Abstract
A bacterial mannitol-1-phosphate dehydrogenase (mtlD) gene driven by the constitutive cauliflower mosaic virus (CaMV) 35S promoter was transferred into tomato plants using an Agrobacterium tumefaciens-mediated transformation protocol in an attempt to improve abiotic stress tolerance in the transformed plants. Transgene integration was confirmed by PCR analysis and Southern blot analysis, and transgene expression was confirmed by reverse transcription (RT)-PCR and direct mtlD (EC 1.1.1.17) activity. Upon exposure to low temperature stress (4°C) in a cold chamber, transgenic plants survived up to 48 h, while non-transformed plants were unable to survive and gradually died. Transgenic plants subjected to the chilling stress showed a significant decrease in electrolyte leakage and increased lipid peroxidation, as assessed by measuring malondialdehyde (MDA) content. Under the cold condition, transgenic plants also showed a significant increase in the activities of antioxidant enzymes (superoxide dismutase and catalase) and in relative water content (RWC) in comparison to non-transformed plants. Drought (polyethylene glycol in medium) and salinity (sodium chloride in medium) tolerance tests revealed that transgenic lines exhibited a higher tolerance for abiotic stresses than non-transformed plants. These findings indicate that the introduction of a bacterial mtlD gene into tomato conferred tolerance to abiotic stresses to the transformed tomato plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental stresses, such as drought, salinity, cold, wind velocity, humidity and heat cause adverse effects on the growth of plants and the productivity and quality of crops (Grover et al. 2003; Djilianov et al. 2005). Abiotic stress is the primary cause of crop loss worldwide, reducing average yields for most major crop plants by more than 50% (Rodriguez et al. 2005). Drought, salinity and cold stress are interconnected and affect the water relations of a plant on the cellular as well as the whole plant level, causing specific as well as unspecific reactions (Beck et al. 2007). These reactions lead to a series of morphological, physiological, biochemical and molecular changes that adversely affect plant growth and productivity (Wang et al. 2001). In response to dehydration stress caused by cold, salinity and drought, the plant cell must adjust their internal osmotic potential to maintain equal water potential with the environment. The modification of plants to enhance stress tolerance is mostly based on the manipulation of genes that protect and maintain the function and structure of cellular components. Present engineering strategies rely on the transfer of a gene that encodes an enzyme present in pathways leading to the synthesis of osmoprotectants (Bartels and Nelson 1994). Plant transformation is an essential tool, both for the experimental investigation of gene function and for the improvement of crops either by enhancing existing traits or introducing new genes, and the genetic engineering of biosynthetic pathways associated with stress responses has emerged as a promising approach to improve tolerance in crops. Adverse environmental conditions stimulate plants to employ various mechanisms, such as shifts in physiology and the increased expression of stress-associated genes leading to the formation of a wide variety of low-molecular-weight metabolites, notably osmolytes and proteins, to cope with the stress condition (Rajam et al. 1998; Zhu 2001; Prabhavathi et al. 2002). These osmolytes include fructans, trehalose, mannitol, myo-inositol, proline, glycine-betaine and polyamines, which accumulate in significant amounts and may be helpful in maintaining osmotic potential, ionic balance, membrane integrity and oxygen free radical level and in protecting chromatin under these conditions (Rajam et al. 1998; Bohnert and Shen 1999; Prabhavathi et al. 2002).
Mannitol, a sugar alcohol and one of the most important osmolytes, is a monosaccharide containing six hydroxyl groups. In higher plants, mannitol, if present at all, is only present in trace amounts (Zimmerman and Zeigler 1975) that are inadequate to contribute to abiotic stress tolerance. The expression of the bacterial mannitol-1-phosphate dehydrogenase (mtlD) gene in transgenic tobacco (Tarczynski et al. 1992, 1993; Karakas et al. 1997) and Arabidopsis (Thomas et al. 1995) led to the accumulation of mannitol in the cytoplasm and increased the tolerance of the transformed plants to salt stress. Studies with transgenic tobacco expressing a chloroplast-localized mtlD also suggest that mannitol protects the cell from oxidative damage from drought and chilling stress by protecting thiol-regulated enzymes, such as phosphoribulokinase, thioredoxin, ferredoxin and glutathione, from hydroxyl radical damage (Shen et al. 1997a). Prabhavathi et al. (2002) demonstrated that transgenic eggplant plants with the mtlD gene exhibited an increased tolerance to salt, drought and cold stresses. Cellular accumulation of mannitol as a result of the expression of the mtlD gene has been reported to protect transgenic wheat (Triticum aestivum L.) from the harmful effects of soil water logging and salinity (Abebe et al. 2003).
