Abstract
Introduction
WNT signaling pathway dysregulation is an important event in the pathogenesis of colorectal cancer (CRC) with APC mutations seen in more than 80% of sporadic CRC. However, such mutations in the WNT signaling pathway genes are rare in inflammatory bowel disease (IBD) associated neoplasia (dysplasia and cancer). This study examined the role of epigenetic silencing of WNT signaling pathway genes in the pathogenesis of IBD-associated neoplasia.
Methods
Paraffin-embedded tissue samples were obtained and methylation of ten WNT signaling pathway genes, including APC1A, APC2, SFRP1, SFRP2, SFRP4, SFRP5, DKK1, DKK3, WIF1 and LKB1, was analyzed. Methylation analysis was performed on 41 IBD samples, 27 normal colon samples (NCs), and 24 sporadic CRC samples.
Results
Methylation of WNT signaling pathway genes is a frequent and early event in IBD and IBD-associated neoplasia. A progressive increase in the percentage of methylated genes in the WNT signaling pathway from NCs (4.2%) to IBD colitis (39.7%) to IBD-associated neoplasia (63.4%) was seen (NCs vs. IBD colitis, p < 0.01; IBD colitis vs. IBD-associated neoplasia, p = 0.01). In the univariate logistic regression model, methylation of APC2 (OR 4.7, 95% CI: 1.1–20.63, p = 0.04), SFRP1 (OR 5.1, 95% CI: 1.1–31.9, p = 0.04), and SFRP2 (OR 5.1, 95% CI: 1.1–32.3, p = 0.04) was associated with progression from IBD colitis to IBD-associated neoplasia, while APC1A methylation was borderline significant (OR 4.1, 95% CI: 0.95–17.5, p = 0.06). In the multivariate logistic regression model, methylation of APC1A and APC2 was more likely to be associated with IBD-associated neoplasia than IBD colitis. (OR APC1A: 6.4, 95% CI: 1.1–37.7 p = 0.04; OR APC2 9.1, 95% CI: 1.3–61.7, p = 0.02).
Summary
Methylation of the WNT signaling genes is an early event seen in patients with IBD colitis and there is a progressive increase in methylation of the WNT signaling genes during development of IBD-associated neoplasia. Moreover, methylation of APC1A, APC2, SFRP1, and SFRP2 appears to mark progression from IBD colitis to IBD-associated neoplasia, and these genes may serve as biomarkers for IBD-associated neoplasia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inflammatory bowel disease (IBD) is a chronic inflammatory disease of the intestines afflicting about one million individuals in the US, with 30,000 new cases each year.1 IBD includes two distinct disease categories, Crohn’s disease (CD) and ulcerative colitis (UC), both of which are associated with an increased risk of CRC.2,3 Additionally, CRC is a major cause of increased mortality in IBD patients.4 The risk of CRC in IBD increases with prolonged disease duration and is greater in those with extensive colitis.5 This risk has been reported as ten- to 20-fold higher in UC patients with disease duration of 20 years or more, although current treatments may have modulated this risk.6–8 As a result of the recognition of this increased risk, regular surveillance colonoscopy at 1–3 years interval is recommended for patients with long-standing disease.9–12
IBD-associated carcinomas arise in areas of dysplasia which are flat and difficult to recognize at colonoscopy.13,14 Consequently, one may have to take 33 or more colonic biopsies to have a 90% confidence of finding dysplasia. To increase the accuracy of colonoscopic surveillance to 95%, nearly twice the number of biopsy specimens are required.15 This makes surveillance labor intensive and expensive. Moreover, some of the recent studies have questioned the effectiveness of surveillance.16–18 In order to improve the effectiveness of surveillance, there is a great need for objective and reliable molecular markers for early detection of IBD-associated neoplastic lesions especially among patients with long-standing disease.
DNA methylation-dependent silencing of cancer related genes is an important and early event in CRC. Methylation of promoter-associated CpG islands leads to binding of various proteins having methyl binding domains like MeCP2, MBD2, and MBD3; these proteins may then initiate a cascade of events, which eventually lead to transcriptional silencing.19 Transcriptional silencing also involves changes in histone tails such as histone H3 lysine 9 methylation and histone H3 lysine 27 methylation as well as recruitment of histone deacetylases.20,21 All of these modifications result in changes to the local chromatin structure, which in turn result in a tightly compacted chromatin, which in turn restricts the access to transcription factors facilitating transcriptional silencing. Aberrant age-related as well as cancer-specific methylation of genes like p16, E-cadherin, hMLH1, p14, HPP1, ER, etc., has already been reported in the setting of IBD-associated neoplasia (dysplasia and cancers).22–26 It is believed that chronic inflammatory states like IBD may predispose to accelerated aberrant methylation and inactivation of tumor suppressor genes, which in turn may accelerate the development of cancer.
