment into a plant genome has become an integral tool in both basic and applied research. Transgenic technology has advanced to such an extent that it is now widely exploited to study various biological phenomena, viz., the influence of biochemical pathways, response to stresses, resistance to pathogens, and, more importantly, to obtain commercial crops with improved herbicide resistance and abiotic stress tolerance (Liénard et al. 2007). Among the different plant transformation methods, Agrobacterium-mediated transformation is an efficient and low-cost system that has been most extensively utilized for stable gene transfer. Agrobacterium-mediated plant transformation exploits the natural ability of Agrobacterium tumefaciens cells to transfer and integrate a fragment of DNA (T-DNA) into the host plant genome.
Tomato (Solanum lycopersicum), a member of the Solanaceae family, is a major fruit crop consumed all over the world. It has also been extensively utilized as a model plant system to understand biological processes and to study functional genomics, proteomics, and metabolomics (Arumuganathan and Earle 1991). Agrobacterium-mediated transformation of tomato has become routine, and tomato plants have been engineered for a variety of purposes, such as improving biotic and abiotic stress tolerance (Jia et al. 2002), characterization of gene functions (Goel et al. 2010; Khare et al. 2010), and production of foreign proteins (Salyaev et al. 2007; Youm et al. 2008). One of the major obstacles in engineering economically important tomato cultivars, however, is the absence of a highly efficient and a reproducible transformation system (Velcheva et al. 2005). Agrobacterium-mediated tomato transformation was first reported in 1985 (Horsch et al. 1985), and since then, several refinements have been reported by different investigators (McCormick et al. 1986; Hamza and Chupeau 1993; van Roekel et al. 1993; Frary and Earle 1996; Ellul et al. 2003; Park et al. 2003; Cortina and Culianez-Macia 2004; Velcheva et al. 2005; Gao et al. 2009; Sharma et al. 2009). Many of these reports, however, do not provide critical details of the transformation process and are restricted to “model” genotypes, which makes it difficult to reproduce and apply the findings from other research groups to additional tomato cultivars (Sun et al. 2006).
The Agrobacterium-mediated transformation process includes preparation of explants, inoculation of explants with Agrobacterium cells, co-cultivation, selection of transformed cells from the bulk of the non-transformed cells, and regeneration of transformed cells to obtain a complete transformed plant. A number of factors involved in genetic transformation greatly influence the overall gene transfer efficiency (Hu and Phillips 2001). The effect of explant type (Davis et al. 1991; Hamza and Chupeau 1993; Park et al. 2003; Wu et al. 2006), different regimes of plant growth regulators (Frary and Earle 1996; Park et al. 2003; Cortina and Culianez-Macia 2004; Wu et al. 2006; Sharma et al. 2009), the effect of pH in co-cultivation medium ( Stachel et al. 1986; Vernade et al. 1988; Gao et al. 2009; Wu et al. 2006), and Agrobacterium cell density in the inoculum (Davis et al. 1991; Wu et al. 2006 Gao et al. 2009) have been amply discussed. However, reports on the influence of parameters including the germination medium, explant age, pre-culture period, pH of pre-culture and co-cultivation media, inoculation medium, washing medium, and initial selection pressure on genetic transformation of tomato are scarce.
As a part of an ongoing program toward the development of transgenic tomato lines for improved water-deficit stress tolerance, various parameters related to Agrobacterium-mediated transformation were evaluated for their role in overall transformation efficiency. The DREB1A/CBF3 gene from Arabidopsis thaliana was used as a marker gene in this study. AtDREB1A is a transcription factor that provides enhanced tolerance to water deficit ( Oh et al. 2005; Behnam et al. 2007). DREB/CBF proteins bind to the cis-acting 5-bp DRE/C-repeat core sequence (CCGAC) present in the promoter region of many downstream water-deficit stress-inducible and cold-regulated (COR) genes in Arabidopsis (Gilmour et al. 1998), Oryza sativa (Oh et al. 2005), and Solanum tuberosum (Behnam et al. 2007). An elite tomato cultivar H-86 (Kashi Vishesh) developed at the Indian Institute of Vegetable Research, Varanasi, India, was used throughout the study. Following optimization experiments, an application of the enhanced protocol was assessed using a further two economically important cultivars, H-24 (Hisar Anmol) and DVRT-1 (Kashi Amrit). In an earlier study, the genotype H-86 was reported to be the most responsive for organogenesis, followed by H-24 and DVRT-1 (Singh et al. 2010), respectively. These three cultivars perform very well in the Indian subcontinent and show field resistance to tomato leaf curl virus. To date, there is no published report on the transformation of these cultivars. In the present study, a highly efficient Agrobacterium-mediated tomato transformation protocol was developed by optimizing factors including explant age, germination medium, pre-culture period, pre-culture and co-culture media, feeder layer, pH, inoculation medium, inoculation duration, washing medium, thiamine concentration, and kanamycin concentration in the initial selective regeneration medium, resulting in successful and equally efficient transformation of all three tomato varieties.
Materials and Methods
Gene construct, Agrobacterium strain, and culture conditions.
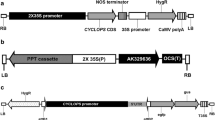
A. tumefaciens strain GV3101 harboring a binary vector based on pCAMBIA2300 (Hajdukiewicz et al. 1994) was used for transformation (Fig. 1). The pCAMBIA2300 vector harbors a neomycin phosphotransferase (npt II) selection marker gene driven by a duplicated Cauliflower Mosaic Virus 35S (CaMV35S) promoter. The entire cassette was fused with AtDREB1A/CBF3 cDNA (GenBank accession no. AF074602; Gilmour et al. 1998) driven by water-deficit stress-inducible rd29A promoter of Arabidopsis (GenBank accession no. D13044; Yamaguchi-Shinozaki and Shinozaki 1993). For the Agrobacterium culture, a single colony from a fresh bacterial culture plate was inoculated in 50 ml liquid Luria–Bertani medium containing 25 mg l−1 kanamycin, 25 mg l−1 gentamycin, and 25 mg l−1 rifampicin and grown overnight at 28°C on a rotary shaker at 200 rpm. Overnight cell cultures were harvested by centrifugation at 5,000×g for 10 min at room temperature, and cells were resuspended in 20 ml inoculation medium (IM; Table 1) to OD600 = 0.8.