Tomato, a member of the Solanaceae family, is a major vegetable crop and consumed all over the world (Mueller et al. 2005). It has a relatively small DNA content and is a genetic model for crop improvement (Arumuganathan and Earle 1991). Tomato is rich in vitamins A, C and lycopene. Lycopene is a powerful antioxidant and protects humans from free radicals that degrade many parts of the body; lycopene is also known to prevent cancer (Gerster 1997; Rao and Agarwal 2000). Tomato is a major agricultural commodity and, therefore, abiotic stress tolerance will facilitate its production, even under adverse climatic conditions. The first report of Agrobacterium-mediated tomato transformation was in 1986 (McCormick et al. 1986), and since then tomato has been engineered for a variety of purposes, including the characterization of gene function, production of insect- and disease-resistant plants, herbicide tolerance, improved fruit quality, delay in fruit ripening and production of foreign proteins (Park et al. 2003; Davuluri et al. 2005; Lin et al. 2004; Youm et al. 2008). Researchers around the world have expended a great deal of effort in attempts to incorporate abiotic stress-tolerant genes in tomato (Gisbert et al. 2000; Jia et al. 2002; Hsieh et al. 2002; Park et al. 2004; Roy et al. 2006). To date, there has been no report of tomato plants transformed with the mtlD gene that confers tolerance against abiotic stresses. Any modification that enables the cultivation of tomato under abiotic stress conditions will represent a major breakthrough in the existing situation.
The objectives of the study reported here were to introduce the bacterial mtlD gene into tomato cv. Pusa Uphar by Agrobacterium mediated-transformation and to screen transgenic plants for enhanced tolerance to abiotic stresses.
Materials and methods
Plant material
Lycopersicon esculentum M. cv. Pusa Uphar seeds were purchased from National Seed Center, Pusa Complex, New Delhi. The seeds were surface sterilized with 70% ethanol and 0.1% HgCl2 with intermittent washes followed by a final rinse with double distilled water. Surface-sterilized seeds were blotted dry and germinated on half-strength MS (Murashige and Skoog 1962) basal medium. Cotyledonary leaves from 7-day-old seedlings were used as explants for transformation.
Bacterial strain and plasmid vector
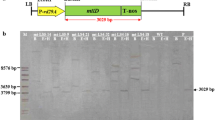
Agrobacterium tumefaciens strain LBA4404 harbouring a binary vector pBinAR was generously provided by Dr. M.V. Rajam (Department of Genetics, University of Delhi, India). This plasmid contains the bacterial (Escherichia coli) mtlD gene with the 35S cauliflower mosaic virus (CaMV) promoter and nopaline synthase (nos) terminator, neomycin phosphotransferase (nptII) gene, a selectable marker with the nopaline synthase promoter and terminator (Fig. 1).
Antibiotic sensitivity test
To determine the optimum concentration of kanamycin to be used for the selection of transformed explants, cotyledonary leaves of non-transformed plants were inoculated on callus induction medium supplemented with different concentrations (50–200 mg/L) of kanamycin, and the cultures were observed to determine the minimum concentration at which explants bleached early and uniformly. Cultures on medium containing different concentrations of the antibiotic cephotaxime (150-400 mg/L) along with callus induction media were also observed to check the overgrowth of Agrobacterium with the regeneration of the explants (data not shown).
Agrobacterium-mediated genetic transformation
Cotyledonary leaf explants were pre-cultured for 2 days on regeneration medium consisting of MS medium supplemented with 0.5 mg/L indole-3-acetic acid (IAA), 2.5 mg/L benzyl 6-aminopurine (BAP), 30 g/L sucrose and 8 g/L agar with the abaxial side touching the medium. The explants were obtained by cutting the edges of the leaves. Single colonies of Agrobacterium harbouring the mtlD and nptII genes were cultured in liquid YEM medium (in g/L: yeast extract 0.4, mannitol 10, NaCl 0.1, MgSO4.7H2O 0.2, K2HPO4 0.5, pH 7.0) supplemented with 50 mg/L kanamycin and 10 mg/L rifampicin. Each cultures was grown overnight (optical density 0.3–0.4 at λ690 nm) and then centrifuged at 5,000 rpm for 10 min at room temperature. The pellet of Agrobacterium cells was dissolved (OD 0.1–0.2 at λ690 nm) in liquid MS medium containing MS salts (MSS). The diluted culture was used to infect the pre-cultured leaf explants for 20 min. The infected explants were blotted dry onto a sterile blotting paper and placed onto a freshly prepared pre-cultured medium for 48 h in the dark at 25ºC. Following co-cultivation, the cotyledons were washed twice in liquid MSS containing 350 mg/L cephotaxime, blotted dry and placed on a selection medium that consisted of the pre-cultured regeneration medium supplemented with 50 mg/L kanamycin and 250 mg/L cephotaxime. Explants were cultured for 7–8 weeks under tissue culture conditions (16/8-h light/dark photoperiod, light intensity 2,000–3,000 lux, temperature 25 ± 1ºC), with subculturing at 3-week intervals onto fresh medium. Regenerated shoots from organogenic callus were then transferred to shoot elongation medium (consisting of MS salts, B5 vitamins, 0.5 mg/L gibberellic acid) fortified with 50 mg/L kanamycin and 250 mg/L cephotaxime. Elongated shoots were then cultured on rooting medium (MS media supplemented with 0.5 mg/L IAA and 50 mg/L kanamycin). The regenerated plantlets were acclimatized in thermocol pots containing a sterile mixture of garden soil, sand and vermiculite in a 1:1:1 ratio. The plants were then transferred to the containment facility for further growth and fruiting.