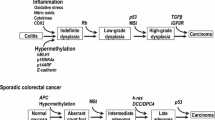
The WNT signaling pathway is crucial to the development of sporadic CRC.27,28 Activating mutations in the WNT signaling pathway are seen in more than 80% of sporadic CRC.29,30 Signaling in the WNT signaling pathway begins when the WNT ligand, a secreted factor binds to the Frizzled (Fz) receptor and LRP 5/6 co-receptor to initiate the signaling cascade.31 Proteins of the disheveled (Dsh or dvl) family then interact with Fz receptors.32 These interactions subsequently lead to inhibition of complex formed by APC, AXIN, and Glycogen synthase kinase-3β (GSK-3β). The APC/AXIN/GSK-3β complex normally plays a role in phosphorylation and degradation of β-catenin. As a result of the inhibition of the APC/AXIN/GSK-3β complex, β-catenin accumulates and is translocated to the nucleus33 where it upregulates the transcription of many cancer-related genes including MYC,34 cyclooxygenase-2 (COX-2),35 and cyclin D1(Fig. 1).36 Powerful developmental regulatory pathways, like the WNT signaling pathway, are controlled by stringent negative regulation, which, if disrupted, can lead to their aberrant activation and tumorogenesis. WNT signaling pathway is normally inhibited by soluble Fz-related proteins (SFRPs) and WNT inhibitory factor (WIF1), which bind to WNT ligands extracellularly37 and by proteins of the dickkopf (DKK) family, which bind to the LRP surface molecule.38 Hence, inactivation of SFRPs, WIF1, and DKKs by mechanisms such as promoter methylation can lead to activation of WNT signaling in cancer cells. Similarly, inactivation of APC by mutation or promoter methylation or both can lead to prevention of degradation of β-catenin as APC normally plays a role in β-catenin degradation by complexing with other proteins as described previously. Although mutations of WNT signaling pathway genes including APC have been described in sporadic CRC, mutations of these genes are rare (0% to 6%) in IBD-associated neoplasia.39
Recent reports have explored the role of WNT signaling in IBD-associated neoplasia, but as yet little is known about the epigenetic regulation of this pathway in IBD-associated neoplasia.40–42 Increased nuclear β-catenin and reduced cytoplasmic APC1A expression has recently been reported in the setting of UC-associated cancers.43,44 Since mutation of WNT signaling genes is a rare event in IBD-associated neoplasia, we hypothesized that methylation of genes in the WNT pathway may be responsible for dysregulation of WNT signaling in these lesions. In the current study, we investigated if promoter methylation of WNT pathway genes occurs during the course of IBD-associated carcinogenesis and furthermore characterize the progression of the methylation events during the course of IBD associated neoplasia. This may help to find potential candidates for investigation as biomarkers for early detection of IBD-associated neoplasia.
We now show that methylation of WNT signaling pathway genes is an early event that can be seen in long-standing IBD colitis and there is a progressive increase in methylation of the WNT signaling pathway genes during the development of IBD-associated neoplasia.
Materials and Methods
Patient Samples
Paraffin-embedded tissue samples were obtained from Johns Hopkins Hospital (JHH) pathology archives in accordance with regulations of the Institutional Review Board and HIPAA compliance. Tissue samples were obtained from 18 IBD patients who underwent colectomy from 1997 to 2006. Eleven of out 18 patients had IBD-associated neoplasia (either dysplasia or cancer) while seven were non-cancer IBD controls. A total of 41 IBD tissue samples were obtained including six IBD cancers, two high-grade dysplasias (HGDs), eight low-grade dysplasias (LGDs), and 25 noncancerous IBD colitis samples. Slides were first reviewed by an expert gastrointestinal pathologist at the JHH and then 5-um-thick sections of the desired paraffin-embedded tissue blocks were procured. In the case of IBD patients with dysplasia/cancer, we studied dysplasia/cancer samples as well as nondysplastic samples from other non-neoplastic areas of the colon. We also investigated methylation of the WNT signaling pathway genes in 27 NCs from 27 patients as well as 24 samples from 24 sporadic CRC patients. Sporadic CRC patients were stage matched to the IBD-associated cancer patients to determine if there were any differences between methylation of these genes between sporadic and IBD-associated cancers.
Methylation Analysis
Methylation analysis was performed using the methylation-specific polymerase chain reaction (MSP) strategy, as previously described.45 DNA was extracted following a standard phenol-chloroform extraction protocol. Bisulfite modification of DNA was done using the EZ DNA methylation Kit™ (Zymo Research) as per manufacturer’s instructions. Methylation-specific PCR was carried out in a 25-μl reaction containing 10× MSP buffer, 10 mM dNTPs, 33 pmol of each of the methylated or unmethylated primers, 0.5 unit of JumpStart™ REDTaq® DNA polymerase and 4 μl of bisulfite-treated DNA. Amplification cycles were as follows: one cycle of 95°C for 5 min followed by 35 cycles of 95°C for 30 s, annealing temp for 30 s, 72°C for 30 s, and a final extension step of 72°C for 5 min. In vitro methylated DNA (IVD) was used as a positive control for MSP. IVD was created by treating cell line DNA with Sassy methylated (New England Bolas) as directed. DKO, which is a double knockout derivative of the CRC cell line Hct116 with knockout of the major DNA methyltransferases (DNMT1−/− and DNMT3b−/−) was used as an additional negative control. DKO lacks methylation at 95% of the known CpG sites.46 Seven and a half microliters of each amplification reaction was loaded and run on 2% agarose gel containing GelStar™ Nucleic Acid Gel Stain (Cambrex Bio Science) and visualized by ultraviolet illumination.
