Abstract
During an investigation of the diversity of aquatic hyphomycetes from southern China, two interesting isolates were collected. These two isolates were cultured and sequenced, and a BLAST search of their LSU sequences against data in GenBank revealed that the closest related taxa were in the genus Microthyrium. Phylogenetic analyses, based on the combined sequence data from the internal transcribed spacer (ITS) and large nuclear subunit ribosomal DNA (LSU), revealed that our isolates belong to the Microthyriaceae. Combined morphological characters allowed us to describe our isolates as two new genera and species in Microthyriaceae, named as: Keqinzhangia aquatica and Pseudocoronospora hainanense. The full descriptions, illustrations, and a phylogenetic tree showing the position of the two new genera were provided in this paper.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microthyriales was introduced by Arnaud in 1918, with the type family Microthyriaceae. Originally, Microthyriales included two families, Microthyriaceae and Micropeltidaceae, based on their flattened ascomata with a poorly developed base. However, Hongsanan and Hyde (2017) excluded Micropeltidaceae from Microthyriales based on their phylogenetic analyses and morphological characteristics. Moreover, their phylogenetic analyses showed that species of Microthyriales cluster together as a distinct clade within Dothideomycetes with high support. Currently, Microthyriales only contains a single family Microthyriaceae (Hongsanan and Hyde 2017; Wijayawardene et al. 2018, 2020).
Saccardo (1883) established the family Microthyriaceae, with the sexual genus Microthyrium Desm. as the type genus. Theissen (1913) included Microthyriaceae in the order Hemisphaeriales. Subsequently, Arnaud (1918) established a new order Microthyriales to accommodate Microthyriaceae and Microthyriopsidaceae. After that, the family has experienced a complicated taxonomic history, and various genera were included, such as foliar epiphytes or saprobes. Wu et al. (2011) reappraised the Microthyriaceae based on examinations of generic types and provided sequence data of several species. They finally accepted seven sexual genera in the Microthyriaceae. In 2017, Wijayawardene et al. (2018) merged both asexual and sexual genera in the outline of Ascomycota and totally accepted nine genera in the family, including eight sexual genera and one asexual genus. In a recent study, Hongsanan et al. (2020) accepted eleven genera in this family based on morphology and phylogeny, including eight sexual genera and three asexual genera. At the same time, they added the definition of an asexual morph on family level.
China has an enormous fungal diversity, and the southwestern region in this country was assessed as one of the world’s 34 biodiversity hotspots (Myers et al. 2000). In recent years, we have been investigating the fungal diversity in China, including soils, submerged leaves, and aquatic plants, and described many new taxa (Qiao et al. 2017, 2018a, b; 2019; 2020; Zheng et al. 2019, 2020a, b, 2021a, b, c). During our ongoing studies of freshwater hyphomycetes in the Yunnan and Hainan provinces, two interesting fungi were collected from submerged leaves of unidentified dicotyledonous plants. These two isolates were cultured and sequenced, and a BLAST search of their LSU sequences against data in GenBank revealed that the closest related taxa were in the genus Microthyrium. To further confirm the position of our isolates, phylogenetic analyses with related taxa within Microthyriaceae were carried out based on complete sequences of internal transcribed spacer (ITS) and partial sequences of the nuclear large subunit ribosomal DNA (LSU) genes. Combining morphological characters, we finally described our isolates as two new genera and species in Microthyriaceae, named as: Keqinzhangia aquatica gen. et sp. nov. and Pseudocoronospora hainanense gen. et sp. nov.