Map of the T-DNA region of binary vector pCAMBIA2300. LB left border, RB right border, nptII neomycin phosphotransferase, CaMV35S cauliflower mosaic virus 35S promoter, 35S poly A CaMV35S poly A signal, Nos Ter terminator sequence of nopaline synthase, AtDREB1A Arabidopsis thaliana dehydration responsive element binding factor 1A, Prd29A promoter rd29A.
Culture conditions and media.
Murashige and Skoog’s (MS; Murashige and Skoog 1962) basal salts supplemented with 3 % (w/v) sucrose was used as the base culture medium. For the aseptic germination of seeds, half-strength MS basal salts were used. The required quantities of salts were mixed and appropriate pH was adjusted using 1 N NaOH or 1 N HCl prior to the addition of 0.5 % (w/v) agar (Duchefa, Haarlem, the Netherlands). Media were autoclaved at 121°C for 20 min. The cultures in all the experiments were incubated at 25 ± 2°C with a 16-h photoperiod provided by cool white fluorescent lights of 40 W (Philips India Ltd.) providing a photosynthetic photon flux density of 80 μmol m−2 s−1. The composition of the media used in different transformation experiments and stages are described in Tables 1 and 2.
MS medium supplemented with 6-benzyladenine (BA; 8.9 μM), kinetin (Kin; 9.3 μM), and kanamycin (50 mg l−1) was used as the basis for the selective regeneration medium (SRM; Table 1). In order to arrest the growth of Agrobacterium cells, cefotaxime (500 mg l−1) was added to the selection medium. For shoot development, shoot buds were cultured on MS medium containing 50 mg l−1 kanamycin and 500 mg l−1 cefotaxime for 15 d. Shoots were then rooted in rooting medium (RM) comprising growth regulator-free MS basal salts containing 100 mg l−1 kanamycin and 400 mg l−1 cefotaxime. Rooted shoots were acclimatized in sterile soil and sand mixture (1:1) and irrigated with sterile water containing 50 mg l−1 kanamycin.
Donor material and explant preparation.
To generate cotyledon explants, seeds of tomato (S. lycopersicum) var. H-86, DVRT-1, and H-24 were extracted from fresh ripe fruits collected from the field. Fruits were washed thoroughly under running water, wiped with ethanol, and then flamed under a laminar airflow cabinet (Klenzaids, India). Surface-sterilized fruits were cut open and the seeds, together with mucilaginous pulp, collected in a sterile beaker and thereafter treated with 3 % (v/v) hydrochloric acid (HCl) for 3 min with gentle shaking to dissolve the pulp. Seeds were again treated with 2 % (v/v) HCl for 3 min and rinsed two to three times with sterile double distilled water. The seeds were finally surface-sterilized using 3 % (v/v) sodium hypochlorite (NaOCl) solution for 3 min and thereafter rinsed four to five times with sterile double distilled water, blotted dry, and inoculated in magenta boxes containing 50 ml germination medium (GM1, GM2, or GM3; Table 1). The cultured seeds were kept in the dark for the initial 3 d and thereafter transferred to the light.
Transformation and plant regeneration.
Cotyledons were cut at the tips as well as at the base, inverted onto the pre-culture and co-culture media (PCM; Table 1) in 90 × 15-mm Petri dishes (Tarsons, Kolkata, India), and pre-cultured for 0–8 d. The tobacco feeder cell layer was prepared by plating 2 ml of actively growing tobacco cell suspension onto 20 ml of solidified medium in Petri dishes. After 1 d of incubation in the growth chamber, dried sterile filter paper discs were placed on top of the tobacco cells and the cotyledon explants then placed on the filter paper. The tobacco feeder cell suspensions were replaced weekly by 1:10 dilutions into fresh liquid medium containing casein hydrolysate (200 mg l−1) and 2,4-dichlorophenoxyacetic acid (0.2 mg l−1), as per the procedures of Hamza and Chupeau (1993) and Cortina and Culianez-Macia (2004). Pre-cultured explants from the PCM plates were dipped in prepared Agrobacterium suspension cultures for different time periods in the dark at 26°C for inoculation. Only inoculation buffer during the transformation was used as the control. After inoculation, excess bacterial suspension was removed by pipetting and the explants kept on sterile filter paper to drain extra bacterial suspension. Agrobacterium-inoculated explants were returned to the same PCM plates for 48 h of co-cultivation. Co-cultivated explants were washed three times using a washing medium (WM; Table 2) with gentle shaking, treated with cefotaxime (5 mg ml−1 in WM for 20 min), rewashed once, and then blotted dry. The washed explants were incubated for the next 10 d on SRM (Table 1) containing 20–50 mg l−1 kanamycin and 500 mg l−1 cefotaxime. The green and regenerating cotyledons were sub-cultured after every 14 d onto fresh SRM of the same type, containing 50 mg l−1 kanamycin. Regenerated shoots from putatively transformed explants were excised and cultured on RM for ∼25 d; no subcultures were performed during this period.
Transformation parameters.
Based on the information from the available literature for the transformation of S. lycopersicum genotypes, a general protocol (non-optimized parameters) was devised, combining various optimal parameters for transformation. In this protocol, cotyledons derived from 6-d-old seedling were germinated on half-strength MS basal medium containing 8.9 μM BA; pre-cultured on MS basal medium containing BA (8.9 μM), Kin (9.3 μM), and thiamine (0.4 mg l−1); and inoculated with Agrobacterium cells for 5 min. Inoculated explants were co-cultivated on the same pre-culture medium for 48 h. After co-cultivation, explants were washed with a medium having only one fourth-strength MS macronutrient salts and finally selected on the regeneration medium containing 0.4 mg l−1 thiamine and 30 mg l−1 kanamycin for 10 d; thereafter, the kanamycin concentration was increased to 50 mg l−1. Later, each parameter was varied, keeping other non-optimized parameters from the general protocol, in a sequential order, one at a time. A particular condition optimized in one experiment was used in the subsequent experiments. In this way, 11 parameters were systematically tested; details of the media composition are given in Table 1.