We tested the promoter methylation of 10 WNT signaling pathway genes including APC1A (adenomatous polyposis coli1a), APC2 (adenomatous polyposis coli2), SFRP1 (secreted frizzled related protein1), SFRP2 (secreted frizzled related protein2), SFRP4 (secreted frizzled related protein4), SFRP5 (secreted frizzled related protein5), DKK1 (dickkopf1), DKK3 (dickkopf3), WIF1 (WNT inhibitory factor1), and LKB1 (serine threonine kinase). All of these genes, except APC2, have been previously described to be hypermethylated and silenced in CRC by candidate gene approaches.47–50 Hypermethylation of APC2 was recently described by Schuebel et al. in their transcriptome-wide approach to find genes hypermethylated in colon cancer.51 Table 1 summarizes the primer sequences used for the MSP reaction and the annealing temperatures used for the respective PCR reactions. Percentage of methylated genes for each tissue type sample was calculated using the following formula: (number of genes methylated) / (number of genes tested) × 100. The means of samples belonging to each tissue type were then compared.
Statistical Analysis
Categorical variables were analyzed using Chi-square tests, while continuous variables were analyzed using Mann–Whitney U test. P values of less than 0.05 were considered significant. Logistic regression was used to calculate the odds ratios (ORs) and 95% confidence interval. All statistical analysis was performed using the STATA 9.2 software package (College Station, TX).
Results
All IBD patients had long-standing disease with median disease duration of 12 years (12.5 years for those with IBD-associated neoplasia vs. 12 years for controls; p = 0.2). The median age of IBD colitis samples was 52 years, while the median age of patients with IBD-associated neoplasia was 54 years (p = 0.4). The cancers from IBD patients were Stages I and II and patients with sporadic CRC were stage-matched to IBD-associated cancers. Family history of cancer was present in 36% of the patients with IBD-associated dysplasia or cancer, while 42% of the patients with IBD colitis without neoplasia had a positive family history of cancer.
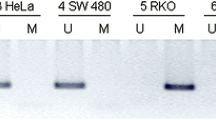
DNA was extracted and successful methylation analysis was performed in 99.2% of samples. Methylation of the WNT signaling pathway genes was seen for all genes in our samples, except for LKB1, which was uniformly unmethylated. LKB1 was therefore excluded from further analyses. Table 2 summarizes the methylation frequencies of WNT signaling pathway genes according to the tissue type analyzed. Methylation of the WNT signaling pathway genes varied according to the tissue type analyzed. NCs were uniformly unmethylated for all the genes, except WIF1, which was methylated in 7%, and SFRP2, which was methylated in 29%. Methylation of some genes can occur in NCs as a process of aging, a process termed age-related methylation. There was no age-related methylation pattern seen for either SFRP2 or WIF1 among the NCs. Methylation of all the WNT signaling pathway genes (except LKB1) was seen frequently in samples with IBD colitis and IBD-associated neoplasia. The frequency of methylation of the WNT signaling pathway genes seen in samples with IBD colitis was significantly higher than those seen in the NCs (see p values in Table 2), except for SFRP2 where the difference was not statistically significant (p = 0.10). Furthermore, the frequency of methylation of the WNT signaling pathway genes (except LKB1, which was unmethylated in all IBD-associated neoplasias) increased in samples with IBD-associated neoplasia compared to samples with IBD colitis. Methylation of APC1A, APC2, SFRP1, and SFRP2 was significantly higher in IBD-associated neoplasia compared to IBD colitis (APC1A 43.8% vs. 16.0%, p = 0.05; APC2 81.3% vs. 48.0%, p = 0.03; SFRP1 87.5% vs. 54.1%, p = 0.03 and SFRP2 86.7% vs. 52.0%, p = 0.03, respectively). Figure 2 shows the methylation profile of WNT signaling pathway genes in one of the IBD patients with cancer as a representative sample. Methylation analysis of the genes in WNT signaling pathway is shown from the cancer and areas of non-cancerous surrounding colitis.
Methylation profile of individual WNT signaling pathway genes in paired samples from one of the IBD-associated cancers showing the increase in methylation from colitis to cancer. Black bars represent methylation, whereas white bars represent unmethylated sample. Note that the patient has acquired methylation of APC1A, SFRP1, SFRP2, and WIF1 during progression to cancer.
