Abstract
During our studies of freshwater fungi from Thailand, a new species Minimelanolocus nonramosus and a new collection of Thysanorea papuana were identified using morphological characters and phylogenetic analyses based on LSU, SSU, and ITS genes. After re-examination of herbarium material, Thysanorea aquatica is synonymized with Th. papuana. The description of the new species is provided with notes. This study increases our understanding of the freshwater hyphomycetes in Eurotiomycetes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Freshwater lignicolous hyphomycetes is a dominant and diverse group, which lives on submerged woody debris and decaying tree leaves (Wong et al. 1998; Shearer et al. 2007; Hyde et al. 2016a). Molecular techniques revealed that most species belong to Sordariomycetes and Dothideomycetes (Cai et al. 2008, 2010; Su et al. 2015; Liu et al. 2016; Luo et al. 2016, 2018; Wei et al. 2018) and rarely to Eurotiomycetes (Liu et al. 2015; Dong et al. 2018). The first molecular report of freshwater fungi in Eurotiomycetes was provided by Liu et al. (2015), in which they obtained sequence data for five species of Minimelanolocus R.F. Castañeda & Heredia in Herpotrichiellaceae and revisited the genus at the molecular level. Their phylogenetic analyses showed that Minimelanolocus species formed a monophyletic clade based on LSU and SSU genes, while Minimelanolocus obscurus (Matsush.) R.F. Castañeda & Heredia was distinct from the Minimelanolocus clade and was related with Exophiala J.W. Carmich species based only on ITS gene (Liu et al. 2015). This conclusion was proven by subsequent studies (Hyde et al. 2016b; Tian et al. 2016). Those studies, however, did not include the genus Thysanorea Arzanlou, W. Gams & Crous, whose morphological characters are very similar to those of Minimelanolocus.
The genus Minimelanolocus was introduced by Ruiz et al. (2001) based on ten new combinations segregated from Pseudospiropes M.B. Ellis, which differs from Minimelanolocus by the presence of distoseptate conidia. Minimelanolocus is characterized by cylindrical and unbranched conidiophores, holoblastic and sympodially proliferating conidiogenous cells, and acrogenous, oblong or clavate to fusiform, hyaline to pale brown or brown, septate conidia (Ruiz et al. 2001; Ma et al. 2011; Liu et al. 2015). In total, 33 species are listed under Minimelanolocus in Index Fungorum (2019), in which seven species were reported from submerged wood (Liu et al. 2015; Hyde et al. 2016b; Dong et al. 2018).
The genus Thysanorea, on the other hand, was established to accommodate Periconiella papuana Aptroot (Arzanlou et al. 2007), which was first isolated in Papua New Guinea (Aptroot and Iperen 1998) and later recorded from India (Pratibha and Prabhugaonkar 2015) and Taiwan (Kirschner 2016). According to published illustrations, Kirschner (2016) suggested that Ramichloridium lignicola C.K.M. Tsui, Goh, K.D. Hyde & Hodgkiss and Alysidiopsis lignicola Mercado, Figueras & J. Mena considered to be possible synonyms of the type of species, Th. papuana (Aptroot) Arzanlou, W. Gams & Crous. Thysanorea papuana was first described as having dimorphic conidiophores with up to five levels of branchlets and typically pyriform, septate conidia with a truncate base and darkened hilum (Arzanlou et al. 2007). Based on re-examination of the material and fresh collections from Taiwan, Kirschner (2016) improved the original description and reported that the conidiophore heads containing branches were easily broken off and could be replaced by regeneration. This character was a good morphological marker not only at the species level but also at the genus level of Thysanorea (Kirschner 2016). It was also reported in A. lignicola and R. lignicola but not in the species Th. aquatica W. Dong, H. Zhang & K.D. Hyde (Dong et al. 2018).
During studies of lignicolous freshwater fungi along a north/south latitudinal gradient in the Asian/Australian region (Hyde et al. 2016a), we collected a novel species of Minimelanolocus and a new specimen of Thysanorea papuana. Both taxa are described here and compared with related species. The holotype of Th. aquatica was re-examined based on morphological and molecular data.