To investigate the effect of the seed germination medium (GM), seeds were germinated on three different germination media: GM1, GM2, or GM3 (Table 1). The effect of the age of explants was evaluated using cotyledon explants isolated from 3- to 9-d-old (post-emergence) seedlings. The explants on the pre-culture medium were maintained for 0–8 d prior to inoculation with Agrobacterium cells. Cotyledons were pre-cultured and co-cultured on one of the six different pre-culture and co-culture media (PCM1–PCM6 media; Table 1) varying in their pH, Kin, and thiamine concentrations. The cotyledon explants were cultured on a layer of tobacco feeder cells, applied onto PCM5 plate either from the pre-culture or from the co-cultivation (a new PCM5 medium plate). To investigate whether pH, BA, and Kin in the IM influence the transformation efficiency, the pre-cultured explants were immersed in 20 ml of different liquid inoculation media (IM1, IM2, IM3, or IM4) containing Agrobacterium cells. To investigate the effect of pH on the transformation efficiency, the pH values of both PCM and IM were adjusted to 4.2, 5.0, or 5.8 using diluted HCl or NaOH solution. The cotyledon explants were inoculated with the A. tumefaciens cell suspension for 5, 10, 15, 30, or 45 min. The growth of Agrobacterium cells after co-cultivation was inhibited by washing with one of six different washing media (WM1–WM6; Table 2). To evaluate the effect of thiamine on the transformation efficiency, two different concentrations of thiamine (0.1 and 0.4 mg l−1) were used in the selective regeneration medium (SRM1 and SRM2, respectively; Table 1). To investigate the effect of kanamycin concentration for first 10 d of selection, kanamycin at concentrations of 20, 30, 40, or 50 mg l−1 was added to the respective SRM (Table 1). Subsequently, the cultures were transferred to fresh medium, the selection pressure was increased to 50 mg l−1 kanamycin (SRM2), and further subcultures were conducted every 14 d to the same medium.
Transformation efficiencies were calculated as the percent co-cultivated explants producing independent transformation events. An independent transformation event is defined as an explant or callus which regenerated after 8 wk culture on the selection medium.
PCR analysis of transgenic plants.
The presence of transgene in transformed shoots was confirmed through PCR amplification of AtDREB1A and nptII genes using gene-specific primers. Genomic DNA was isolated from leaves of putative transformed shoots by the Purelink plant DNA purification kit (Invitrogen, Carlsbad, CA, USA). The primer pair for the amplification of the AtDREB1A gene was F-5′-ttggctccgattacgagtcttcg-3′ and R-5′-aacgatacgtcgtcatcatcg-3′, which produced an amplified product of 607 bp. The expected product size for nptII gene-specific primer pair F-5′-gcgataccgtaaagcacgaggaa-3′ and R-5′-gatggattgcacgcaggttctc-3′ was 728 bp.
Southern blot analysis.
The Southern (DNA) blot was prepared by digesting 15 μg of genomic DNA with HindIII, which was expected to generate a T-DNA junction fragment of size ≥4.05 kb. Digested DNA samples were separated on a 0.8 % agarose gel and transferred to a positively charged nylon membrane (Hybond-N+, Amersham Biosciences, Ltd., Little Chalfont, UK). The membrane was hybridized with a digoxigenin-11-dUTP-labeled AtDREB1A probe; the probe was prepared following PCR amplification of the internal fragment of the AtDREB1A gene. Hybridization was carried out in DIG Easy Hyb buffer overnight at 50°C following the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche, Mannheim, Germany) application manual. The membrane was washed twice in 2× solution of sodium chloride/sodium citrate (SSC) + 0.1 % sodium dodecyl sulfate (SDS) at 25°C for 5 min and twice in 0.5× SSC + 0.1 % SDS at 68°C for 15 min with constant agitation. Hybridized DNA bands were visualized following the DIG immunological detection protocol.
Inheritance patterns of the transgene.
Segregation of the AtDREB1A as well as the nptII transgenes in T1 progeny seedlings of 56 independent transformation events was analyzed. For each of the 56 independent transformed plants, 120 T1 generation seeds from self-fertilized T0 fruits were germinated in pots filled with sand and soil mixture (1:1). After 25 d growth, leaf tissue was taken for genomic DNA isolation and further PCR analysis was conducted using AtDREB1A and nptII gene-specific primers.
Statistical analyses.
Each experiment was set up in a randomized block design with three replications per treatment, where each replicate had minimum 50 explants. Data were analyzed using ANOVA appropriate for the design using SPSS 11.5; the differences among the treatment means were compared using Fisher’s least significant difference test (LSD) at the 5 % probability level. Tukey’s test at the 5 % probability level was used to separate treatment mean values by SPSS 11.5. Chi-square analysis of the goodness of fit was performed to analyze the T1 segregation ratios.
Results and Discussion
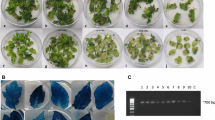
The tomato cultivar H-86 was used in the experiments to optimize transformation factors. A transformation frequency of 22.05 % was recorded when using the protocol with non-optimized parameters. During the standardization of the parameters, at least 150 explants were used to test each treatment; the initial response of explants could be scored as early as 5 wk on the selection medium in terms of dark green swelling and incipient regeneration from the embryo axis (Fig. 2c ). Pre-cultured control explants which did not receive co-cultivation became necrotic and died on the selection medium after 5 wk (Fig. 2d ). Adventitious shoot regeneration in cotyledons predominantly occurred at the cut ends resulting from the broken embryo axis (Fig. 2c ). The putative transformed shoots rooted on 100 mg l−1 kanamycin rooting medium were found to be transformed (Fig. 2g–i ). The presence and transmission of the AtDREB1A gene was confirmed by PCR assays of leaves randomly chosen from putatively transformed shoots (Fig. 3), Southern blot analysis (Fig. 4), and PCR assays of progeny (T1) from T0 transgenic plants (Fig. 5b , c). Transformation efficiencies were calculated as the percent co-cultivated explants producing independent transformation events. An independent transformation event is defined as an explant or callus which regenerated after 8 wk culture on the selection medium.
Different stages of Agrobacterium-mediated transformation of tomato var. H-86. In vitro-germinated seedlings on GM2. (a) Pre-cultured cotyledon explants on PCM. (b) Shoot initiation on selection medium after 5 wk. (c) Non-transformed cotyledons on selection medium after 4 wk. (d) Shoot development from transformed callus on selection medium after 12 wk of culture. (e) Multiple shoots from regenerating callus on selection medium. (f) Well-rooted plantlets on selection medium. (g) Acclimatization of a transgenic plant. (h) Mature transgenic tomato T0 plants under containment conditions bearing normal fruits.
Southern blot analysis of ten randomly selected transgenic events using the AtDREB1A gene-specific probe. Transgenic plants showed transgene integration, whereas no hybridization signal was detected in the untransformed WT control. Signals corresponding to transgenic events represent the T-DNA left border/plant junction fragments ≥4.05 kb. P plasmid pCAMBIA2300. Numbers 30, 34, 39, 44, 53, 68, 71, 79, 63, and 96 represent independent transgenic events.