Molecular analysis of transformants
PCR analysis
Genomic DNA was isolated from young leaves of transgenic and non-transformed plants using the cetyl tri-ammonium bromide (CTAB) method of Doyle and Doyle (1987). About 100 ng of DNA from each sample was used for the 25-μl PCR reaction mixture. PCR cycling was carried out using two sets of gene-specific primers: the mtlD forward primer (5′-TTGGCGCAGGTAATATCG GTCGTG-3′) and reverse primer (5′-GCAGCGGCAATACCTTCAATCAGGT-3′) and the nptII forward primer (5′-GAGGCTATTCGGCTATGACTG-3′) and reverse primer (5′-ATCCGGAGGGGCGATACCGGA-3′). PCR amplification was carried out under the following conditions: an initial denaturation cycle at 94°C for 5 min, followed by 35 cycles of denaturation (94°C for 30 s), annealing (nptII 55°C and mtlD 63°C for 1 min) and synthesis at 72°C, with a final extension step at 72°C for 5 min. The amplified products were separated by electrophoresis on 1% agarose gel containing ethidium bromide, and the bands were visualized using an AlphaImager EC (Alpha Innotech, San Leandro, CA).
Southern hybridization
PCR-positive plants were then confirmed for the presence of the mtlD gene by southern blot analysis. Genomic DNA was restricted with HindIII to check for mtlD gene integration, and the southern blots were prepared according to the procedure of Sambrook et al. (1989). The 976-bp amplicon of the mtlD gene, labeled with [32P]-dCTP using the Hexalab DNA Labeling kit (Fermentas, Burlington, ON, Canada), was used as the probe and hybridized to a Hybond N+ nylon membrane (0.45 μM; Amersham, Arlington Heights, IL). An autoradiograph was obtained by exposing the X-ray film to the membrane.
Reverse transcription-PCR analysis
Total RNA of the transgenic plants was isolated using the Qiagen RNeasy Plant Mini kit (cat. no. 74904; Qiagen, Hilden, Germany), and the reverse-transcription (RT)-PCR was performed as follows. The reaction mixture [5× RT buffer (Fermentas), 10 mM dNTP mix, oligo dT primer] was heated to 65°C, following which 1 μg RNA and MMLV RT were added, and reverse transcription was performed at 37°C for 60 min. The first-strand cDNA was used for the PCR analysis using gene (mtlD)-specific primers to amplify the 976-bp fragment of the mtlD gene. The PCR cycling conditions consisted of an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation (95°C for 30 s), annealing (63°C for 1 min), and extension (72°C for 1 min), and a final extension at 72°C for 7 min. The amplified products were separated by electrophoresis on a 1% agarose gel containing ethidium bromide, and the bands were visualized using an AlphaImager EC (Alpha Innotech).
mtlD activity assay
The mtlD assay was performed using a spectrophotometer at 25°C according to the method of Ikawa et al. (1972) with some modifications (Iwamoto et al. 2003). Fructose-6-phosphate (F-6-P) reducing activity was measured by monitoring the disappearance of NADH at 340 nm in a 1-ml reaction mixture containing 50 mM HEPES–KOH (pH 7.0), 3 mM Na2-EDTA, 3 mM F-6-P, 0.2 mM NADH, 100 mM NaCl and 50 μL of the enzyme extract. The reaction was initiated by adding the F-6-P. Protein concentration was estimated using the method of Bradford (1976).
Bioassay for cold stress tolerance
Cold stress tolerance test
Plants of the T1 transgenic lines and non-transformed plants were incubated at 4°C for 48 h, and their ability to withstand under such conditions was monitored carefully. Comparative analyses of the various important physiological and biochemical parameters related to abiotic stress tolerance (see below) were assayed in both transgenic and non-transformed plants under normal (non-stress) and stress conditions.
Electrolyte leakage
Electrolytes are contained within the membranes of plant cells. Unstressed, undamaged plant cells maintain electrolytes within the membrane. As the cells are subjected to stress, electrolytes leak into surrounding tissues. An estimation of cell damage and hardiness can be made by comparing the conductivity of the leaked contents from injured to uninjured tissues in water (Mattsson 1996; McNabb and Takahashi 2000). Electrolyte leakage was measured using a conductivity meter (Cond 3151/SET; WTW, Weilheim, Germany) according to the operating instructions. Ten leaf discs of equal size were placed in 25 ml water and assessed under the following conditions: (a) after a 15-min vacuum filtration and (b) after being autoclaved at 121°C for 30 min. The conductivity of the solution was measured as follows: EL(%) = value a/value b × 100.
Relative water content
Relative water content (RWC) was measured as reported by Roberto et al. (2001). Ten leaf discs from fully expanded leaves were weighed (fresh weight) immediately after harvest and placed in autoclaved distilled water for 3 h at 4°C, following which their turgid weight was recorded. The samples were dried in an oven at 80°C overnight to obtain their dry weight. The RWC was calculated by the formula: RWC = fresh weight − dry weight/turgid weight − dry weight × 100.