Materials and methods
Isolation and morphological study of strains
Submerged dicotyledonous leaves were collected from Yunnan and Hainan Provinces. Samples were preserved in zip-lock plastic bags, labeled, and transported to the laboratory. The decomposed leaves were cut into several 2–4 × 2–4 cm sized fragments in the laboratory and then were incubated on corn meal agar (CMA, 20 g cornmeal, 18 g agar, 40 mg streptomycin, 30 mg ampicillin, 1000 ml distilled water) medium for 10 days at room temperature. Single conidia were isolated with a sterilized needle under an Olympus BX51 microscope and cultivated on. Morphological observations were made from CMA after incubation at 25 °C for 1 week, and microscopic photographs were taken on an Olympus BX51 microscope under differential interference contrast model and captured with an Olympus DP 10 digital camera using Olympus DP controller (V.3,1,1208) software. Measurements were based on 30 random conidia and 10 conidiophores.
Pure cultures were deposited in the Herbarium of the Laboratory for Conservation and Utilization of Bio-Resources, Yunnan University, Kunming, Yunnan, P.R. China (YMF, formerly Key Laboratory of Industrial Microbiology and Fermentation Technology of Yunnan), the China Center for type Culture Collection (CCTCC), and the China General Microbiological Culture Collection Center (CGMCC).
DNA extraction, PCR amplification, and sequencing
Pure cultures were grown on potato dextrose agar (PDA, 200 g potato, 20 g dextrose, 18 g agar, 1000 ml distilled water) medium for 7 days at 25 °C. Actively growing mycelium was scraped off from the surface of the culture and transferred to 2 ml Eppendorf micro-centrifuge tubes. Total genomic DNA was extracted according to the procedures in Turner et al. (1997). Primers used for PCR amplification and sequencing of the nuclear large subunits ribosomal DNA (LSU) and the internal transcribed spacer (ITS) were LROR/LR7 (White et al. 1990) and ITS1/ITS4 (Vilgalys and Hester 1990), respectively. Each 25 μL PCR reaction volume consisted of 12.5 μL T5 Super PCR Mix (containing Taq polymerase, dNTP and Mg2+, Beijing TsingKe Biotech Co., Ltd., Beijing, China), 1 μL of forward primer (10 μM), 1 μL of reverse primer (10 μM), 1 μL DNA template, 5 μL of PCR buffer, and 4.5 μL sterile water. PCR reactions were run in an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) following the PCR thermal cycle programs described by Qiao et al. (2020). PCR products were purified by using the PCR product purification kit (Biocolor BioScience and Technology Co., Shanghai, China), and forward and reverse sequenced on an ABI 3730 XL DNA sequencer (Applied Biosystems, Foster City, CA, USA) with the same primers, using a Thermo Sequenase Kit as described by Kindermann et al. (1998). These sequences were deposited in the GenBank database at the National Center for Biotechnology Information (NCBI) and the accession numbers are listed in Table 1.
Sequence alignment and phylogenetic analyses
Preliminary BLAST searches with the LSU sequences of our isolates against the GenBank nucleotide database determined the closely related species, it showed that their closest related taxon is in the genus Microthyrium. Based on this information, related sequences at the two marker loci of ITS and LSU, which include 13 representatives belonging to Microthyriaceae, two representatives belonging to Natipusillales, two representatives belonging to Phaeotrichales, three representatives belonging to Venturiales, and three representatives belonging to Zeloasperisporiales, were downloaded according to recent studies (Crous et al. 2019; Gonzalez et al. 2020; Hongsanan et al. 2020). Kirschsteiniothelia lignicola Boonmee & K.D. Hyde was used as the outgroup.
The sequences, together with the newly generated sequences, were manually aligned with ClustalX 1.83 (Thompson et al. 1997). The resulting alignments were subsequently checked and refined using BioEdit version v. 7.0.4.1 (Hall 1999). The two alignments were combined with BioEdit and then converted to a NEXUS file using the programme MEGA6 (Tamura et al. 2013). The resulting combined sequence matrix contained 1,215 nucleotide positions from two loci (855 from LSU, 360 from ITS), and it was uploaded to TreeBASE (www.treebase.org; accession number: S28478).