Material and methods
Isolation and morphology
Pieces of decaying wood were collected from several small streams in Chiang Rai Province, Thailand, following the procedures described in Kurniawati et al. (2010). The samples were placed in Ziploc plastic bags with moist sterile tissue, taken to the laboratory, and incubated at room temperature (25 °C). After 1–2 weeks, specimens were examined using a stereomicroscope (SMZ-171) to locate fruiting bodies (Taylor and Hyde 2003). Photomicrographs were taken using a Cannon EOS 600D camera attached to a Nikon ECLIPSE Ni compound microscope. The fungal structures were measured using Tarosoft (R) Image Frame Work program, and images were processed using Adobe Photoshop CS6 Extended version 13.0 software (Adobe Systems, USA). Isolations were made from single conidia as described by Chomnunti et al. (2014). Water agar (WA) was used for conidial germination and incubated overnight in an incubator at room temperature. Single germinating conidia were selected and transferred to a new potato dextrose agar (PDA) plate to obtain pure cultures. The colonies were checked every 3 days. Herbarium specimens are deposited in the herbarium of Mae Fah Luang University (MFLU), Chiang Rai, Thailand. Living cultures are deposited in the Mae Fah Luang University Culture Collection (MFLUCC). Facesoffungi and Index Fungorum numbers are registered as described by Jayasiri et al. (2015).
DNA extraction, PCR amplification, and sequencing
Cultures were grown on PDA at 25 °C until enough mycelia were obtained, and a Biospin Fungus Genomic DNA Extraction Kit (Bioer Technology Co., Ltd., Hangzhou, P.R. China) was used to extract total genomic DNA from the fresh mycelia following the manufacturer’s instructions. DNA amplification was performed using the polymerase chain reaction (PCR). Fragments of three loci, LSU, SSU, and ITS, were used for phylogenetic analyses, and the following primer pairs, LROR/LR5, NS4/NS5, and ITS5/ITS4, were used for amplification and sequencing (Vilgalys and Hester 1990; White et al. 1990). Amplifications were performed in a 25 μL reaction containing 9.5 μL ddH2O, 12.5 μL 2× PCR Master Mix, 1 μL of DNA template, and 1 μL of each primer (10 μM). The PCR thermal cycles for amplification of the gene regions followed Su et al. (2015). The PCR products were examined on 1.0% agarose electrophoresis gels stained with ethidium bromide. Sequencing reactions were conducted by Shanghai Sangon Biological Engineering Technology and Services Co., Shanghai, P.R. China.
Phylogenetic analyses
Phylogenetic analyses were conducted to illustrate the phylogeny of Minimelanolocus and Thysanorea following Tian et al. (2016) and Dong et al. (2018). The newly generated sequences together with other sequences obtained from GenBank (Table 1) were initially aligned using MAFFTv.7 (Katoh and Standley 2013) on the online server (http://maffTh.cbrc.jp/alignment/server/) and optimized manually when needed. A maximum likelihood analysis was performed using RAxMLGUI v. 1.3 (Silvestro and Michalak 2011). For both analyses, the optimal ML tree search was conducted with 1000 separate runs using the default algorithm of the program from a random starting tree for each run. The final tree was selected among suboptimal trees from each run by comparing the likelihood scores using the GTR + GAMMA substitution model. Maximum likelihood bootstrap values equal to or greater than 80% are presented as the first set of numbers above the nodes in the resulting ML tree. Bayesian analysis was conducted using MrBayes v. 3.1.2 (Ronquist and Huelsenbeck 2003) to evaluate posterior probabilities (BYPP) (Rannala and Yang 1996) with Markov Chain Monte Carlo sampling (BMCMC). The best-fit model was GTR + I + G for LSU, SYM + I + G for ITS, and HKY + I + G for SSU. Bayesian posterior probabilities equal to or greater than 0.90 are shown above each node of the resulting consensus tree. These trees were viewed in Treeview (Page 1996) and edited further using Adobe Illustrator CS v.5. Newly generated sequences are deposited in GenBank. The alignment was deposited in TreeBASE (https://www.treebase.org/) under the accession number 23791. In this study, we follow the recommendations of Jeewon and Hyde (2016) to determine new taxa and introduce the new species Minimelanolocus nonramosus.
Results
Phylogenetic study
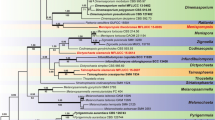
The placement of the two isolates within the family Herpotrichiellaceae was determined based on the phylogenetic analyses of LSU, SSU, and ITS sequences. The maximum likelihood tree included 31 isolates belonging to Herpotrichiellaceae species, and two outgroups Cyphellophora oxyspora (W. Gams) Réblová & Unter. (CBS 698.73) and C. sessilis (de Hoog) Réblová & Unter. (CBS 243.85) with an alignment length of 2349 characters. The best scoring RAxML tree with a final likelihood value of − 9697.482313 was selected to represent the relationships among the taxa and is presented in Fig. 1. For Bayesian analysis, six simultaneous Markov chains were run for 1,171,000 generations, and trees were sampled every 1000th generation and 1171 trees were obtained. The first 293 trees, representing the burn-in phase of the analyses, were discarded, while the remaining 878 trees were used to calculate posterior probabilities in the majority rule consensus tree (the critical value for the topological convergence diagnostic was 0.01).