T1 seedlings derived from self-fertilized primary transformed plants (T 0 ) germinated in seed trays under containment conditions (a). PCR amplification products of AtDREB1A (b) or nptII (c) genes from genomic DNA of T1 seedling progenies. Lanes P, plasmid DNA; NT, DNA from non-transformed seedling; numbers in lanes b and c correspond to individual T1 seedlings; M, 1-kb DNA ladder.
Germination medium.
There were significant differences in the transformation frequencies obtained using cotyledon explants (∼200) derived from seeds germinated on the different GM (Table 3). GM supplemented with 8.9 μM BA (GM2; Table 1) was found to improve the transformation efficiency to 22.05 % compared to 6.22 % for growth regulator-free germination medium (GM1). In general, the seedlings germinated on GM2 were more vigorous than those from GM1. The explants from seedlings grown on BA-supplemented medium produced the highest frequency of regenerated plants when sub-cultured to media also containing BA. The regeneration of explants obtained from seedlings grown on media lacking BA was subsequently inhibited when transferred to BA-containing regeneration medium. These results are in agreement with the findings of Newman et al. (1996) who specifically studied the effects of BA on tomato transformation. In the present study, GA3 was found to compromise the transformation rates by reducing the number of independent transformation events (to 15.67 %) when GA3 (5.8 μM) and BA (8.9 μM) were added to the medium (GM3; Table 3). GA3 has been reported to accelerate seed germination in tomato (Andreoli and Khan 1999). However, the incorporation of 2.89 μM GA3 in the shoot regeneration medium along with 8.9 μM BA has been reported to adversely affect the regeneration response of tomato hypocotyl and cotyledon explants (Shivakumar et al. 2007). GA3 partially inhibits starch accumulation in tomato cotyledons (Branca et al. 1994), a prerequisite for the development of shoots in vitro. It has been reported that pretreatment of source plants with anti-GA3 compounds such as 2-chloroethyl trimethyl ammonium chloride caused an increase in the frequency of shoot regeneration in tomato (De Langhe and De Bruijne 1976).
Age of explants.
It is well established that the age of the explants greatly influences their transformation and regeneration potential (Davis et al. 1991). Although transformation was achieved with cotyledons of all ages (3–9 d), significant differences were noted among different explant ages (Fig. 6a ). An increase in the transformation frequency was observed with increasing age of the explant until 6 d, when the maximum transformation frequency of 22.44 % (44/193) was achieved, after which it gradually declined. The higher frequencies observed for up to 6-d-old seedlings may be attributed to the fact that active plant cell metabolism is required for the synthesis of vir-inducing factors (Stachel et al. 1986), leading to effective plant transformation. Mazumdar et al. (2010) reported that in the case of Jatropha curcas, the cotyledonary explants excised from freshly in vitro-germinated seedlings were the most amenable to Agrobacterium-mediated transformation as compared to older explants. Similarly, Famiani et al. (1994) demonstrated that explants derived from young apple leaves showed more regeneration potential than older leaves. Developing leaves have less differentiated and more metabolically active cells, and show improved plant regeneration responses, when provided with suitable hormonal and nutritional conditions. In contrast, Davis et al. (1991) found a higher percentage of transformation and less necrosis when using older tomato leaves. The majority of reports for tomato transformation suggest using cotyledons from 6- to 10-d-old seedlings for transformation (Davis et al. 1991; Frary and Earle 1996; Park et al. 2003; Sun et al. 2006; Wu et al. 2006; Shivakumar et al. 2007; Gao et al. 2009).
Effect of different parameters on the transformation frequencies of cotyledon explants from tomato var. H-86: explant age (a); pre-culture period (b); pre-culture and co-culture medium (c); tobacco feeder cell layer: No FL no feeder layer, Pre-cul + co-cul FL feeder layer during pre-culture and co-cultivation, Co-cul FL feeder layer during co-cultivation only (d); washing medium (e); kanamycin concentration in initial selection medium (f). Data represent the mean values ± SE of three different experiments, where a minimum of 50 explants were taken in each experiment. Differences between means were compared by Fisher’s least significant difference test (LSD; P = 0.05). Means were separated by Tukey’s test (P = 0.05); mean values having the same letter are not significantly different.
Pre-culture period.
The transformation frequency was significantly affected by the pre-culture period (Fig. 6b ). During pre-culture, most of the explants swelled substantially. Lower transformation frequencies (0.45 % for explants without pre-culture and 6.32 % for those with 2 d pre-culture) were recorded with short pre-culture periods (1–2 d), whereas pre-culture beyond 2 d significantly improved transformation (18.49 and 22.48 % for 3 and 6 d, respectively). These results are in contrast to the reports of pre-culture for 1 d (10.6 %: Frary and Earle 1996; 20.0 %: Park et al. 2003) and 2 d (12.5 %: Cortina and Culianez-Macia 2004; 11.3 %: Ellul et al. 2003). Actively dividing cells, particularly those in the S phase of the cell division cycle, are the most amenable to genetic transformation by Agrobacterium (Villemont et al. 1997; Gordon-Kamm et al. 2002). Over the past two decades, several studies revealed the existence of a “window of competence,” a time during which the cells are susceptible to Agrobacterium transformation, although the basis for this response has not yet been completely established (Sunilkumar et al. 1999; Costa et al. 2002). Pre-culture facilitates active cell division, and pre-culture treatment of tissues has been observed to improve the transformation frequency in tomato (Park et al. 2003; Cortina and Culianez-Macia 2004). It has also been reported that a pre-culture period increases the regeneration percentage and cell or tissue vitality to overcome the stress resulting from the co-cultivation with Agrobacterium (Arcos-Ortega et al. 2010). Tomato tissues have also been transformed without a pre-culture phase, but the transformation frequency was reported to be low (2–15.5 %; Sigareva et al. 2004).
Pre-culture and co-culture media.