Furthermore, the methylation of all the WNT signaling pathway genes was tested in 24 sporadic CRC who were matched in stage (stages 1 and 2) to the corresponding IBD-associated cancers. The methylation frequencies of the WNT signaling pathway genes seen in the sporadic CRC was similar to those seen in IBD-associated neoplasias (including dysplasias and cancers) except for SFRP4 and WIF1. SFRP4 was significantly more methylated in IBD-associated neoplasia (81.3%) compared to sporadic CRC (37.5%) (p < 0.01). On the contrary, WIF1 was significantly more methylated in sporadic CRC(100%) compared to IBD-associated neoplasia (68.8%), p < 0.01. The remainder of the frequencies were comparable (p = ns; data not shown).
We then calculated the percentage of methylated genes in the WNT signaling pathway for each sample and compared the means for samples belonging to each tissue type. A progressive increase in the percentage of methylated genes was seen from NCs (mean = 4.2%) to IBD colitis (mean = 39.7%) to IBD-associated neoplasia (mean = 63.4%) was seen (NC vs. IBD colitis, p < 0.01, IBD colitis vs. IBD-associated neoplasia, p = 0.01) (Fig. 3). There was no significant difference in the percentage of methylated genes between sporadic CRC and IBD-associated neoplasia (61.3% vs. 63.4% respectively; p = 0.42). However, methylation is an early event in IBD-associated neoplasia as relatively early lesions like LGDs show a high level of percentage of methylated genes (mean = 75%).
Percentage of methylated genes in different tissue types. Percentage of methylated genes increased from normal colons (n = 24, mean = 4.2%) to IBD colitis (n = 25, mean = 39.7%) to IBD-associated neoplasia (n = 16, mean = 63.4%) was seen (NC vs. IBD colitis, p < 0.01, IBD colitis vs. IBD-associated neoplasia, p = 0.01). There was no significant difference between IBD neoplasia (mean = 63.4%) and sporadic CRC (mean = 61.3%) p = 0.42.
We next used logistic regression model to find out whether methylation of these genes can be used to predict the presence of IBD-associated neoplasia, i.e., dysplasia or cancer. In the univariate analysis methylation of APC2 (odds ratio [OR] 4.7 [95% CI 1.1–20.63], p = 0.04), SFRP1 (OR 5.1 [95% CI 1.1–31.9], p = 0.04), and SFRP2 (OR 5.1 [95% CI 1.1–32.3], p = 0.04) was significantly associated with higher risk for IBD-associated neoplasia compared to IBD colitis. Methylation of APC1A was found to be borderline significant (OR 4.1 [95% CI 0.95–17.5], p = 0.06). Moreover, in the multivariate model using these four genes, including APC1A, APC2, SFRP1, and SFRP2, methylation of APC2 and APC1A was found to be significantly associated with higher risk for IBD-associated neoplasia [OR for methylated APC2 9.1 [95% CI 1.3–61.7], p = 0.02 and OR for methylated APC1A 6.4 [96% CI 1.1–37.7], p = 0.04) with an ROC value of 77.36%.
Discussion
Our study demonstrates for the first time epigenetic involvement of the WNT signaling pathway in IBD-associated neoplasia. We have shown that methylation of the WNT signaling pathway genes first begins in patients with long-standing IBD colitis and that the frequency of methylation of these genes increases progressively during the development of IBD-associated neoplasia. The percentage of methylated genes increases significantly from NCs to IBD colitis to IBD-associated neoplasia. A limitation of our current study is the relatively small sample size and the lack of expression and immunohistochemical data to fully confirm the silencing of the methylated genes.
However, the frequent methylation of the WNT signaling pathway genes in patients with IBD colitis suggests that chronic inflammation may play an important role in the methylation of WNT signaling genes. Further increase in the methylation of these genes in neoplastic lesions (dysplasia and cancer) suggests a progressive role of methylation of WNT signaling pathway genes in the pathogenesis of IBD-associated neoplasia. This is further supported by the fact that relatively early lesions like LGDs in IBD patients show high levels of methylation of the WNT signaling pathway genes. In the current study, we did not analyze non-inflamed normal colons from patients with IBD. Recent data suggest that some degree of methylation can occur in the non-inflamed terminal ileum, but it is still significantly lower than that seen in inflamed mucosa.52
Chronic inflammation can lead to chronic injury, which may predispose tumor-related genes to methylation and silencing. This process has also been described in other chronic inflammatory states like Barrett’s esophagus, which predisposes to esophageal cancer,53 chronic gastritis, which predisposes to gastric cancer,54 and cirrhosis, which predisposes to hepatocellular cancer.55 WNT signaling pathway genes are some of the genes affected by methylation in patients with IBD. Methylation of p16, E-cadherin, hMLH1, p14, HPP1, ER, etc. has already been reported by us and others in the setting of IBD-associated neoplasia (dysplasia and cancers).22–26 We studied the current panel of genes because of their importance to the biology of colorectal cancers.27,28
Dysplasia is regarded not only as the precursor, but also a marker, of coexisting malignancy in IBD patients. However, there is a great deal of controversy in the diagnosis of dysplasia. Inflammatory epithelial changes can mimic dysplasia, and there is a significant degree of intra- and inter-observer variability in the pathological diagnosis.56 This is not limited to less experienced pathologists, since even expert pathologists can differ in the opinion too.57,58 This has led to the recommendation that two pathologists, one of whom is an experienced gastrointestinal pathologist, should independently review biopsy specimens from cases of IBD-associated dysplasia.59–61 This criterion was also used when we procured samples for the current study. Our findings of early methylation of APC2, SFRP1, SFRP2, and APC1A in IBD-associated dysplasia may provide a more objective marker for the diagnosis of these lesions. In fact, all patients with LGD and HGD showed 100% methylation of the APC2, which suggests that this could be used as a marker for dysplasia.