Maximum likelihood tree generated by combining LSU, SSU, and ITS. Bootstrap support values for maximum likelihood (ML, first value) equal to or greater than 90% are shown above the nodes. The values of the Bayesian posterior probabilities from MCMC analyses (BYPP, second value) equal or higher than 0.90 are shown above the nodes. Hyphen (“-”) indicates a value lower than 90% for ML and a posterior probability lower than 0.90 for Bayesian. Ex-type isolates are in bold, and newly generated sequences are indicated in red. The tree was rooted to Cyphellophora sessilis CBS 243.85 and C. axyspora CBS 698.73 (Cyphellophoraceae)
The relationships among various isolates of Th. papuana were determined based on the phylogenetic analyses of ITS sequences and is presented in Fig. 2. The maximum likelihood tree included 6 isolates of Th. papuana, and 8 Minimelanolocus species as outgroups based on the above tree. The best scoring RAxML tree with a final likelihood value of − 1759.781121 was selected (Fig. 2). For the Bayesian analysis, six simultaneous Markov chains were run for 1,540,000 generations, and trees were sampled every 1000th generation and 1540 trees were obtained. The first 385 trees, representing the burn-in phase of the analyses, were discarded, while the remaining 1155 trees were used to calculate posterior probabilities in the majority rule consensus tree.
Maximum likelihood tree generated by ITS gene. Bootstrap support values for maximum likelihood (ML, first value) equal to or greater than 90% are shown above the nodes. The values of the Bayesian posterior probabilities from MCMC analyses (BYPP, second value) equal or higher than 0.90 are shown above nodes. Hyphen (“-”) indicates a value lower than 90% for ML and a posterior probability lower than 0.90 for Bayesian. Ex-type isolates are in bold, and newly generated sequences are indicated in red. The tree was rooted to Minimelanolocus species
Taxonomy
Minimelanolocus nonramosus X.D. Yu, G.N. Wang & H. Zhang, sp. nov.
Index Fungorum number: IF 555776; Facesoffungi number: FoF 04784; Fig. 3.
Minimelanolocus nonramosus (holotype, MFLU 17–1736). a, b Colonies on submerged wood. c, d Conidiophores. e–g Conidiogenous cells with pigmented conidiogenous loci. The arrow shows them becoming suddenly slimmer and paler, which probably implies a new percurrent proliferation after breaking off. h–m Conidia. n Germinating conidium. o Colony on PDA (front view). p Colony on PDA (reverse). Scale bars: a, b = 100 μm; c, d = 50 μm, e–g = 10 μm, h–n = 5 μm
Holotype: MFLU 17–1736.
Etymology: referring to the unbranched conidiophores
Saprobic on decaying, submerged wood in freshwater. Sexual morph: Undetermined. Asexual morph: Colonies superficial, effuse, scattered, pale brown to dark brown. Mycelium mostly immersed, composed of septate, pale brown, smooth hyphae. Conidiophores macronematous, mononematous, erect, cylindrical, slender, mostly flexuous, brown to dark brown, gradually paler and becoming pale brown to subhyaline at the upper part, unbranched, septate, slightly constricted at the septa, thick-walled, smooth, upside of the stipe sometimes becoming suddenly slimmer and paler, which probably implies a new percurrent proliferation after the break offs, (160–) 270–360 μm long, 3–7 μm wide (\( \overline{x} \) = 290.6 × 5 μm, n = 15). Conidiogenous cells holoblastic, terminal, discrete, sympodially proliferating, pale brown to subhyaline, thin-walled, smooth, subcylindrical, slightly thickened, covered by several pigmented, minutely denticulate conidiogenous loci, 5–23 μm long, 1.5–4.0 μm wide. Conidia acrogenous, cylindrical, rounded at the apex, narrow and truncate at the base with a darkened hilum (ca 0.5–1.0 μm diameter), 1–2(− 3)-septate, slightly or not constricted at the septa with several minute guttula, solitary, hyaline, thin-walled, smooth, 11–17 μm long, 1.5–4.0 μm wide (\( \overline{x} \) = 13.6 × 2.7 μm, n = 20), with a ratio of length/width of approximately 4:1.