Bayesian inference (BI) and maximum likelihood (ML) were used in this study for phylogenetic analyses. BI analysis was conducted with MrBayes v3.2.2 (Ronquist et al. 2012) with NEXUS files. The Akaike information criterion (AIC) implemented in jModelTest 2.0 (Posada 2008) was used to select the best fit models after likelihood score calculations were done. GTR + F + I + G4 was estimated as the best-fit model under the output strategy of AIC. Me-tropolis-coupled Markov chain Monte Carlo (MCMCMC) searches were run for 1,000,000 generations sampling every 500th generation. Two independent analyses with four chains each (one cold and three heated) were run until stationary distribution was achieved. The initial 25% of the generations of CMC sampling were excluded as burn-in. The refinement of the phylogenetic tree was used for estimating Bayesian inference posterior probability (BIPP) values. ML analysis was computed by RAxML (Stamatakis 2006) with the PHY files generated with ClustalX 1.83, using the GTR-GAMMA model. Maximum likelihood bootstrap proportions (MLBP) were computed with 1000 replicates. Trees were visualized in FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/Figtree/, July 2021). Bayesian inference posterior probabilities (BIPP) ≥ 0.95 and maximum likelihood bootstrap proportions (MLBP) ≥ 75% are indicated at nodes.
Results
Phylogenetic analyses
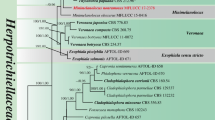
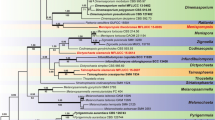
BLAST analyses with LSU sequences revealed Microthyrium as closely related taxon to our isolates but with relatively low percentage of identities. The combined analysis of the two loci (ITS and LSU), which was analyzed by BI and ML approaches, confirmed the status of our isolates in the family Microthyriaceae. In this tree, YMF 1.04626 and YMF 1.04517 grouped into the Microthyriaceae with good support. YMF 1.04626 was clustered together with the sexual genus Microthyrium with good support (MLBP/BIPP = 92%/1.0), and the clade was close to the asexual genus Neoanungitea Crous. YMF 1.04517 formed an isolated clade, close to Hamatispora L.T.H. Yen, K. Yamag. & K. Ando, Neoanungitea, and Microthyrium with good support (MLBP/BIPP = 80%/0.98). Combined with morphological differences, we described YMF 1.04626 and YMF 1.04517 as two new asexual genera and species in Microthyriaceae, named as Keqinzhangia aquatica and Pseudocoronospora hainanense (Fig. 1).
Phylogenetic tree generated by maximum likelihood analyses using combined sequences of the nuclear large subunit (LSU) and the internal transcribed spacers (ITS) loci. Bootstrap support values for maximum likelihood (ML) over 75% and Bayesian posterior probabilities greater than 0.95 are indicated above or below the nodes as MLBP/BIPP. Kirschsteiniothelia lignicola strain MFLUCC10-0036 was used as the outgroup. The strain numbers are noted after the species names with ex-type strains indicated by T. Novel species are indicated in bold
Taxonomy
Keqinzhangia Z.F.Yu, M.Qiao & R.F. Castañeda, gen. nov.
Etymology: Named in honor of Prof. Keqing Zhang of Yunnan University for his contribution on biological control of pathogenic nematodes.
MycoBank number: MB 840,430.
Asexual morph hyphomycetous. Vegetative hyphae cylindrical, branched, microguttulate, septate, hyaline, smooth-walled. Fertile hyphae cylindrical-obclavate, inflated and subulate at the tip, macroguttulate, dark septate, hyaline, smooth-walled. Conidiophores prostrate, not differentiated. Conidiogenous cells holothallic, narrow cylindrical to cylindrical, discrete, indeterminate, forming conidia by random thallic-arthric conidial ontogeny. Conidial secession schizolytic. Conidia thallic-arthric, solitary, polymorphic, cylindrical, cylindrical-obclavate, obclavate, bacilliform, fusiform, sub-oblecythiform or cuneiform, unicellular to septate, hyaline. Chlamydospores globose, terminal, solitary or short catenulate, subhyaline. Sexual state: Unknown.