APC1A (classical APC) is a classical tumor suppressor gene. Its gene product forms a complex with GSK-3β, axin/conductin, and β-catenin. Subsequently, β-catenin is phosphorylated and degraded. Mutation of APC1A prevents this degradation and causes accumulation of β-catenin in the cell. The β-catenin then translocates to the nucleus where it upregulates the transcription of many cancer-related genes. APC1A is mutated in 80% of sporadic CRC.29 Additionally, APC1A mutations usually occur early in sporadic colorectal carcinogenesis since lesions like tubular adenomas have been found to harbor APC1A mutations. Earlier studies, which looked into the mutation of APC1A in IBD, found a 0–6% mutation rate, and that too in advanced lesion like HGD and cancers.39,62 In contrast to its low mutation frequency, APC1A protein expression was found to be abnormal in 67% of the UC-associated neoplasia, which cannot be explained by low mutation frequency.44 Our results suggest that methylation of APC1A is an early and frequent event in IBD-associated neoplasia, and our results now suggest that methylation-associated silencing may account for the decreased protein expression of APC1A in these lesions. Since methylation of APC1A is more common than mutation, epigenetic inactivation may be a predominant mechanism for inactivation of APC1A and subsequent development of cancer in IBD patients.
APC2, identified more recently, has a high degree of homology with APC1A.63APC2 can also modulate WNT signaling like APC1A.63,64 Apart from mutations in APC1A, IBD-associated neoplasias also differ in the timing and frequency of mutation of other tumor suppressor gene P53. P53 mutations occur late in sporadic CRC, being more frequent in cancers compared to adenomas, whereas P53 mutations occur early and are more frequent in IBD-associated neoplasia. Early lesions like LGDs and sometimes even non-dysplastic mucosa in ulcerative colitis can have P53 mutations.65,66 Interestingly, APC2 has been shown to interact with a protein, 53BP2, which in turn interacts with P53 and an anti-apoptotic gene Bcl2. This suggests a mechanistic role of APC2 in the P53/Bcl2-linked pathway of cell cycle progression and cell death.67 Interaction of APC2 with P53 pathway is very interesting if viewed in the context of IBD-associated neoplasia. Early dysplastic lesions show a high level of P53 mutation and Bcl2 overexpression.65,68,69 Similarly, we have found APC2 to be methylated to high levels in early dysplastic lesions. Therefore, APC2 methylation, P53 mutation, and Bcl2 overexpression might play a synergistic role in the pathogenesis of these lesions. This will require further investigation.
The frequent inactivation of SFRP1 and SFRP2 in IBD-associated neoplasias further highlights the importance of canonical WNT pathway in the pathogenesis of IBD-associated neoplasia. SFRPs can block the WNT signaling pathway either by interacting with WNT proteins or by forming nonfunctional complexes with frizzled receptors Fz.70 Methylation of SFRPs can thus lead to activation of WNT signaling pathway even in the absence of mutations in APC and β-catenin.48
Recently, there has been increasing interest in developing stool DNA based biomarkers. In fact, the current ACS guidelines concluded that there is now sufficient data to include stool DNA as an acceptable option for CRC screening.71 DNA shed into stool theoretically provides a more comprehensive sampling of abnormal cells than random punch biopsies for cancer surveillance among IBD patients. Stool DNA testing for hypermethylation of the SFRP-1 promoter has already been shown to be a sensitive and specific screening tool for sporadic CRC.72 Our findings of methylation of APC1A, APC2, SFRP1, and SFRP2, if validated in prospective studies, could be used to devise stool-based DNA methylation tools for early detection of dysplasia and cancer in IBD patients.
Conclusions
Methylation of the WNT signaling pathway genes is seen in patients with IBD colitis and the frequency of methylation of the WNT signaling pathway genes increases progressively during development of IBD-associated neoplasia. Moreover, the findings of early methylation of APC1A, APC2, SFRP1, and SFRP2 in IBD-associated neoplasia may provide objective markers and a method for early detection of IBD-associated neoplasia. In IBD surveillance, strategies for CRC prevention would be strengthened by the addition of objective markers for dysplasia and cancer, which will be independent of the expertise of the pathologist. Further studies will be required to validate the findings of the current study.
References
Hanauer SB. Inflammatory bowel disease: epidemiology, pathogenesis, and therapeutic opportunities. Inflamm Bowel Dis 2006;12(Suppl 1):S3–S9. doi:10.1097/01.MIB.0000195385.19268.68.