Material examined: Thailand, Phang Nga, Thap Put, saprobic on decaying wood submerged in a stream, 1 September 2017, X.D. Yu, V9 (MFLU 17–1736, holotype), ex-type living culture MFLUCC 17–2378.
Culture characteristics: Conidia germinating on PDA; colonies slow growing, reaching 30 mm diameter in 20 days at 25 °C, brown to gray, reverse dark brown, circular, velvety, dry, fairly dense, margin entire.
Notes: The phylogenetic tree of LSU, SSU, and ITS sequence data shows the new collection clusters within the Minimelanolocus sensu lato clade. It fits well with the characters of Minimelanolocus in having mononematous and unbranched conidiophores, terminal conidiogenous cells with sympodial, denticulate conidiogenous loci, and septate conidia (Ruiz et al. 2001; Liu et al. 2015). Therefore, it is described as a new species of Minimelanolocus. Minimelanolocus nonramosus is distinguished from the phylogenetically close species Minimelanolocus obscurus (Matsush.) R.F. Castañeda & Heredia by cylindrical, hyaline, and smaller conidia (\( \overline{x} \) = 13.6 × 2.7 μm), while the latter has clavate, hyaline to pale brown, and larger conidia (\( \overline{x} \) = 21.5 × 4.6 μm). Minimelanolocus aquatilis L.B. Conc., M.F.O. Marques, Gusmão & R.F. Castañeda, which has no DNA sequence available yet, differs from M. nonramosus in having shorter conidiophores (43–78 μm in M. aquatilis vs. 270–360 μm in M. nonramosus) and obclavate, 3–5-septate, verruculose, pale brown to subhyaline, and larger conidia (21–30 × 5–8 μm in M. aquatilis vs. 13.6 × 2.7 μm in M. nonramosus) (Fiuza et al. 2017).
Minimelanolocus is morphologically close to Pseudospiropes in having polyblastic and integrated conidiogenous cells with sympodial percurrent proliferations (Ruiz et al. 2001). However, the conidiogenous loci in Pseudospiropes are broad, protuberant, thickened, and strongly melanized, apparently with several layers, forming a discoid black scar and markedly different from the loci in Minimelanolocus. In addition, Pseudospiropes species have distoseptate conidia, which differ from the conidia of M. nonramosus (Ruiz et al. 2001). The genus Pleurophragmium Costantin resembles Minimelanolocus in having single unbranched conidiophores with sympodial, denticulate conidiogenous cells (Hughes 1958; Ellis 1971). However, conidial color of Pleurophragmium species is dematiaceous except for P. acutum (Grove) M.B. Ellis, P. malaysianum Matsush. and P. naviculiforme Matsush. (D’Souza and Bhat 2012). M. nonramosus differs from these species in having cylindrical, 1–2-septate conidia and conidial size.
The upper part of some conidiophores in M. nonramosus (e in Fig. 3) becomes suddenly slimmer and paler, which seems to be a new percurrent proliferation after the head secedes. However, this phenomenon is not very common. We also observed this phenomenon in the plate of Minimelanolocus melanicus H.Y. Su, Udayanga & K.D. Hyde, which was not described by the authors (Liu et al. 2015).
Thysanorea papuana (Aptroot) Arzanlou, W. Gams & Crous. Stud. Mycol. 58: 80 (2007) Fig. 4.
Thysanorea papuana (form MFLU 17–1657). a, b Colonies on submerged wood. c Conidiophore. d Long regenerative extension on the conidiophore with percurrent proliferation. e Heads and branches breaking off from the conidiophore. f–h Conidiogenous cells with denticulate conidiogenous loci. i–l Conidia. m Germinating conidium. n Colony on PDA (front view). o Colony on PDA (reverse). Scale bars: a, b = 80 μm, c = 30 μm, d–h = 10 μm, i–m = 5 μm
≡ Periconiella papuana Aptroot, Nova Hedwigia 67: 491 (1998)
= Ramichloridium lignicola KM. Tsui, Goh, KD. Hyde & Hodgkiss, Cryptog. Mycol. 22: 141 (2001)
? = Alysidiopsis lignicola Mercado, Figueras & J. Mena, Mycotaxon 60: 444 (1996). Facesoffungi Number: FoF02731
= Thysanorea aquatica W. Dong, H. Zhang & K.D. Hyde, Mycol. Progr. 17: 625 (2018), syn. nov. Fig. 5.