Type species: Keqinzhangia aquatica Z.F. Yu, M. Qiao & R.F. Castañeda.
Keqinzhangia aquatica Z.F. Yu, M. Qiao & R.F. Castañeda, sp. nov. (Figs. 2, 3, 4).
Etymology: Epithet refers to the collection from stream water.
MycoBank number: MB 840,432.
Asexual morph hyphomycetous. Colonies flat, growing slowly on CMA, attaining about 2.4 cm diam. after 20 days at 25 °C. Pale mouse grey, reverse mouse grey. Mycelium mostly immersed, composed of cylindrical, branched, densely micro-guttulate, septate, subhyaline to hyaline vegetative hyphae and cylindrical-obclavate, inflated and subulate at the tip, macroguttulate, dark septate, hyaline, smooth-walled fertile hyphae. Conidiophores prostrate, undifferentiated. Conidiogenous cells holothallic, narrowly cylindrical, frequently undifferentiated, hyaline, forming conidia by random thallic-arthric disarticulation. Conidia thallic-arthric, solitary, polymorphic, cylindrical-obclavate, long obclavate, cylindrical, bacilliform, fusiform, narrow doliiform, subdolabriform, suboblecythiform or cuneiform, truncate at the ends or truncate at the base and obtuse or rounded at the apex, 0–6(-7)-septate, slightly or strongly constricted at the dark septa, sinuate, macroguttulate, smooth, hyaline, 12–76.5 × 3–6.2 μm, arising after random disarticulation of fertile hyphae at the darker septa. Clamydospores solitary or catenate, broad globose, subglobose to ellipsoidal, terminal, slightly or densely guttulate, smooth, subhyaline, 8–12.6 × 4.1–5.4 μm. Sexual state: Unknown.
Holotype: YMF 1.04262, isolated from leaves of an unidentified dicotyledonous plant submerged in a stream, E’mei National Conservation Area, Sichuan Province, China, 29°35′1′′N, 103°17′3′′E, ca. 1750 m elev., Jun 2014, Zefen Yu, permanently preserved in a metabolically inactive state (deep freezing) in the Conservation and Utilization of Bio-Resources in Yunnan. Ex-type culture CCTCC AF 2,021,070 = CGMCC 3.16100.
Notes: In Keqinzhangia aquatica, the fertile hyphae are located at the margin of the colony arise laterally from vegetative hyphae forming aerial mycelium with narrow cylindrical, cylindrical, long cylindrical-obclavate, obclavate, inflated or globose, subulate cellular structures, that include the tip growth. The thallic-arthric conidia are formed by random fission at the darker septa of preexisting cells of the fertile hyphae in a similar holothallic mode described by Cole (1986) and Seifert et al. (2011).
Pseudocoronospora Z.F.Yu, M. Qiao & R.F. Castañeda, gen. nov.
Etymology: Name refers to the morphological similarity to the genus coronospora.
MycoBank number: MB 840,431.
Asexual morph hyphomycetous. Conidiophores macronematous, mononematous, erect, straight, septate, unbranched, smooth, brown. Conidiogenous cells polyblastic, sympodial extended, integrated, terminal, indeterminate, denticulate. Conidial secession rhexolytic. Conidia solitary, acropleurogenous, obclavate, crowned, with mammiform protuberances arranged near the apex; septate, smooth or verruculose, hyaline, fringed at the base. Sexual state: Unknown.
Type species: Pseudocoronospora hainanense Z.F. Yu, M. Qiao & R.F. Castañeda.