Ekbom A, Helmick C, Zack M, Adami HO. Ulcerative colitis and colorectal cancer. A population-based study. N Engl J Med 1990;323(18):1228–1233.
Ekbom A, Helmick C, Zack M, Adami HO. Increased risk of large-bowel cancer in Crohn’s disease with colonic involvement. Lancet 1990;336(8711):357–359. doi:10.1016/0140-6736(90)91889-I.
Langholz E, Munkholm P, Davidsen M, Binder V. Colorectal cancer risk and mortality in patients with ulcerative colitis. Gastroenterology 1992;103(5):1444–1451.
Greenstein AJ, Sachar DB, Smith H, Pucillo A, Papatestas AE, Kreel I, et al. Cancer in universal and left-sided ulcerative colitis: factors determining risk. Gastroenterology 1979;77(2):290–294.
Harpaz N, Talbot IC. Colorectal cancer in idiopathic inflammatory bowel disease. Semin Diagn Pathol 1996;13(4):339–357.
Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut 2001;48(4):526–535. doi:10.1136/gut.48.4.526.
Rutter MD, Saunders BP, Wilkinson KH, Rumbles S, Schofield G, Kamm MA, et al. Thirty-year analysis of a colonoscopic surveillance program for neoplasia in ulcerative colitis. Gastroenterology 2006;130(4):1030–1038. doi:10.1053/j.gastro.2005.12.035.
Farrell RJ, Peppercorn MA. Ulcerative colitis. Lancet 2002;359(9303):331–340. doi:10.1016/S0140-6736(02)07499-8.
Hata K, Watanabe T, Kazama S, Suzuki K, Shinozaki M, Yokoyama T, et al. Earlier surveillance colonoscopy programme improves survival in patients with ulcerative colitis associated colorectal cancer: results of a 23-year surveillance programme in the Japanese population. Br J Cancer 2003;89(7):1232–1236. doi:10.1038/sj.bjc.6601247.
Karlen P, Kornfeld D, Brostrom O, Lofberg R, Persson PG, Ekbom A. Is colonoscopic surveillance reducing colorectal cancer mortality in ulcerative colitis? A population based case control study. Gut 1998;42(5):711–714.
Woolrich AJ, DaSilva MD, Korelitz BI. Surveillance in the routine management of ulcerative colitis: the predictive value of low-grade dysplasia. Gastroenterology 1992;103(2):431–438.
Chambers WM, Warren BF, Jewell DP, Mortensen NJ. Cancer surveillance in ulcerative colitis. Br J Surg 2005;92(8):928–936. doi:10.1002/bjs.5106.
Ullman TA. Preventing neoplastic progression in ulcerative colitis. J Clin Gastroenterol 2005;39(4Suppl 2):S66–S69. doi:10.1097/01.mcg.0000155554.01336.ff.
Rubin CE, Haggitt RC, Burmer GC, Brentnall TA, Stevens AC, Levine DS, et al. DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology 1992;103(5):1611–1620.
Axon AT. Colonic cancer surveillance in ulcerative colitis is not essential for every patient. Eur J Cancer 1995;31A(7–8):1183–1186. doi:10.1016/0959-8049(95)00131-2.
Choi PM, Nugent FW, Schoetz DJ Jr, Silverman ML, Haggitt RC. Colonoscopic surveillance reduces mortality from colorectal cancer in ulcerative colitis. Gastroenterology 1993;105(2):418–424.
Lynch DA, Lobo AJ, Sobala GM, Dixon MF, Axon AT. Failure of colonoscopic surveillance in ulcerative colitis. Gut 1993;34(8):1075–1080. doi:10.1136/gut.34.8.1075.
Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet 2000;16(4):168–174. doi:10.1016/S0168-9525(99)01971-X.
Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2005;2(Suppl 1):S4–S11. doi:10.1038/ncponc0354.
Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349(21):2042–2054. doi:10.1056/NEJMra023075.
Azarschab P, Porschen R, Gregor M, Blin N, Holzmann K. Epigenetic control of the E-cadherin gene (CDH1) by CpG methylation in colectomy samples of patients with ulcerative colitis. Genes Chromosomes Cancer 2002;35(2):121–126. doi:10.1002/gcc.10101.
Fleisher AS, Esteller M, Harpaz N, Leytin A, Rashid A, Xu Y, et al. Microsatellite instability in inflammatory bowel disease-associated neoplastic lesions is associated with hypermethylation and diminished expression of the DNA mismatch repair gene, hMLH1. Cancer Res 2000;60(17):4864–4868.
Hsieh CJ, Klump B, Holzmann K, Borchard F, Gregor M, Porschen R. Hypermethylation of the p16INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res 1998;58(17):3942–3945.
Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res 2001;61(9):3573–3577.