Material examined: Thailand, Prachuap Khiri Khan, on submerged wood in a stream, 30 July 2015, W. Dong, 29C (MFLU 15–2695), living culture: MFLUCC 15–0966; Thailand, Chiang Rai, on submerged wood in a stream, 4 August 2017, G.N. Wang, G3 (MFLU 17–1657), living culture: MFLUCC 17–2315.
Notes: Molecular evidence supports that our new collection (MFLUCC 17–2315) belongs to Thysanorea papuana (Fig. 2). Morphologically, our collection is similar to the holotype in conidiophores and conidial morphology, but they differ in the levels of branchlets (2–3 in our collection vs. 3–5 in the holotype) and the length of regenerative extension (35–65 μm in our collection vs. 15–30 μm in the holotype) (Arzanlou et al. 2007; Pratibha and Prabhugaonkar 2015; Kirschner 2016). These differences are considered not significant, since Kirschner (2016) stated that more complex branching appeared to be an artifact of prolonged cultivation in Th. papuana.
Thysanorea aquatica (MFLUCC 15–0966) was recently introduced by Dong et al. (2018) based on only two levels of branchlets and hyaline conidia with guttules and constricted septate, which they considered to be distinct enough from Th. papuana. The character of the conidiophores was not clearly described in the original description. We re-examined the herbarium material and re-described the conidiophores as “composed of unbranched stipe and branched head, with stipe sometimes percurrently proliferating below the branched head which is paler than stipe, branched head sometimes breaking off and replaced by a regenerative extension, each branch composed of (1–)2(–6) cells, each cell 4–6 μm long, 1–3 μm wide.” The significant character is that conidiophore heads contain branches which could break off and be replaced by regeneration, a feature consistent with the holotype. In addition, only elder conidia have guttulae and more constricted septa. The others are morphologically identical to the holotype (f in Fig. 5). Therefore, we synonymize Th. aquatica under Th. papuana.
The morphology of Th. papuana is variable (Table 2). Branches were up to four levels in the specimen from Taiwan (Kirschner 2016) and up to three levels in the specimen from India (Pratibha and Prabhugaonkar 2015), while they were up to 5–6 levels from the culture of Arzanlou et al. (2007). Septation ranges from 0 to 3, but are mostly 1. The conidial size is between 4 and 11 in length and 2–4 μm in width. The ITS divergence of 1–2% between the ex-type, MFLUCC 15–0966 and MFLUCC 17–2315 are within the generally accepted 1.5% nucleotide differences in the ITS regions that may be indicative of conspecificity (Jeewon and Hyde 2016).
Discussion
The generic delimitation of Thysanorea with one single species Th. papuana was discussed by Kirschner (2016) who suspected that more complex branches appeared to be an artifact of a degenerated culture, because the conidiophore heads found on the natural substrate were not as complex as those in culture. This difference was attributed to prolonged growth of the fungus in culture (Kirschner 2016). Unfortunately, our collection did not sporulate in culture. The conidiophore character “branched head sometimes or often breaks off from the stipe and with a percurrent proliferation” was easily observed in all the collections of Th. papuana. This character is an important morphological marker in Thysanorea at the genus level and morphologically significant to distinguish it from other related hyphomycetous taxa, such as Dactylaria.
The genus Minimelanolocus has a strong resemblance to Thysanorea in having mononematous and cylindrical conidiophores, holoblastic and sympodial proliferating conidiogenous cells with denticulate conidiogenous loci, as well as mostly septate, hyaline to pale brown conidia. The only difference between the two genera is the branched conidiophores in Thysanorea and the unbranched conidiophores in Minimelanolocus (Liu et al. 2015; Kirschner 2016; Dong et al. 2018). In this study, as well as in Dong et al.’s 2018, the type species, Th. papuana, was added to the phylogenetic tree. Both analyses showed that Thysanorea is closely related with Minimelanolocus and nested within a clade corresponding to Minimelanolocus which is revealed to be paraphyletic (Fig. 1). There is no coding gene reported for any Minimelanolocus species. We sequenced the TEF gene for Th. papuana (GenBank No. 392331 for strain MFLUCC15–0966 and GenBank No. 392330 for strain MFLUCC 17–2315) and Minimelanolocus thailandensis (GenBank No. MK392329). The similarity between MFLUCC15–0966 and MFLUCC 17–2315 is 98%, while that between Th. papuana and M. thailandensis is 92%. More sequence data, particularly protein coding genes, together with the recollection and sequencing of M. navicularis (R.F. Castañeda) R.F. Castañeda, the generic type, will help to clarify phylogenetic relationships between these closely related genera. Once its placement is known, the phylogenetic status of both genera will be further clarified and some generic rearrangement might be needed in the future.