Notes: The genus Coronospora was established by Ellis (1971) with C. dendrocalami M.B. Ellis as the type species, in which after the conidiogenous events the cicatrized loci are produced following sympodial extensions of the polyblastic conidiogenous cells disposed in geniculate conidiophores and the conidia are liberated via schizolytic conidial secession (Seifert et al. 2011; Zhang and Zhang 2004; Ellis 1971), but in Pseudocoronospora hainanense the conidiogenous loci are tiny or conspicuous denticles and the conidial basal cells are fringed after the rhexolytic conidial secession. Matsushima (2001) observed the Coronospora morph in the culture of Ascoronospora Matsush., so he thought that Coronospora is the asexual state of Ascoronospora. Then Kirk et al. (2008) and Wijayawardene et al. (2018) accepted the link between two genera.
Pseudocoronospora hainanense Z.F. Yu, M. Qiao & R.F. Castañeda, sp. nov. (Figs. 5 and 6).
Etymology: Epithet refers to the region Hainan where the type strain was collected.
MycoBank number: MB 840,433.
Asexual morph hyphomycetous. Colonies on CMA attaining 3 cm diam. after 20 days at 25 °C, effuse, white to pale flesh, reverse buff. Hyphae thin-walled, septate, hyaline, smooth. Conidiophores macronematous, mononematous, straight or slightly flexuous, somewhat geniculate toward the apex, septate, unbranched, smooth, mid brown or pale brown below, pale brown to subhyaline towards the apex, 16.5–49 × 3.5–5.0 μm. Conidiogenous cells polyblastic, sympodial extended, integrated, terminal, sometimes intercalary, indeterminate, pale brown to subhyaline, denticulate, denticle conspicuous, narrowly cylindrical. Conidial secession rhexolytic. Conidia solitary, acropleurogenous, obclavate, crowned with 2–3 broadly mammiform protuberances, radially arranged near the rounded to obtuse apex; 2-septate, smooth or slightly verruculose at the basal and central cells, hyaline, 27.2–33 × 3.7–8.0 μm, with a minute basal frill. Sexual state: Unknown.
Holotype: YMF 1.04517, isolated from leaves of an unidentified dicotyledonous plant submerged in a stream, Diaoluoshan National Forest Park, Hainan Province, China, 18°42′11′′N, 109°53′16′′E, ca. 1124 m elev., April 2014, Zefen Yu, permanently preserved in a metabolically inactive state (deep freezing) in the Conservation and Utilization of Bio-Resources in Yunnan. Ex-type culture CCTCC AF 2,021,129 = CGMCC 3.18823.
Discussion
In recent years, more and more molecular data of species in Microthyriaceae has become available. Hongsanan et al. (2020) accepted 11 genera, which include three asexual genera Hamatispora, Neoanungitea, and Pseudopenidiella Crous & Koukol, in Microthyriaceae based on morphological characteristics and sequence analyses of the ITS and LSU barcodes. In this study, our phylogenetic analyses determined two isolates to belong to the Microthyriaceae. Combined with morphological characteristics, we finally described them as two new asexual genera and species in Microthyriaceae, named as Keqinzhangia aquatica and Pseudocoronospora hainanense.
The new genus Keqinzhangia is phylogenetically close to the sexual genus Microthyrium and the asexual genus Neoanungitea. Although we observed cultures for a long time, we did not see any sexual reproductive structures in K. aquatica. Besides, their LSU sequence similarity is relatively low (90%). Therefore, we cannot determine the connection between them. Although Neoanungitea is an asexual genus, Keqinzhangia is obviously different from Neoanungitea in conidiogenesis (holothallic vs. holoblastic) and the shape of conidia (cylindrical-obclavate, bacilliform, fusiform vs. fusoid-ellipsoid) (Crous et al. 2019).
Our other new genus, Pseudocoronospora, is phylogenetically close to the asexual genus Hamatispora and Neoanungitea. Hamatispora is a hyphomycetous genus with staurospores that are question-mark-shaped or hook-shaped with 3 arms developing from each cell on the helicoid part (Yen et al. 2018). Therefore, Pseudocoronospora species is easily distinguished from Hamatispora and Neoanungitea by morphology.