Moriyama T, Matsumoto T, Nakamura S, Jo Y, Mibu R, Yao T, et al. Hypermethylation of p14 (ARF) may be predictive of colitic cancer in patients with ulcerative colitis. Dis Colon Rectum 2007;50(9):1384–1392. doi:10.1007/10350-007-0302-x.
de Lau W, Barker N, Clevers H. WNT signaling in the normal intestine and colorectal cancer. Front Biosci 2007;12:471–491. doi:10.2741/2076.
Pinto D, Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp Cell Res 2005;306(2):357–363. doi:10.1016/j.yexcr.2005.02.022.
Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, et al. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet 1992;1(4):229–233. doi:10.1093/hmg/1.4.229.
Potter JD. Colorectal cancer: molecules and populations. J Natl Cancer Inst 1999;91(11):916–932. doi:10.1093/jnci/91.11.916.
Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev 1997;11(24):3286–3305. doi:10.1101/gad.11.24.3286.
Klingensmith J, Nusse R, Perrimon N. The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes Dev 1994;8(1):118–130. doi:10.1101/gad.8.1.118.
Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 1996;382(6592):638–642. doi:10.1038/382638a0.
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science 1998;281(5382):1509–1512. doi:10.1126/science.281.5382.1509.
Howe LR, Subbaramaiah K, Chung WJ, Dannenberg AJ, Brown AM. Transcriptional activation of cyclooxygenase-2 in Wnt-1-transformed mouse mammary epithelial cells. Cancer Res 1999;59(7):1572–1577.
Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci USA 1999;96(10):5522–5527. doi:10.1073/pnas.96.10.5522.
Bafico A, Gazit A, Pramila T, Finch PW, Yaniv A, Aaronson SA. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J Biol Chem 1999;274(23):16180–16187. doi:10.1074/jbc.274.23.16180.
Zorn AM. Wnt signalling: antagonistic Dickkopfs. Curr Biol 2001;11(15):R592–R595. doi:10.1016/S0960-9822(01)00360-8.
Tarmin L, Yin J, Harpaz N, Kozam M, Noordzij J, Antonio LB, et al. Adenomatous polyposis coli gene mutations in ulcerative colitis-associated dysplasias and cancers versus sporadic colon neoplasms. Cancer Res 1995;55(10):2035–2038.
You J, Nguyen AV, Albers CG, Lin F, Holcombe RF. Wnt pathway-related gene expression in inflammatory bowel disease. Dig Dis Sci 2008;53(4):1013–1019. doi:10.1007/s10620-007-9973-3.
You XJ, Bryant PJ, Jurnak F, Holcombe RF. Expression of Wnt pathway components frizzled and disheveled in colon cancer arising in patients with inflammatory bowel disease. Oncol Rep 2007;18(3):691–694.
van Dekken H, Wink JC, Vissers KJ, Franken PF, Ruud Schouten W, Hop WCJ, Kuipers EJ, Fodde R, Janneke van der Woude C. Wnt pathway-related gene expression during malignant progression in ulcerative colitis. Acta Histochem 2007;109(4):266–272. doi:10.1016/j.acthis.2007.02.007.
Aust DE, Terdiman JP, Willenbucher RF, Chang CG, Molinaro-Clark A, Baretton GB, et al. The APC/beta-catenin pathway in ulcerative colitis-related colorectal carcinomas: a mutational analysis. Cancer 2002;94(5):1421–1427. doi:10.1002/cncr.10334.
Aust DE, Terdiman JP, Willenbucher RF, Chew K, Ferrell L, Florendo C, et al. Altered distribution of beta-catenin, and its binding proteins E-cadherin and APC, in ulcerative colitis-related colorectal cancers. Mod Pathol 2001;14(1):29–39. doi:10.1038/modpathol.3880253.
Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996;93(18):9821–9826. doi:10.1073/pnas.93.18.9821.
Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 2002;416(6880):552–556. doi:10.1038/416552a.
Arnold CN, Goel A, Niedzwiecki D, Dowell JM, Wasserman L, Compton C, et al. APC promoter hypermethylation contributes to the loss of APC expression in colorectal cancers with allelic loss on 5q. Cancer Biol Ther 2004;3(10):960–964.
Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 2004;36(4):417–422. doi:10.1038/ng1330.
Aguilera O, Fraga MF, Ballestar E, Paz MF, Herranz M, Espada J, et al. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene 2006;25(29):4116–4121. doi:10.1038/sj.onc.1209439.
Taniguchi H, Yamamoto H, Hirata T, Miyamoto N, Oki M, Nosho K, et al. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene 2005;24(53):7946–7952. doi:10.1038/sj.onc.1208910.
Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 2007;3(9):1709–1723. doi:10.1371/journal.pgen.0030157.
Wang FY, Arisawa T, Tahara T, Takahama K, Watanabe M, Hirata I, et al. Aberrant DNA methylation in ulcerative colitis without neoplasia. Hepatogastroenterology 2008;55(81):62–65.
Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, et al. Fields of aberrant CpG island hypermethylation in Barrett’s esophagus and associated adenocarcinoma. Cancer Res 2000;60(18):5021–5026.