A significant difference in the transformation frequency of cultivar H-86 was noted due to the different pre-culture and co-culture media (PCM1–PCM6; Table 1), which varied mainly in terms of Kin (0.0, 2.3, and 9.3 μM) and thiamine (0.1 and 0.4 mg l−1) concentrations and the pH (5.8, 5.0, and 4.2) of the medium (Fig. 6c ). In general, the transformation efficiency (based on ∼200 explants per treatment) increased with an increase in Kin and thiamine concentrations. In the case of Kin, the transformation efficiency increased from 8.02 % (2.3 μM Kin, PCM2) to 13.32 % (9.3 μM Kin, PCM3). This effect may be attributed to the ability of Kin to induce cell division leading to enhanced regeneration (An 1985). Chateau et al. (2000) reported the positive roles of growth regulators since they stimulate cell division and dedifferentiation, and set the phase of the plant cell cycle for efficient Agrobacterium-mediated transformation. Based on the results of preliminary regeneration experiments (data not shown), a considerable degree of regeneration from cotyledon explants was obtained on MS medium supplemented with 9.3 μM Kin and 8.9 μM BA (PCM3). The addition of 0.4 mg l−1 thiamine in MS medium (PCM4) caused a dramatic increase in the transformation efficiency to 23.53 % compared to that of 0.1 mg l−1 thiamine (13.32 %). However, among the different PCMs, the highest transformation frequency (30.89 %) was found on MS medium supplemented with 9.3 μM Kin, 8.9 μM BA, and 0.4 mg l−1 thiamine (PCM5), pH 5.0.
Tobacco feeder cell layer.
The effect of tobacco feeder cell layer during pre-culture and co-cultivation or only during the co-cultivation of tomato cotyledon transformation (Fig. 6d ) was studied; this experiment was conducted on MS medium containing BA (8.9 μM), kinetin (9.3 μM), and thiamine (0.4 mg l−1) having pH 5.0 (PCM5). The results indicated that the use of tobacco feeder cells during pre-culture and co-cultivation is beneficial to the transformation rate (24.23 %), but not essential (19.47 %; Fig. 6d ). Furthermore, a comparable rate of transformation could be achieved even in the absence of a feeder layer, which is consistent with that of Sharma et al. (2009). Previous reports indicated the necessity of tobacco or petunia cells as a feeder layer during the pre-culture and Agrobacterium co-cultivation of tomato explants, which may be attributed to the phenolic compounds that enhance Agrobacterium-mediated DNA delivery (McCormick et al. 1986; van Roekel et al. 1993; Frary and Earle 1996; Cortina and Culianez-Macia 2004; Frary and Van Eck 2005). Interestingly, a sharp decline was noted in the transformation frequency (to 8.94 %) when tobacco feeder cells were used during co-cultivation only. However, the reasons for this response require further investigation.
Inoculation medium.
The composition of the IM had a significant effect on the transformation efficiency of cotyledon explants. The use of sterile water (Wu et al. 2006) or MS basal salts (Davis et al. 1991) for Agrobacterium inoculation of explants has been previously reported. Three different pH values (5.8, 5.0, and 4.2) and two concentrations of thiamine (0.1 and 0.4 mg l−1), along with 8.9 μM BA and 9.3 μM Kin, were tested in IM, and their effect was recorded on the transformation efficiency. No significant difference in the transformation frequency was recorded between IM1 (30/208, 14.4 %) and IM2 (32/186, 17.16 %). However, IM2 effected a slight increase in the transformation frequency (Table 3), whereas a sharp improvement was recorded with IM3 (24.86 %). It is worthwhile to note that IM3 differed from IM2 and IM4 only with respect to pH. In addition, the medium composition of IM3 was similar to PCM5, except the addition of agar in PCM5 which used for pre-culture and co-cultivation. These results are in agreement with the report of Vernade et al. (1988) which indicates that a significant difference between the pre-induction medium and the wound site environment may result to the inactivation of vir gene expression.
Influence of medium pH.
It has been reported that the surrounding pH of the medium is crucial for Agrobacterium virulence (Vernade et al. 1988). Three different pH values (4.2, 5.0, and 5.8) were evaluated both for IM as well as the PCM using approximately 200 explants per treatment. The highest transformation frequencies of 30.89 and 24.86 % were obtained using the pH value of 5.0 in media PCM5 and IM3, respectively. A lower pH value (4.2) either in PCM (PCM6) or in IM (IM4) drastically reduced the transformation frequency (Table 3). These results are in agreement with the findings of Wu et al. (2006) where the highest transformation frequency was obtained with inoculation and co-cultivation media having pH 5.0 coupled with acetosyringone. Similarly, Gao et al. (2009) reported that pH 5.2 was the best for the co-cultivation of tomato cotyledons. The dependence of vir genes induction on pH values between 5.0 and 6.0 has also been noted by other investigators during the co-cultivation of A. tumefaciens with plant cells (Stachel et al. 1986; Usami et al. 1988; Vernade et al. 1988). Stachel et al. (1986) obtained a maximal level of vir induction at pH 5.1, while Vernade et al. (1988) reported a fall in vir induction when the pH increased from 5.1 to 5.6. The pH control of vir gene expression is known to act at the level of expression of virG (Vernade et al. 1988). In addition, induction of the vir cascade absolutely requires an intact virA gene, which takes place only under acidic pH conditions (Usami et al. 1988). Our study confirms that a lower pH (5.0) of inoculation and pre-culture and co-cultivation media produced the highest transformation frequency.
Period of inoculation.
The time period provided for A. tumefaciens cells to adhere to the plant tissue surface also affects the transformation rates. Different inoculation periods (5, 10, 15, 30, or 45 min) produced significantly variable results. The maximum transformation frequency of 25.34 % (47/184; Table 3) was obtained when the tissue was inoculated with A. tumefaciens cells for 5 min. Inoculation beyond 5 min resulted in a gradual decrease in the frequency of transformation, and most of the explants died due to Agrobacterium overgrowth and necrosis. A similar response for tomato explants was observed by both Frary and Earle (1996) and Shivakumar et al. (2007). In contrast, a longer inoculation period (30 min) has been also reported to give a comparable (22 %) transformation frequency (Cortina and Culianez-Macia 2004); however, such a result might be attributed to a different genotype used in the study.
Washing medium.
Removal of the excess Agrobacterium cells from the explant tissue after co-cultivation using a WM is critical as it prevents Agrobacterium overgrowth during subsequent subcultures. The washing medium (WM5) consisting of one third-strength MS macronutrient salts resulted in the highest transformation efficiency, using 200 explants per treatment (36.6 %; Fig. 6e ), while quarter-strength MS macronutrient salts (WM6) also produced a reasonable rate of transformed plants (26.15 %). Washing of the co-cultivated explants with sterile water was ineffective, leading to a significantly reduced transformation frequency, to just 1.04 %. In contrast, washing with full-strength MS medium (WM1) attracted fungal contamination and caused Agrobacterium overgrowth. Previous reports on tomato transformation used only full-strength MS basal salts for washing of co-cultivated explants (Davis et al. 1991; Sharma et al. 2009).
Effect of thiamine on transformation.