Malondialdehyde estimation
The comparative rate of lipid peroxidation was assayed from the leaves of transgenic and non-transformed plants by determining the level of malondialdehyde (MDA) production after exposure to cold stress (4°C) for 48 h, using the thiobarbituric acid (TBA) method as described by Wang et al. (2008) with slight modifications of our own. Leaf samples (0.1 g) were homogenized with a mortar and pestle in 5 ml of 0.5% (v/v) TBA solution in 10% trichloroacetic acid (TCA) and centrifuged at 12,000 g for 15 min at 4°C. The supernatant was heated in a boiling water bath for 25 min and allowed to cool in an ice bath. The supernatant was then again centrifuged at 12,000 g for 15 min at 4°C. The resulting supernatant was collected and used for spectrophotometric determination of MDA. MDA content was calculated by multiplying the difference between absorbance at 535 nm and that at 600 nm by the extinction co-efficient of 6.45 μM/L and then subtracting the absorbance at 435 nm multiplied by the extinction co-efficient of 0.56 μM/L from the data.
Antioxidant enzyme activity
The level of the antioxidant enzyme superoxide dismutase (SOD) was estimated in transgenic and non-transformed plants before and after exposure to cold stress (4°C for 48 h) according to the method of Moaed et al. (2006). The enzyme extract was prepared by grinding 0.5 g leaf sample first with liquid nitrogen and then with 10 ml of 100 mM potassium phosphate buffer, pH 7.5, containing 0.5 mM EDTA. The extract was centrifuged at 14,000 g at 4°C for 20 min, and the supernatant was used as the enzyme preparation. The protein concentration was estimated according to the method of Bradford (1976).
The assay is based on the formation of the blue-coloured formazone by nitro-blue tetrazolium chloride (NBT) dye and the O2 − radical; formazone absorbs light maximally at 560 nm, and SOD decreases this absorbance by reducing the formation of the O2 − radical (Dhindsa et al. 1981). The reaction mixture (3 ml) contained 13 mM methionine, 25 mM NBT/0.1 mM EDTA, 50 mM phosphate buffer (pH 7.1), 50 mM sodium carbonate and 0.1 ml enzyme extract. The reaction was started by adding 2 μM riboflavin and placing the tube under 30 W fluorescent lamps for 15 min. A complete reaction mixture without enzyme, which gave maximal color, served as the control. The reaction was stopped by switching off the light and placing the tubes in the dark. A non-irradiated complete reaction mixture served as the blank. The absorbance was recorded at 560 nm, and 1 U of enzyme activity was taken as that amount of enzyme that reduced the absorbance reading to 50% in comparison with the tube lacking the enzyme. The enzyme was expressed in specific activity (U/mg protein).
The catalase assay is based on the absorbance of H2O2 at 240 nm in the UV range. A decrease in the absorbance is recorded over a time period. Catalase (EC 1.11.1.6) was measured as described by Jaleel et al. (2007) with a slight modification of our own. The assay mixture (3 ml) contained 0.1 M potassium phosphate buffer, pH 7.0, 15 mM H2O2 and 100 μl of enzyme extract. The enzyme activity was expressed μmole/mg protein/min.
Bioassay for salinity and drought tolerance
Drought tolerance bioassay using the polyethylene glycol method
Leaf explants of non-transformed and transgenic plants were cultured on callusing medium containing polyethylene glycol (PEG) 4000 (10%) after surface sterilization. Ten explants were used per treatment, and the experiment was repeated three times. The fresh weight (FW) and dry weight (DW) of the leaf explants along with the developing calli and shoots were scored after 30 days of culture. The dry weight of the tissue was obtained after oven drying at 80°C overnight. The water content of the tissue was calculated as the (FW–DW)/DW ratio.
Salinity tolerance bioassay using NaCl
The seeds from non-transformed to transgenic T0 plants were surface sterilized and inoculated onto MS basal medium supplemented with 50, 100 and 200 mM NaCl, respectively, for the salt tolerance assay. Tolerance was based on percentage seed germination.
Statistical analysis
Data obtained from different sets of experiments were analysed for significance (P ≤ 0.05) using the t test.
Results and discussion
The objective of this study was to investigate the role of the mtlD gene in imparting abiotic stress tolerance in transgenic tomato plants. The mtlD gene encodes mannitol-1-phosphate dehydrogenase, which utilizes F-6-P and NADH to form mannitol-1-phosphate and NAD+, and mannitol-1-phosphate in turn dephosphorylates into mannitol with the help of phosphatase (Tarczynski et al. 1992). Thus, the accumulation of mannitol depends directly on the availability and/or activity of the mtlD enzymes in plants. To study the transgenic tomato plants transformed with the mtlD gene, we used an effective in vitro regeneration protocol reported earlier for tomato cv. Pusa Uphar (Khare et al. 2008). Transformed cotyledonary leaf disc explants began to differentiate green translucent callus at their cut ends within 2–3 weeks of culture in selection medium supplemented with kanamycin and cephotaxime. However, the non-transformed explants became bleached and ultimately died in selection media. The selection of transformed explants was obtained best with 50 mg/l kanamycin supplemented to the medium, and the bacterial overgrowth was controlled with 250 mg/l cephotaxime supplemented to the MS medium. Adventitious shoots were regenerated from callus within 6–8 weeks. Shoots 2–3 cm long were transferred from the shoot elongation media to the rooting media. A 95% rooting rate was observed 8–10 days after transfer. The regenerated plants were hardened and transferred to pots and placed in the containment facility for further analysis and advancement of generations (Fig. 2a–h).