Tamura G, Yin J, Wang S, Fleisher AS, Zou T, Abraham JM, et al. E-Cadherin gene promoter hypermethylation in primary human gastric carcinomas. J Natl Cancer Inst 2000;92(7):569–573. doi:10.1093/jnci/92.7.569.
Okochi O, Hibi K, Sakai M, Inoue S, Takeda S, Kaneko T, et al. Methylation-mediated silencing of SOCS-1 gene in hepatocellular carcinoma derived from cirrhosis. Clin Cancer Res 2003;9(14):5295–5298.
Collins RH Jr, Feldman M, Fordtran JS. Colon cancer, dysplasia, and surveillance in patients with ulcerative colitis. A critical review. N Engl J Med 1987;316(26):1654–1658.
Connell WR, Lennard-Jones JE, Williams CB, Talbot IC, Price AB, Wilkinson KH. Factors affecting the outcome of endoscopic surveillance for cancer in ulcerative colitis. Gastroenterology 1994;107(4):934–944.
Melville DM, Jass JR, Morson BC, Pollock DJ, Richman PI, Shepherd NA, et al. Observer study of the grading of dysplasia in ulcerative colitis: comparison with clinical outcome. Hum Pathol 1989;20(10):1008–1014. doi:10.1016/0046-8177(89)90273-6.
Dixon MF, Brown LJ, Gilmour HM, Price AB, Smeeton NC, Talbot IC, et al. Observer variation in the assessment of dysplasia in ulcerative colitis. Histopathology 1988;13(4):385–397. doi:10.1111/j.1365-2559.1988.tb02055.x.
Riddell RH, Goldman H, Ransohoff DF, Appelman HD, Fenoglio CM, Haggitt RC, et al. Dysplasia in inflammatory bowel disease: standardized classification with provisional clinical applications. Hum Pathol 1983;14(11):931–968. doi:10.1016/S0046-8177(83)80175-0.
Rubin DT, Kavitt RT. Surveillance for cancer and dysplasia in inflammatory bowel disease. Gastroenterol Clin North Am 2006;35(3):581–604. doi:10.1016/j.gtc.2006.07.001.
Kern SE, Redston M, Seymour AB, Caldas C, Powell SM, Kornacki S, et al. Molecular genetic profiles of colitis-associated neoplasms. Gastroenterology 1994;107(2):420–428.
Nakagawa H, Murata Y, Koyama K, Fujiyama A, Miyoshi Y, Monden M, et al. Identification of a brain-specific APC homologue, APCL, and its interaction with beta-catenin. Cancer Res 1998;58(22):5176–5181.
van Es JH, Kirkpatrick C, van de Wetering M, Molenaar M, Miles A, Kuipers J, et al. Identification of APC2, a homologue of the adenomatous polyposis coli tumour suppressor. Curr Biol 1999;9(2):105–108. doi:10.1016/S0960-9822(99)80024-4.
Brentnall TA, Crispin DA, Rabinovitch PS, Haggitt RC, Rubin CE, Stevens AC, et al. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology 1994;107(2):369–378.
Burmer GC, Rabinovitch PS, Haggitt RC, Crispin DA, Brentnall TA, Kolli VR, et al. Neoplastic progression in ulcerative colitis: histology, DNA content, and loss of a p53 allele. Gastroenterology 1992;103(5):1602–1610.
Nakagawa H, Koyama K, Murata Y, Morito M, Akiyama T, Nakamura Y. APCL, a central nervous system-specific homologue of adenomatous polyposis coli tumor suppressor, binds to p53-binding protein 2 and translocates it to the perinucleus. Cancer Res 2000;60(1):101–105.
Yin J, Harpaz N, Tong Y, Huang Y, Laurin J, Greenwald BD, et al. p53 point mutations in dysplastic and cancerous ulcerative colitis lesions. Gastroenterology 1993;104(6):1633–1639.
Bronner MP, Culin C, Reed JC, Furth EE. The bcl-2 proto-oncogene and the gastrointestinal epithelial tumor progression model. Am J Pathol 1995;146(1):20–26.
Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci 2003;116(Pt 13):2627–2634. doi:10.1242/jcs.00623.
Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Dash C, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Cancer J Clin 2008;58:130–160.
Zhang W, Bauer M, Croner RS, Pelz JO, Lodygin D, Hermeking H, et al. DNA stool test for colorectal cancer: hypermethylation of the secreted frizzled-related protein-1 gene. Dis Colon Rectum 2007;50(10):1618–1626, 1626–1617. discussion, doi:10.1007/s10350-007-0286-6.
Acknowledgments
We thank Marco A. Riojas for technical assistance.
NA is supported by the American Surgical Association Fellowship Award, Wendy Will Cancer Fund, and the Richard Ross Clinician Scientist Award.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Dhir, M., Montgomery, E.A., Glöckner, S.C. et al. Epigenetic Regulation of WNT Signaling Pathway Genes in Inflammatory Bowel Disease (IBD) Associated Neoplasia. J Gastrointest Surg 12, 1745–1753 (2008). https://doi.org/10.1007/s11605-008-0633-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11605-008-0633-5