Thysanorea was ecologically widespread from Oceania to Asia occurring on dead woody substrates to submerged wood (Table 2). In Thailand, Th. papuana has been reported not only on submerged wood (Dong et al. 2018; this study) but also on terrestrial baits (as Ramichloridium lignicola, Kodsueb et al. 2016). This study increases our understanding of freshwater hyphomycetes in Eurotiomycetes.
References
Aptroot A, Iperen AV (1998) New ascomycetes and ascomycete records from Papua New Guinea. Nova Hedwigia 67:481–497. https://doi.org/10.1111/j.1756-1051.1998.tb01555.x
Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin HD, Crous PW (2007) Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud Mycol 58:57–93. https://doi.org/10.3114/sim.2007.58.03
Cai L, Guo XY, Hyde KD (2008) Morphological and molecular characterisation of a new anamorphic genus Cheirosporium, from freshwater in China. Persoonia 20:53–58. https://doi.org/10.3767/003158508X314732
Cai L, Kurniawati E, Hyde KD (2010) Morphological and molecular characterization of Mariannaea aquaticola sp. nov. collected from freshwater habitats. Mycol Prog 9:337–343. https://doi.org/10.1007/s11557-009-0641-1
Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu JC, Liu XZ, Stadler M (2014) The sooty moulds. Fungal Divers 66:1–36
D’Souza MA, Bhat DJ (2012) A new species of Pleurophragmium from India. Mycotaxon 119:477–482. https://doi.org/10.5248/119.477
Diederich P, Ertz D, Lawrey JD, Sikaroodi M, Untereiner WA (2013) Molecular data place the hyphomycetous lichenicolous genus Sclerococcum close to Dactylospora (Eurotiomycetes) and S. parmeliae in Cladophialophora (Chaetothyriales). Fungal Divers 58:61–72. https://doi.org/10.1007/s13225-012-0179-4
Dong W, Hyde KD, Bhat DJ, Zhang H (2018) Introducing Aculeata aquatica gen. et sp. nov., Minimelanolocus thailandensis sp. nov. and Thysanorea aquatica sp. nov. (Herpotrichiellaceae, Chaetothyriales) from freshwater in northern Thailand. Mycol Prog 17:617–629. https://doi.org/10.1007/s11557-018-1389-2
Ellis MB (1971) More dematiaceous hyphomycetes. Mycologia 83:69–439
Fiuza PO, Conceição LB, Marques MFO, Castañeda-Ruiz RF (2017) Dictyotrichocladium aquaticum gen. & sp. nov. and Minimelanolocus aquatilis sp. nov. from freshwater in Brazil’s semiarid region. Mycotaxon 132:433–440. https://doi.org/10.5248/132.433
Geiser DM, Gueidan C, Miadlikowska J, Lutzoni F, Kauff F, Hofstetter V, Fraker E, Schoch CL, Tibell L, Untereiner WA (2006) Eurotiomycetes: Eurotiomycetidae and Chaetothyriomycetidae. Mycologia 98:1053–1064. https://doi.org/10.1080/15572536.2006.11832633
Gueidan C, Villaseñor CR, Hoog GSD, Gorbushina AA, Untereiner WA, Lutzoni F (2008) A rock-inhabiting ancestor for mutualistic and pathogen-rich fungal lineages. Stud Mycol 6:111–119
Hughes SJ (1958) Revisiones hyphomycetum aliquot cum appendice de nominibus rejiciendis. Can J Bot 36:727–836
Hyde KD, Fryar S, Tian Q, Bahkali AH, Xu JC (2016a) Lignicolous freshwater fungi along a north/south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol 19:190–200. https://doi.org/10.1016/j.funeco.2015.07.002
Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Abdel-Aziz FA, Abdel-Wahab MA, Banmai S, Chomnunti P, Cui BK, Daranagama DA, Das K, Dayarathne MC, de Silva NL, Dissanayake AJ, Doilom M, Ekanayaka AH, Gibertoni TB, Góes-Neto A, Huang SK, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li WJ, Lin CG, Liu JK, Lu YZ, Luo ZL, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, de Santiago ALMA, Drechsler-Santos ER, Senanayake IC, Tanaka K, Tennakoon TMDS, Thambugala KM, Tian Q, Tibpromma S, Thongbai B, Vizzini A, Wanasinghe DN, Wijayawardene NN, Wu