Mycrothyriales is a poorly known order. Previously accepted species in this order were based mostly on morphological characters; little molecular data were available. For the past few years, new molecular data are available (Crous et al. 2016, 2017, 2019; Hongsanan et al. 2020; Wu et al. 2011, 2014). A recent study showed that species of Microthyriales cluster together as a distinct clade within Dothideomycetes with high support based on sequence analyses of LSU and ITS (Hongsanan et al. 2020). This study shows the importance of obtaining pure cultures and gene sequences in order to identify the origins and phylogenetic positions of fungal species.
Availability of data and material
These new generated sequences were uploaded to the GenBank database at the National Center for Biotechnology Information (NCBI), and are available.
Code availability
Not applicable.
References
Abarca GH, Castañeda-Ruiz RF, Mota RMA, Hernandez CIB, Gomez S et al (2011) A new species of Heliocephala from Mexico with an assessment of the systematic position of the anamorph genera Heliocephala and Holubovaniella. Mycologia 103:631–640. https://doi.org/10.3852/10-230
Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KTW et al (2015) Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 75:27–274. https://doi.org/10.1007/s13225-015-0346-5
Arnaud G (1918) Lés Asterinées. Ann Éc Natl Agric Montp 2(16):1–288
Boonmee S, Ko Ko TW, Chukeatirote E, Hyde KD, Chen H et al. (2012) Two new Kirschsteiniothelia species with Dendryphiopsis anamorphs cluster in Kirschsteiniotheliaceae fam. nov. Mycologia 104(3):698–714. https://doi.org/10.3852/11-089
Cole GT (1986) Models of cell-differentiation in conidia fungi. Microbiol Rev 50(2):95–132
Crous PW, Wingfield MJ, Richardson DM, Le Roux JJ, Strasberg D et al (2016) Fungal planet description sheets: 400–468. Persoonia 36:316–458. https://doi.org/10.3767/003158516X692185
Crous PW, Wingfield MJ, Burgess TI, Carnegie AJ, Hardy GESJ et al (2017) Fungal Planet description sheets: 625–715. Persoonia 39:270–467. https://doi.org/10.3767/persoonia.2017.39.11
Crous PW, Luangsa-Ard JJ, Wingfield MJ, Carnegie AJ, Hernández-Restrepo M et al (2018) Fungal Planet description sheets: 785–867. Persoonia 41:238–417. https://doi.org/10.3767/persoonia.2018.41.12
Crous PW, Wingfield MJ, Lombard L, Roets F, Swart WJ et al (2019) Fungal Planet description sheets: 951–1041. Persoonia 43:223–425. https://doi.org/10.3767/persoonia.2019.43.06
Ellis MB (1971) Dematiaceous Hyphmycetes x. Mycol Paper 125:1–30
Ferrer A, Miller AN, Shearer CA (2011) Minutisphaera and Natipusilla: two new genera of freshwater Dothideomycetes. Mycologia 103:411–423. https://doi.org/10.3852/10-177
Gonzalez II, Garcia D, Guarro J, Gene J (2020) Heliocephala variabilis and Pseudopenidiella vietnamensis: two new Hyphomycetous species in the Microthyriaceae (Dothideomycetes) from Vietnam. Microorganisms 8(4):478. https://doi.org/10.3390/microorganisms8040478
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41(41):95–98. https://doi.org/10.1021/bk-1999-0734.ch008
Hongsanan S, Hyde KD (2017) Phylogenetic placement of Micropeltidaceae. Mycosphere 8(10):1930–1942. https://doi.org/10.5943/mycosphere/8/10/15
Hongsanan S, Chomnunti P, Crous PW, Chukeatirote E, Hyde KD (2014) Introducing Chaetothyriothecium, a new genus of Microthyriales. Phytotaxa 161:157–164