Two different concentrations of thiamine (0.1 and 0.4 mg l−1) were tested both in PCM and SRM (Table 1) based on Cortina and Culianez-Macia (2004). Thiamine at 0.4 mg l−1 in the PCM4 and SRM2 media significantly improved the transformation efficiency to 37.89 %, which was approximately double that of SRM1 containing 0.1 mg l−1 thiamine (18.85 %; Table 3). When SRM1 was tested, PCM3 medium containing 0.1 mg l−1 thiamine was used for pre-culture and co-cultivation. A higher concentration (0.4 mg l−1) of thiamine in the medium resulted in a decrease in the number and size of necrotic lesions and promoted cell growth that resulted in a two fold increase in the transformation efficiency. In contrast, a four fold increase in the transformation frequency was reported by Cortina and Culianez-Macia (2004), but this may be attributed to the genotypic differences.
Kanamycin concentration in selection media.
An effective selection regime is very important for developing an efficient plant transformation procedure. Among the different concentrations (20, 30, 40, or 50 mg l−1) of kanamycin tested for the initial selection period of 10 d, the highest (44.30 %, 82/185 explants) transformation efficiency was recorded with medium containing 20 mg l−1 kanamycin. A further increase in kanamycin concentration resulted in a reduced rate of transformation: 35.85, 30.17, and 29.59 % for 30, 40, and 50 mg l−1, respectively (Fig. 6f ). A kanamycin sensitivity test of untransformed tomato cotyledons revealed complete yellowing and bleaching of explants on medium containing 30 mg l−1 kanamycin (Fig. 2d ; data not shown). At a high initial selection pressure (40 or 50 mg l−1 kanamycin), non-transformed cells died rapidly and it is possible that the dead tissues may have poisoned the selection medium, leading to slow or non-proliferation of transformed cells (Pacurar et al. 2008). In the case of Brassica oleracea, delays in the transfer of co-cultivated explants to a higher kanamycin concentration in the selection medium by 7–10 d have been recommended (Chakrabarty et al. 2002). A similar result was also reported with the transformation of Brassica campestris hypocotyls, where 30 mg l−1 kanamycin was used for initial 4 d before transferring the explants to a medium containing 50 mg l−1 kanamycin (Xiang et al. 2000). In contrast, Shivakumar et al. (2007) applied no selection pressure for the initial 5 d after the co-cultivation of tomato cotyledons and then transferred the shoots to a selection medium containing 75 mg l−1 kanamycin. A rather low selection pressure early in the selection process might allow transformed cells to reach the phase of high regeneration capacity, and plants can be regenerated from transformed cells with the support of cross-feeding from surrounding non-transformed tissue (D’Halluin et al. 1992).
Southern analysis of primary transformants (T0 plants).
Southern blot analysis of the transgenic events using AtDREB1A probe confirmed the integration of the target gene into the genome of tomato plants (Fig. 4). Genomic DNA isolated from leaves of ten randomly selected events of the H-86 genotype was analyzed. Genomic DNA of the putative transformed and non-transformed wild-type (WT) control plants was digested with HindIII which cuts once within the T-DNA between the right border and upstream of the rd29A promoter. Since the HindIII site is unique in the T-DNA and the other site originates from the flanking plant genomic sequence, a T-DNA left border (LB)/plant junction fragment(s) of size ≥4.05 kb is expected. As shown in Fig. 4, all ten transformed plants analyzed revealed single or multiple insertions of the transgene. Interestingly, nine of ten transformed plants showed a single band, indicative of being single-copy T-DNA insertions. The sizes of the LB junction fragments in the plants analyzed were greater than 4.05 kb. As expected, no signal was detected in the non-transformed control.
Inheritance of transgene.
T1 seeds from putative transformed plants were grown in a glasshouse and all were observed to be normal in development and morphology (Fig. 5a ). The presence of the transgene in the T1 progeny plants of 56 independently transformed lines was confirmed through PCR amplification of AtDREB1A (Fig. 5b ) and nptII genes (Fig. 5c ). PCR analysis revealed that 71.5 % of the seedling progenies from independent events showed a good fit to typical Mendelian segregation ratios (64.3 % with a 3:1 ratio indicating one transgene locus, 5.4 % indicating two transgene loci, and 1.8 % indicating three transgene loci; Table 4). Some distortion (28.5 %) from Mendelian segregation ratios was also observed in transformed lines (10.7 % for 1:3 and 17.8 % for 1:1; Table 4). The segregation data permit the estimation of the number of unlinked T-DNA inserts present in the transformed lines (Katavic et al. 1994). Taken together, the results from the Southern analysis and inheritance studies indicate that stable transformation with single-copy insertion events occurred in the majority of transformed lines, but few multiple-copy transformation and chimeric events were also obtained. Interestingly, progenies of 17.8 % (10 out of 56) plants segregated as a 1:1 ratio in the T1 generation, suggesting that the transgene was inherited exclusively through one gamete with the insertional mutation compromising the viability of the other gamete (Cooley et al. 1995; Mohanty et al. 1999). The non-Mendelian segregation ratios recorded in the present study may be alternatively attributed to chimeras in the primary transformed plants containing both transformed and non-transformed cells (McHughen and Jordan 1989).
Optimizing the transformation parameters resulted in a two fold increase in the transformation efficiency of the H-86 genotype to 44.3 %. To determine the adaptability of the optimized transformation protocol for S. lycopersicum var. H-86 for other tomato genotypes, varieties H-24 and DVRT-1 were also tested. A similar level of increase was recorded for the transformation rates for the other two varieties, which is obvious since all the three varieties share a common lineage involving Lycopersicon hirsutum f. grabratum. In the case of H-24, a transformation frequency of 42.7 % was obtained compared to 18.7 % with non-optimized factors, while for DVRT-1, a 35.3 % frequency was recorded compared to 16.7 % for non-optimized parameters (Table 5). The higher transformation efficiencies are probably attributable to the cumulative effects of optimized factors for tomato cultivar H-86 and to the better regeneration capability of the genotype H-86 (Singh et al. 2010). The results also indicate that the protocol can be successfully applied to other tomato varieties. In previous studies with other varieties of tomato, transformation efficiencies of 6 % (var. Pusa Ruby; Vidya et al. 2000), 9 % (var. Moneymaker; van Roekel et al. 1993), 11 % (var. Moneymaker; Frary and Earle 1996), 14 % (var. UC82B; Hamza and Chupeau 1993), 25 % (UC82; Hu and Phillips 2001), 37 % (var. Moneymaker; Ling et al. 1998), and 41.4 % (var. Pusa Ruby; Sharma et al. 2009) have been achieved.