Different stages in the transformation of tomato (Lycopersicon esculentum L. var. Pusa Uphar). a In vitro germinated seedlings, b explants in preculture media, c cotyledonary explants on selection medium after co-cultivation with Agrobacterium harbouring the pBinAR binary vector (l regenerated explants, II bleached explants, all in selection media), d regenerated callus from explant, e callus with regenerating shoot buds growing on selection medium, f shoot elongation, g young plantlet with well-developed roots on selection medium, h hardened transformed T0 plants
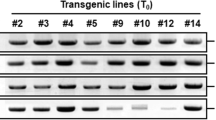
The 970-bp mtlD and 750-bp nptII gene fragments were amplified in 85% of the putative transformants by PCR (Fig. 3a, b). Southern blot analysis was performed using the mtlD gene as a probe. Twenty-two plants were found to be positive for transgene integration (Fig. 4). Expression of the transgene was assessed using RT-PCR, five plants of transformed putative T0 plants were found to be positive for transgene expression (Fig. 5). These five promising transgenic T0 plants, selected on the basis of gene expression and a preliminary cold stress analysis (data not shown), were advanced to the T1 generation. The T1 transgenic lines were then germinated and studied for cold, drought and salinity tolerance by assaying a number of important abiotic stress-related biochemical and physiological parameters that may be regulated due to the introgression of the mtlD gene into the tomato genome.
PCR products from the DNA of leaves of putative transgenic plants of the tomato var. Pusa Uphar. a Amplification of the nptII gene (750 bp), b amplification of the mtlD gene (970 bp). Lanes: M 100-bp ladder, P plasmid DNA, NT DNA sample from non-transformed plant, 1–10 DNA samples from putative transgenic plants
To elucidate the cold stress tolerance, we exposed both transgenic lines and non-transformed plants to 4°C in a cold chamber. The transgenic plants could clearly withstand the cold treatment very well up to 48 h; however, the non-transformed plants were not able to tolerate this cold stress for this period of time and ultimately died during recovery (Fig. 6). The results of the mtlD assay in T1 transgenic lines also suggested the expression of the transgene in tomato plants: the activity of MtlD was much higher in transgenic lines and almost negligible in non-transformed plants (Fig. 7). Abiotic stresses damage the structure of the cell membrane, making it leaky and resulting in the outflow of ions that are vital for proper cell functioning (Jia et al. 2002). The cell membrane is one of the first targets of many plant stresses (Roy et al. 2006). In our study, transgenic plants showed less electrolyte leakage than the non-transformed plants under stress condition. The electrolyte leakage was higher in non-transformed plants under the stress condition (25%), while the difference between stressed and non-stressed transgenic lines was negligable(Fig. 8). This lower electrolyte leakage in transgenic lines reflect the membrane integrity of these plants (Uemura et al. 1995), suggesting that integration of the mtlD gene in the tomato genome contributes to the maintenance of membrane integrity and thus contributes to the plant’s ability to resist low temperature stress. Mannitol, a compatible solute, prevents water loss from the cell and protects the cellular proteins because it facilitates osmotic adjustment. The accumulation of mannitol has the effect of lowering the water potential inside the cell and thus preventing intracellular water loss (Mahajan and Tuteja 2005). Relative water content is a parameter often used to assess the water retention capacity of plants. We assayed RWC in both transgenic and non-transformed plants under normal and cold stress conditions and found that the RWC of non-transformed plants was comparatively less as compared to transgenic plants under both conditions. The magnitude of the RWC level was observed lower under the cold stress condition (Fig. 9).