H, Yang J, Zeng XY, Zhang H, Zhang JF, Bulgakov TS, Camporesi E, Bahkli A, Amoozegar AM, Araujo-Neta LS, Ammirati JF, Baghela A, Bhatt RP, Bojantchev S, Buyck B, da Silva GA, de Lima CLF, de Oliveira RJV, de Souza CAF, Dai YC, Dima B, Duong TT, Ercole E, Mafalda-Freire F, Ghosh A, Hashimoto A, Kamolhan S, Kang JC, Karunarathna SC, Kirk PM, Kytövuori I, Lantieri A, Liimatainen K, Liu ZY, Liu XZ, Lücking R, Medardi G, Mortimer PE, TTT N, Promputtha I, KNA R, Reck MA, Lumyong S, Shahzadeh-Fazeli SA, Stadler M, Soudi MR, Su HY, Takahashi T, Tangthirasunun N, Uniyal P, Wang Y, Wen TC, Xu JC, Zhang ZK, Zhao YC, Zhou JZ, Zhu L (2016b) Fungal diversity notes 367–491: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 80:1–270. https://doi.org/10.1007/s13225-016-0366-9
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822. https://doi.org/10.1038/nature05110
Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat DJ, Buyck B, Cai L, Dai YC, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu JK, Luangsa-Ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo JM, Ghobad-Nejhad M, Nilsson H, Pang KL, Pereira OL, Phillips AJL, Raspe O, Rollins AW, Romero AI, Etayo J, Selcuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen TC, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li WJ, Perera RH, Phookamsak R, De Silva NI, Thambugala KM, Tian Q, Wijayawardene NN, Zhao RL, Zhao Q, Kang JC, Promputtha I (2015) The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers 74:3–18. https://doi.org/10.1007/s13225-015-0351-8
Jeewon R, Hyde KD (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7:1669–1677. https://doi.org/10.5943/mycosphere/7/11/4
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kirschner R (2016) Revision of the morphology and biogeography of Thysanorea papuana. Mycosphere 7:820–827. https://doi.org/10.5943/mycosphere/7/6/13
Kodsueb R, Lumyong S, McKenzie EHC, Bahkali AH, Hyde KD (2016) Relationships between terrestrial and freshwater lignicolous fungi. Fungal Ecol 19:155–168. https://doi.org/10.1016/j.funeco.2015.09.005
Kurniawati E, Zhang H, Chukeatirote E, Sulistyowati L, Moslem MA, Hyde KD (2010) Diversity of freshwater ascomycetes in freshwater bodies at Amphoe Mae Chan, Chiang Rai. Cryptogam Mycol 31:323–331
Liu XY, Udayanga D, Luo ZL, Chen LJ, Zhou DQ, Su HY, Hyde KD (2015) Backbone tree for Chaetothyriales with four new species of Minimelanolocus from aquatic habitats. Fungal Biol 119:1046–1062. https://doi.org/10.1016/j.funbio.2015.08.005
Liu JK, Yang J, Maharachchikumbura SSN, McKenzie EHC, Jones EBG, Hyde KD, Liu ZY (2016) Novel chaetosphaeriaceous hyphomycetes from aquatic habitats. Mycol Prog 15:1157–1167. https://doi.org/10.1007/s11557-016-1237-1
Luo ZL, Bao DF, Bhat DJ, Yang J, Chai HM, Li SH, Bahkali AH, Su HY, Hyde KD (2016) Sporoschisma from submerged wood in Yunnan, China. Mycol Prog 15:1145–1155. https://doi.org/10.1007/s11557-016-1236-2
Luo ZL, Hyde KD, Bhat DJ, Jeewon R, Maharachchikumbura SSN, Bao DF, Li WL, Su XJ, Yang XY, Su HY (2018) Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycol Prog 17:511–530. https://doi.org/10.1007/s11557-018-1377-6
Ma J, Ma LG, Zhang YD (2011) Pseudospiropes linderae sp. nov. and notes on Minimelanolocus (both anamorphic Strossmayeria) new to China. Nova Hedwigia 93:465–473. https://doi.org/10.1127/0029-5035/2011/0093-0465
Mercado SA, Figueras MJ, Mena PJ (1996) A new species of Alisidiopsis from Mexico. Mycotaxon 60:443–448
Page RDM (1996) Tree view: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Pratibha J, Prabhugaonkar A (2015) New record of Thysanorea papuana from India. Mycosphere 6:480–485