In summary, this present study revealed that the parameters including cotyledon age, pre-culture and co-cultivation on the regeneration medium, pre-culture duration, pH of the medium, inoculation medium, washing medium, thiamine concentration, and kanamycin concentration for initial selection are critical to achieve high transformation rates. The improved tomato transformation system described here is reliable, suited for small-scale as well as large-scale transformation experiments generating a large number of transgenic lines. However, still there is need to test the protocol with other popular tomato lines, especially which are completely unrelated to those tested in the present experiment.
References
An G (1985) High efficiency transformation of cultured tobacco cells. Plant Physiol 79:568–570
Andreoli C, Khan A (1999) Matriconditioning integrated with gibberellic acid to hasten seed germination and improve stand establishment of pepper and tomato. Pesqui Agropecuá Bras 34:1953–1958
Arcos-Ortega GF, Chan-Kuuk RA, González-Kantún WA, Souza-Perera R, Nakazawa-Ueji YE, Avilés-Berzunza E, Godoy-Hernández G, Lawton MA, Aguilar JJZ (2010) Agrobacterium tumefaciens-transient genetic transformation of Habanero pepper (Capsicum chinense Jacq.) leaf explants. Elect J Biotech 13. doi:10.2225/vol13-issue4-fulltext-10 Available at: http://www.ejbiotechnology.cl/content/vol13/issue4/full/10ISSN0717-3458
Arumuganathan K, Earle E (1991) Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep 9:208–218
Behnam B, Kikuchi A, Celebi-Toprak F, Kasuga M, Yamaguchi-Shinozaki K, Watanabe KN (2007) Arabidopsis rd29A::DREB1A enhances freezing tolerance in transgenic potato. Plant Cell Rep 26:1275–1282
Branca C, Torelli A, Fermi P, Altamura MM, Bassi M (1994) Early phases in in vitro culture of tomato cotyledons: starch accumulation and protein pattern in relation to the hormonal treatment. Protoplasma 182:59–64
Chakrabarty R, Viswakarma N, Bhat SR, Kirti PB, Singh BD, Chopra VL (2002) Agrobacterium-mediated transformation of cauliflower: optimization of protocol and development of Bt-transgenic cauliflower. J Biosci 27:495–502
Chateau S, Sangwa RS, Sangwan-Norreel BS (2000) Competence of Arabidopsis thaliana genotypes and mutants for Agrobacterium tumefaciens-mediated gene transfer: role of phytohormones. J Exp Bot 51:1961–1968
Cooley J, Ford T, Christou P (1995) Molecular and genetic characterization of elite transgenic rice plants produced by electric discharge particle acceleration. Theor Appl Genet 90:97–104
Cortina C, Culianez-Macia FA (2004) Tomato transformation and transgenic plant production. Plant Cell Tissue Organ Cult 76:269–275
Costa MGC, Otoni WC, Moore GA (2002) An evaluation of factors affecting the efficiency of Agrobacterium-mediated transformation of Citrus paradise (Macf.) and production of transgenic plants containing carotenoid biosynthetic genes. Plant Cell Rep 21:365–373
D’Halluin K, De Block M, Denecke J, Janssens J, Leemans J, Reynaerts A, Botterman J (1992) The bar gene as selectable and screenable marker in plant engineering. Methods Enzymol 216:415–426
Davis ME, Lineberger RD, Miller AR (1991) Effects of tomato cultivar, leaf age, and bacterial strain on transformation by Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult 24:115–121
De Langhe F, De Bruijne E (1976) Continuous propagation of tomato plants by means of callus culture. Sci Hortic 4:221–227
Ellul P, Garcia-Sogo B, Pineda B, Ríos G, Roig L, Moreno V (2003) The ploidy level of transgenic plants in Agrobacterium-mediated transformation of tomato cotyledons (Lycopersicon esculentum L. Mill.) is genotype and procedure dependent. Theor Appl Genet 106:231–238
Famiani F, Ferradini N, Staffolani P, Standardi A (1994) Effect of leaf excision time and age, BA concentration and dark treatments on in vitro shoot regeneration of M26 apple rootstock. J Hort Sci 69:679–685
Frary A, Earle ED (1996) An examination of factors affecting the efficiency of Agrobacterium-mediated transformation of tomato. Plant Cell Rep 16:235–240
Frary A, Van Eck J (2005) Organogenesis from transformed tomato explants. Meth Mol Biol 286:141–150
Gao N, Shen W, Cao Y, Su Y, Shi W (2009) Influence of bacterial density during pre-culture on Agrobacterium-mediated transformation of tomato. Plant Cell Tissue Organ Cult 98:321–330
Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16:433–442
Goel D, Singh AK, Yadav V, Babbar SB, Bansal KC (2010) Overexpression of osmotin gene confers tolerance to salt and drought stresses in transgenic tomato (Solanum lycopersicum L.). Protoplasma 245:133–141
Gordon-Kamm W, Dilkes BP, Lowe K, Hoerster G, Sun X, Ross M, Church L, Bunde C, Farrell J, Maddock S, Snyder J, Sykes L, Li Z, Woo YM, Bidney D, Larkins BA (2002) Stimulation of the cell cycle and maize transformation by disruption of the plant retinoblastoma pathway. Proc Natl Acad Sci USA 99:11975–11980
Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25:989–994
Hamza S, Chupeau Y (1993) Re-evaluation of conditions for plant regeneration and Agrobacterium-mediated transformation from tomato (Lycopersicon esculentum). J Exp Bot 44:1837–1845
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Hu W, Phillips GC (2001) A combination of overgrowth-control antibiotics improves Agrobacterium tumefaciens-mediated transformation efficiency for cultivated tomato (L. esculentum). In Vitro Cell Dev Biol Plant 37:12–18
Jia GX, Zhu ZQ, Chang FQ, Li Y (2002) Transformation of tomato with the BADH gene from Atriplex improves salt tolerance. Plant Cell Rep 21:141–146
Katavic V, Haughn GW, Reed D, Marilyn M, Kunst L (1994) In planta transformation of Arabidopsis thaliana. Mol Gen Genet 245:363–370
Khare N, Goyary D, Singh NK, Shah P, Rathore M, Anandhan S, Sharma D, Arif M, Ahmed Z (2010) Transgenic tomato cv. Pusa Uphar expressing a bacterial mannitol-1-phosphate dehydrogenase gene confers abiotic stress tolerance. Plant Cell Tissue Organ Cult 103:267–277