Mannitol-1-phosphate dehydrogenase (mtlD) activities in non-transformed (C) and T1 transgenic lines (T1–T5) from stressed (4°C for 48 h) plants. Values are given as the mean ± standard deviation (SD) of five experiments in each line. Bars: SD. Asterisks (*) indicate a statistically significant change relative to control plants (P ≤ 0.05) as determined using the t test
Electrolyte leakage from transgenic (T1–T5) and non-transformed (NT) plants under non-stress (a) and stress (b) conditions. Values are given as the mean ± SD of five experiments (replicates) in each line. Bars: SD. Asterisks (*) indicate a statistically significant change in transgenic lines relative to control plants (P ≤ 0.05) as determined using the t test
Relative water content (RWC) of T1 transgenic lines (T1–T5) compared to non-transformed (NT) tomato plants under non-stress (a) and stress (b) conditions after 48 h of stress. Values are given as the mean ± SD of five experiments in each line. Bars: SD. Asterisks (*) indicate a statistically significant change in the transgenic lines relative to the non-transformed plants (P ≤ 0.05) as determined using t test
The MDA concentration in transgenic and non-transformed plants did not change significantly prior to the cold treatment. However, there was a significant increase in the MDA concentration in non-transformed tomato plants after 48 h of cold treatment, suggesting that non-transformed plants suffered higher oxidative lipid injury than transgenic plants during the cold stress period (Fig. 10). The superoxide radical is regularly synthesized in the chloroplast and mitochondria, and it has also been reported to be produced in limited amounts in microbodies (Sairam and Aruna 2004). The antioxidant enzymes SOD and catalase ensure the removal of the superoxide radical and the H2O2 produced during abiotic stresses, such as chilling, drought and salt stress (Fadzilla et al. 1997; Dionisio-Sese and Tobita 1998; Baek et al. 2006). We measured the levels of these two antioxidant enzymes in non-transformed and transgenic plants under both normal conditions and following exposure to 4°C for 48 h. The activities of both enzymes were found to be significantly increased (P ≤ 0.05) in transgenic plants relative to non-transformed plants under the cold stress condition (Fig. 11A, B), supporting previous reports that the accumulation of mannitol increases stress tolerance through the scavenging of reactive oxygen species (ROS) and the stabilization of macromolecular structures (Shen et al. 1997b). Mannitol as a scavenger of the hydroxyl radical has been reported both in vitro (Smirnoff and Cumbes 1989) and in vivo (Shen et al. 1997a) in transgenic tobacco. Thus, our findings also suggest the effective detoxification of free radicals/H2O2 by the elevated level of antioxidant enzymes, in this case, SOD and catalase, in transgenic plants.
Malondialdehyde (MDA) concentration in T1 transgenic lines in comparison to non-transformed tomato at 0 and 48 h cold stress at 4°C. Values are given as mean ± SD of five experiments in each line. Bars represent standard deviation. Asterisks (*) indicate a statistically significant change in transgenic lines relative to non-transformed plants (P ≤ 0.05) as determined using the t test
Antioxidant enzymes assay. A Superoxide dismutase (SOD) activity in T1 transgenic lines (T1–T5) in comparison to non-transformed (control, C) tomato plants under non-stress (a) and stress (b) conditions. B Catalse activity in T1 transgenic lines in comparison to nontransformed (C) tomato plants under non-stress (a) and stress (b) conditions. Values are given as mean ± SD of five experiments in each line. Bars: SD. Asterisks (*) indicate a statistically significant change in transgenic lines relative to non-transformed plants (P ≤ 0.05) as determined using the t test
Fresh (a) and dry (b) weights of non-transformed and the transgenic lines cultured on 0 and 10% polyethylene glycol (PEG) 400-supplemented regeneration medium for 30 days. Values are given as the mean ± SD of five experiments in each line. Bars: SD. Asterisks (*) exhibit statistically significant change in transgenic lines as compared to non-transformed plants at the level of P ≤ 0.05 as determined using t test
PEG is a non-penetrating and non-ionic stress-inducing compound. It is an extremely hydrophilic organic polymer that is able to remove much of the free water within a solution. Therefore, it does not cause any ionic stress but only osmotic stress. During the drought tolerance assay, the explants of transgenic plants were observed to be more regenerating than the non-transformed plants on the basis of fresh and dry weight. In each experiment, the water content of transgenic plants was observed to be higher than that of non-transformed plants. The higher weight of transgenic plants in terms of FW and DW in the PEG assay could also be attributed to the expression of the mtlD gene (Fig. 12a, b).
When the seeds of both non-transformed and transgenic tomato plants were cultured on MS medium supplemented with different concentrations of NaCl, the germination of non-transformed seeds decreased drastically with increasing concentrations of NaCl. The germination of the seeds of transgenic lines also decreased, but the germination percentage was higher than that of the salt-stressed non-transformed plants. At a concentration of 200 mM NaCl, non-transformed seeds did not germinate at all, while the transgenic seeds had a germination percentage of 24–30%, revealing that the latter had a higher resistance for salinity stress (Table 1).
Our findings suggest that mtlD introgression into the tomato cv. Pusa Uphar genome imparts abiotic stress tolerance, thereby corroborating results from previous studies on mtlD transgenic tobacco (Tarczynski et al. 1993), Arabidopsis (Thomas et al. 1995), eggplant (Prabhavathi et al. 2002) and wheat (Abebe et al. 2003). The survival of transgenic tomato exposed to such abiotic stresses may be due to the protection conferred by the accumulation of mannitol, which leads to various mechanisms, such as osmotic adjustment, stabilization of macromolecules, proteins/enzymes and membranes, detoxification of ROS and an increased concentration of low molecular osmolytes (Shen et al. 1997a, b; Sakamoto and Murata 2001; Prabhavathi et al. 2002). In conclusion, the introgression of mtlD gene provided tolerance to transgenic tomato plant against abiotic stresses.