Rannala B, Yang Z (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol 43:304–311. https://doi.org/10.1007/BF02338839
Réblová M, Untereiner WA, Réblová K (2013) Novel evolutionary lineages revealed in the Chaetothyriales (Fungi) based on multigene phylogenetic analyses and comparison of ITS secondary structure. PLoS One 8:1–28. https://doi.org/10.1371/journal.pone.0063547
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ruiz RFC, Heredia G, Reyes M, Arias RM, Decock C (2001) A revision of the genus Pseudospiropes and some new taxa. Cryptogam Mycol 22:3–18. https://doi.org/10.1016/S0181-1584(01)01057-0
Shearer CA, Descals E, Kohlmeyer B, Kohlmeyer J, Marvanová L, Padgett D, Porter D, Raja HA, Schmit JP, Thorton HA (2007) Fungal biodiversity in aquatic habitats. Biodivers Conserv 16:49–67. https://doi.org/10.1007/s10531-006-9120-z
Silvestro D, Michalak I (2011) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337. https://doi.org/10.1007/s13127-011-0056-0
Su HY, Udayanga D, Luo ZL, Manamgoda DS, Zhao YC, Yang J, Liu XY, Mckenzie EHC, Zhou DQ, Hyde KD (2015) Hyphomycetes from aquatic habitats in Southern China: species of Curvularia (Pleosporaceae) and Phragmocephala (Melannomataceae). Phytotaxa 226:201–216. https://doi.org/10.11646/phytotaxa.226.3.1
Taylor JE, Hyde KD (2003) Microfungi of tropical and temperate palms. Fungal Divers Res Ser 12:1–459
Tian Q, Doilom M, Luo ZL, Chomnunti P, Bhat DJ, Xu JC, Hyde KD (2016) Introducing Melanoctona tectonae gen. et sp. nov. and Minimelanolocus yunnanensis sp. nov. (Herpotrichiellaceae, Chaetothyriales). Cryptogam Mycol 37:477–492. https://doi.org/10.7872/crym/v37.iss4.2016.477
Tsui CKM, Hyde KD, Hodgkiss IJ (2001) Colonization patterns of wood-inhabiting fungi on baits in Hong Kong rivers, with reference to the effects of organic pollution. Antonie van Leeuwenhoek 79:33–38. https://doi.org/10.1023/a:1010210631215
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA froms everal Cryptococcus species. J Bacteriol 172:4238–4246
Wei MJ, Zhang H, Dong W, Boonmee S, Zhang D (2018) Introducing Dictyochaeta aquatica sp. nov. and two new species of Chloridium (Chaetosphaeriaceae, Sordariomycetes) from aquatic habitats. Phytotaxa 362:187–199. https://doi.org/10.11646/phytotaxa.362.2.5
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18:315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Wong MKM, Goh TK, Hodgkiss IJ, Hyde KD, Ranghoo VM, Tsui CKM, Ho WH, Wong WSW, Yuen TK (1998) Role of fungi in freshwater ecosystems. Biodivers Conserv 7:1187–1206. https://doi.org/10.1023/A:1008883716975
Acknowledgments
Gen-Nuo Wang thanks Zong-Long Luo, Yong-Zhong Lu, Shaun Pennycook, Dan-Feng Bao, De-Ping Wei, Nimali de Silva, and Mingkwan Doilom for their valuable suggestions and help.
Funding
This study is primarily supported by the National Natural Science Foundation of China (project ID: NSF 31500017 to Huang Zhang), the Yunnan Young and Middle Aged Academic and Technical Leaders Reserve Talents (2018HB008), and the Graduate Research and International Exchange Program of Kunming University of Science and Technology 2017. Saranyaphat Boonmee would like to thank the National Research Council of Thailand (projects No. 61215320023).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Section Editor: Roland Kirschner
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, GN., Yu, XD., Dong, W. et al. Freshwater hyphomycetes in Eurotiomycetes: a new species of Minimelanolocus and a new collection of Thysanorea papuana (Herpotrichiellaceae). Mycol Progress 18, 511–522 (2019). https://doi.org/10.1007/s11557-019-01473-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-019-01473-7