Liénard D, Sourrouille C, Gomord V, Faye L (2007) Pharming and transgenic plants. Biotechnol Annu Rev 13:115–147
Ling H-Q, Kriseleit D, Ganal MG (1998) Effects of ticarcillin/potassium clavulanate on callus growth and shoot regeneration in Agrobacterium-mediated transformation of tomato (Lycopersicon esculentum Mill). Plant Cell Rep 17:843–847
Mazumdar P, Basu A, Paul A, Mahanta C, Sahoo L (2010) Age and orientation of the cotyledonary leaf explants determine the efficiency of de novo plant regeneration and Agrobacterium tumefaciens-mediated transformation in Jatropha curcas L. South African J Bot 76:337–344
McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R (1986) Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep 5:81–84
McHughen A, Jordan MC (1989) Recovery of transgenic plants from ‘escape’ shoots. Plant Cell Rep 7:611–614
Mohanty A, Sarma NP, Tyagi AK (1999) Agrobacterium-mediated high frequency transformation of an elite indica rice variety Pusa Basmati 1 and transmission of the transgenes to R2 progeny. Plant Sci 147:127–137
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol 15:473–497
Newman PO, Krishnaraj S, Saxena PK (1996) Regeneration of tomato (Lycopersicon esculentum Mill.): somatic embryogenesis and shoot organogenesis from hypocotyl explants induced with 6-benzyladenine. Intl J Plant Sci 157:554–560
Oh S-J, Song SI, Kim YS, Jang H-J, Kim SY, Kim M, Kim Y-K, Nahm BH, Kim J-K (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138:341–351
Pacurar DL, Thordal-Christensen H, Nielsen KK, Lenk I (2008) A high-throughput Agrobacterium-mediated transformation system for the grass model species Brachypodium distachyon L. Transgenic Res 17:965–975
Park SH, Morris JL, Park JE, Hirschi KD, Smith RH (2003) Efficient and genotype-independent Agrobacterium-mediated tomato transformation. J Plant Physiol 160:1253–1257
Salyaev RK, Rekoslavskaya NI, Stolbikov AS, Hammond RW, Shchelkunov SN (2007) Synthesis of hepatitis B virus surface antigen in tomato plants transgenic for the preS2-S gene. Doklady Biochem Biophysics 416:290–293
Sharma MK, Solanke AU, Jani D, Singh Y, Sharma AK (2009) A simple and efficient Agrobacterium-mediated procedure for transformation of tomato. J Biosci 34:423–433
Shivakumar B, Mythili JB, Anand L, Saiprasad GVS (2007) Influence of genotype on Agrobacterium-mediated transformation of tomato. Indian J Hort 64:251–257
Sigareva M, Spivey R, Willits MG, Kramer CM, Chang YF (2004) An efficient mannose selection protocol for tomato that has no adverse effect on the ploidy level of transgenic plants. Plant Cell Rep 23:236–245
Singh A, Singh M, Singh BD (2010) Comparative in vitro shoot organogenesis and plantlet regeneration in tomato genotypes. Indian J Hort 67:37–42
Stachel SE, Nester EW, Zambryski PC (1986) A plant cell factor induces Agrobacterium tumefaciens vir gene expression. Proc Natl Acad Sci USA 83:379–383
Sun H-J, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47:426–431
Sunilkumar G, Vijayachandra K, Veluthambi K (1999) Preincubation of cut tobacco leaf explants promotes Agrobacterium-mediated transformation by increasing vir gene induction. Plant Sci 141:51–58
Usami S, Okamoto S, Takebe I, Machida Y (1988) Factor inducing Agrobacterium tumefaciens vir gene expression is present in monocotyledonous plants. Proc Natl Acad Sci USA 85:3748–3752
van Roekel JSC, Damm B, Melchers LS, Hoekema A (1993) Factors influencing transformation frequency of tomato (Lycopersicon esculentum). Plant Cell Rep 12:644–647
Velcheva M, Faltin Z, Flaishman M, Eshdat Y, Perl A (2005) A liquid culture system for Agrobacterium -mediated transformation of tomato (Lycopersicon esculentum). Plant Sci 168:121–130
Vernade D, Herrera-Estrella A, Wang K, Van Montagu M (1988) Glycine betaine allows enhanced induction of the Agrobacterium tumefaciens vir genes by acetosyringone at low pH. J Bacteriol 170:5822–5829
Vidya CSS, Manoharan M, Kumar CTR, Savithri HS, Sita GL (2000) Agrobacterium-mediated transformation of tomato (Lycopersicon esculentum var. Pusa Ruby) with coat-protein gene of physalis mottle tymovirus. J Plant Physiol 156:106–110
Villemont E, Dubois F, Sangwan RS, Vasseur G, Bourgeois Y, Sangwan-Norreel BS (1997) Role of the host cell cycle in the Agrobacterium-mediated genetic transformation of petunia: evidence of an S-phase control mechanism for T-DNA transfer. Planta 201:160–172
Wu YF, Chen Y, Liang XM, Wang XZ (2006) An experimental assessment of the factors influencing Agrobacterium-mediated transformation. Russ J Plant Physiol 53:252–256
Xiang Y, Wong W-KR, Ma MC, Wong RSC (2000) Agrobacterium-mediated transformation of Brassica campestris ssp. parachinensis with synthetic Bacillus thuringiensis cry1Ab and cry1Ac genes. Plant Cell Rep 19:251–256
Yamaguchi-Shinozaki K, Shinozaki K (1993) Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol 101:1119–1120
Youm JW, Heung J, Jeon JH, Kim H, Kim YH, Ko K, Joung H, Kim HS (2008) Transgenic tomatoes expressing human beta-amyloid for use as a vaccine against Alzheimer’s disease. Biotechnol Lett 30:1839–1845
Acknowledgments
The authors are grateful to Dr. K. C. Bansal, Director, National Bureau of Plant Genetic Resources (NBPGR), New Delhi, India, for providing the AtDREB1A gene construct. The support from National Project on Transgenic Crops (NPTC), ICAR, New Delhi, India, is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: John W. Forster
Rights and permissions
About this article
Cite this article
Rai, G.K., Rai, N.P., Kumar, S. et al. Effects of explant age, germination medium, pre-culture parameters, inoculation medium, pH, washing medium, and selection regime on Agrobacterium-mediated transformation of tomato. In Vitro Cell.Dev.Biol.-Plant 48, 565–578 (2012). https://doi.org/10.1007/s11627-012-9442-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-012-9442-3