Abbreviations
- BAP:
-
Benzyl 6-aminopurine
- CTAB:
-
Cetyl tri-ammonium bromide
- EDTA:
-
Ethylenediaminetetraacetic acid
- IAA:
-
Indole 3-acetic acid
- MDA:
-
Malondialdehyde
- mtlD:
-
Mannitol-1-phosphate dehydrogenase
- NBT:
-
Nitro-blue tetrazolium chloride
- nptII :
-
Neomycin phosphotransferase II
- ROS:
-
Reactive oxygen species
- SOD:
-
Superoxide dismutase
- TBA:
-
Thiobarbituric acid
- TCA:
-
Trichloroacetic acid
References
Abebe T, Guenzi AC, Martin B, Cushman JC (2003) Tolerance of mannitol accumulating transgenic wheat to water stress and salinity. Plant Physiol 131:1748–1755
Arumuganathan K, Earle E (1991) Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep 9:208–218
Baek K, Skinner DZ, Ling P, Chen X (2006) Molecular structure and organization of the wheat genomic manganese superoxide dismutase gene. Genome 49:209–218
Bartels D, Nelson D (1994) Approaches to improve stress tolerance using molecular genetics. Plant Cell Environ 17:659–667
Beck EH, Heim R, Hansen J (2007) Plant resistance to cold stress: Mechanisms and environmental signals triggering frost hardening and dehardening. J Biosci 29:449–459
Bohnert HJ, Shen B (1999) Transformation and compatible solutes. Sci Hortic 78:237–260
Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem 72:248–254
Davuluri GR, van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Jhlig J, Bramley PM, Pennings HMJ, Bowler C (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23:890–895
Dhindsa RA, Plumb-Dhindsa P, Thorpe TA (1981) Leaf senescence: correlated with increased permeability and lipid peroxidation and decreased levels of SOD and Catalase. J Exp Bot 126:93–101
Dionisio-Sese ML, Tobita S (1998) Antioxidant responses of rice seedlings to salinity stress. Plant Sci 135(1):1–9
Djilianov D, Georgieva T, Moyankova D, Atanassov A, Shinozaki K, Smeeken SCM, Verma DPS, Murata N (2005) Improved abiotic stress tolerance in plants by accumulation of osmoprotactants—gene transfer approach. Biotechnol Biotechnol Eq 19(3):63–71
Doyle JJ, Doyle JI (1987) Isolation of plant DNA from fresh tissue. Phytochem Bull Bot Soc Am 19:11–15
Fadzilla NM, Finch PR, Burdon RH (1997) Salinity, oxidative stress and antioxidant responses in shoot culture of rice. J Exp Bot 48(307):325–331
Gerster H (1997) The potential role of lycopene for human health. J Am College Nutr 16:109–126
Gisbert C, Rus MA, Bolarin CM, Lopez CMJ, Arrillaga I, Montesinos C, Caro M, Serrano R, Morenov V (2000) The yeast HAL1 gene improves salt tolerance of transgenic tomato. Plant Physiol 123:393–402
Grover A, Aggarwal PK, Kapoor A, Katiyar-Agarwal S, Agarwal M, Chandramouli A (2003) Addressing abiotic stresses in agriculture through transgenic technology. Curr Sci 84(3):355–367
Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, Wang YC, Chan MT (2002) Heterology expression of the Arabidopsis C-repeat/Dehydration element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol 129:1–9
Ikawa T, Watanabe T, Nisizawa K (1972) Enzymes involved in the last steps of the biosynthesis of mannitol in brown algae. Plant Cell Physiol 13:1017–1029
Iwamoto K, Kawanobe H, Ikawa T, Shiraiwa Y (2003) Characterization of salt-regulated mannitol-1-phosphate dehydrogenase in the red alga Caloglossa continua. Plant Physiol 133:893–900
Jaleel CA, Gopi R, Manivannan P, Panneerselvam R (2007) Antioxidative potentials as a protective mechanism in Catharanthus roseus (L.) G. Don. Plants under salinity stress. Turk J Bot 31:245–251
Jia GX, Zhu ZQ, Chang FQ, Li YX (2002) Transformation of tomato with the BADH gene from Atriplex improves salt tolerance. Plant Cell Rep 21:141–146
Karakas B, Ozias Akins P, Stushnoff C, Suefferheld M, Rieger M (1997) Salinity and drought tolerance of mannitol accumulation transgenic tobacco. Plant Cell Environ 20:609–616
Khare N, Shah P, Singh NK, Rathore M, Goyary D, Anandhan S, Gupta SM, Arif M, Lakhera PC, Ahmed Z (2008) In vitro regeneration protocol for tomato (Lycopersicon esculentum M var. Pusa Uphar). J Appl Biosci 34(2):199–202
Lin WC, Lu CF, Wu JW, Cheng ML, Lin YM, Yang NS, Black L, Green SK, Wang JF, Cheng CP (2004) Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res 13:567–581
Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444:139–158
Mattsson A (1996) Predicting field performance using seedling quality assessment. New Forests 13:223–248
McCormick S, Niedermeyer J, Fry J, Barnason A, Horsh R, Fraley R (1986) Leaf disc transformation of cultivated tomato (Lycopersicon esculentum) using Agrobacterium tumefaciens. Plant Cell Rep 5:81–84
McNabb K, Takahashi E (2000) Freeze damage to loblolly pine seedlings as indicated by conductivity measurements and outplanting survival. Research Report 00-4. Auburn University Southern Forest Nursery Management Cooperative, Auburn
Moaed A, Deshmukh PS, Sairam RK, Kushwaha SR, Singh TP (2006) Protective role of antioxidant enzymes under high temperature stress. Plant Sci 117:382–388
Mueller LA, Tanskley SD, Giovannoni JJ, van Eck J, Stack S, Choi D, Kim D, Chen M (2005) The tomato sequencing project, the first corner stone of the international Solanaceae project (SOL). Comp Funct Genom 6:153–158
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol 15:473–497
Park SH, Morris JL, Park JE, Hirschi KD, Smith RH (2003) Efficient and genotype-independent Agrobacterium-mediated tomato transformation. J Plant Physiol 160:1253–1257
Park EJ, Jeknic Z, Sakamoto A, Denoma J, Yuwansiri R, Murata N, Chen TH (2004) Genetic engineering of glycine-betaine synthesis in tomato protects seeds, plants, and flowers from chilling damage. Plant J 40:474–487
Prabhavathi V, Yadav JS, Kumar PA (2002) Abiotic stress tolerance in transgenic egg plant (Solanum melongena L.) by introduction of bacterial mannitol phospho dehydrogenase gene. Mol Breed 9:137–147
Rajam MV, Dagar S, Waie B, Yadav JS, Kumar PA, Shoeb F (1998) Genetic engineering of polyamines and corbohydrate metabolism for osmotic stress tolerance in higher plants. J Biosci 23:473–482
Rao A, Agarwal S (2000) Role of antioxidant lycopene in cancer and heart disease. J Am College Nutr 19:563–569
Roberto A, Li GJ, Undurraga S, Dang LM, Allen GJ, Alper LS, Fink GR (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H1-pump. Proc Nat Acad Sci USA 98(20):11444–11449
Rodriguez M, Canales E, Borras-Hidalgo O (2005) Molecular aspects of abiotic stress in plants. Biotecnol Apl 22:1–10
Roy R, Purty RS, Agrawal V, Gupta SC (2006) Transformation of tomato cultivar “Pusa Ruby” with bspA gene from Populus tremula for drought tolerance. Plant Cell Tissue Org Cult 84:55–67
Sairam RK, Aruna T (2004) Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci 86(3):407–421
Sakamoto A, Murata N (2001) The use of bacterial choline oxidase, a glycinebetaine-synthesizing enzyme, to create stress resistant transgenic plants. Plant Physiol 125:180–188
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, New York
Shen B, Jensen RG, Bohnert HJ (1997a) Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to chloroplasts. Plant Physiol 113:1177–1183
Shen B, Jensen RG, Bohnert HJ (1997b) Mannitol protects against oxidation by hydroxyl radicals. Plant Physiol 115:527–532
Smirnoff N, Cumbes QJ (1989) Hydroxyl radical scavenging activity of compatible solutes. Phytochem 28:1057–1060
Tarczynski MC, Jensen RG, Bohnert HJ (1992) Expression of a bacterial mtlD gene in transgenic tobacco leads to production and accumulation of mannitol. Proc Natl Acad Sci USA 89:2600–2604
Tarczynski MC, Jensen RG, Bohnert HJ (1993) Stress protection of transgenic tobacco by production of the osmolite mannitol. Science 259:508–510
Thomas JC, Sepahi M, Arendall B, Bohnert HJ (1995) Enhancement of seed germination in high salinity by engineering mannitol expression in Arabidopsis thaliana. Plant Cell Environ 18:801–806
Uemura M, Joseph RA, Steponkus PL (1995) Cold acclimation of Arabidopsis thaliana: Effect on plasma membrane lipid composition and freeze-induced lesion. Plant Physiol 109:15–30
Wang WX, Vinocur B, Shoseyov O, Altman A (2001) Biotechnology of plant osmotic stress tolerance: physiological and molecular considerations. Acta Hortic 560:285–292
Wang Y, Qiu L, Dai C, Wang J, Luo J, Zhang F, Ma J (2008) Expression of insect (Microderma puntipennis dzungarica) antifreeze protein MpAFP149 confers the cold tolerance to transgenic tobacco. Plant Cell Rep 27(8):1349–1358
Youm JW, Heung J, Jeon JH, Kim H, Kim YH, Ko K, Joung H, Kim HS (2008) Transgenic tomatoes expressing human beta-amyloid for use as a vaccine against Alzheimer’s disease. Biotechnol Lett 30:1839–1845
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Zimmerman MH, Zeigler H (1975) List of sugars and sugar alcohols in sieve tube exudates. Encyc Plant Physiol 1:480–503
Acknowledgements
The authors are grateful to the Life Sciences Research Board, DRDO, Ministry of Defence, Govt. of India for the financial support. Authors are also grateful to Dr. M. V. Rajam, Delhi University, South Campus, India for providing the gene constructs.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khare, N., Goyary, D., Singh, N.K. et al. Transgenic tomato cv. Pusa Uphar expressing a bacterial mannitol-1-phosphate dehydrogenase gene confers abiotic stress tolerance. Plant Cell Tiss Organ Cult 103, 267–277 (2010). https://doi.org/10.1007/s11240-010-9776-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-010-9776-7