Hongsanan S, Hyde KD, Phookamsak R, Wanasinghe DN, McKenzie EHC et al (2020) Refined families of Dothideomycetes: orders and families incertae sedis in Dothideomycetes. Fungal Diversity 105(1):17–318. https://doi.org/10.1007/s13225-020-00462-6
Hongsanan S, Tian Q, Bahkali AH, Yang J-B, McKenzie EHC et al. (2015) Zeloasperisporiales ord. nov., and two new species of Zeloasperisporium. Cryptogamie Mycologie 36(3):301–317. https://doi.org/10.7872/crym/v36.iss3.2015.301
Kindermann J, El-Ayouti Y, Samuels GJ, Kubicek CP (1998) Phylogeny of the genus Trichoderma based on sequence analysis of the internal transcribed spacer region 1 of the rDNA cluster. Fungal Genet Biol 24(3):298–309. https://doi.org/10.1006/fgbi.1998.1049
Kirk P, Cannon P, Minter D, Stalpers J (2008) Dictionary of the fungi, 10th edn. CAB International, Wallingford, UK
Lumbsch HT, Lindemuth R, Schmitt I (2000) Evolution of filamentous Ascomycetes inferred from LSU rDNA sequence data. Plant Biol 2:525–529. https://doi.org/10.1055/s-2000-7472
Matsushima T (2001) Matsushima Mycological Memoirs No. 10. Matsushima fungus collection, Published by author, Kobe, Japan.
Myers N, Mittermeier RA, Mittermeier CG, Fonseca G, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403(6772):853–858. https://doi.org/10.1038/35002501
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25(7):1253–1256. https://doi.org/10.1093/molbev/msn083
Qiao M, Zheng H, Lv R, Yu Z (2020) Neodactylariales, Neodactylariaceae (Dothideomycetes, Ascomycota): new order and family, with a new species from China. Mycokeys 73(6):69–85. https://doi.org/10.3897/mycokeys.73.54054
Qiao M, Huang Y, Deng C, Yu ZF (2017) Tripospermum sinense sp. nov. from China. Mycotaxon 132(3):513–517. https://doi.org/10.5248/132.513
Qiao M, Guo JS, Tian WG, Yu ZF (2018a) Ellisembia hainanensis sp. nov. from Hainan, China. Mycotaxon 133(1):97–10. https://doi.org/10.5248/133.97
Qiao M, Du X, Bian ZH, Peng J, Yu ZF (2018b) Ellisembia pseudokaradkensis sp. nov. from Hainan, China. Mycotaxon 132(4):813–817. https://doi.org/10.5248/132.813
Qiao M, Zheng H, Zhang Z, Yu ZF (2019) Seychellomyces sinensis sp. nov. from China. Mycotaxon 134(2):391–398. https://doi.org/10.5248/134.391
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A et al. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. System Biol 61(3):539–542. https://doi.org/10.1093/sysbio/sys029
Saccardo PA (1883) Sylloge Pyrenomycetum, vol II. Sylloge Fungorum 2:1–815
Samerpitak K, Van der Linde E, Choi HJ, Gerrits van den Ende AHG, Machouart M et al (2014) Taxonomy of Ochroconis, genus including opportunistic pathogens on humans and animals. Fungal Diversity 65:89–126. https://doi.org/10.1007/s13225-013-0253-6
Schoch CL, Crous PW, Groenewald JZ, Boehm EW, Burgess TI et al (2009) A class-wide phylogenetic assessment of Dothideomycetes. Stud Mycol 64:1–15. https://doi.org/10.3114/sim.2009.64.01
Seifert K, Morgan-Jones G, Gams W, Kendrick B (2011) The genera of hyphomycetes. CBS Biodiversity Series 9:997
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690. https://doi.org/10.1093/bioinformatics/btl446
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30(12):2725–2729.
Theissen F (1913) Hemisphaeriales. Annales Mycologici 11(5):425–467
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882. https://doi.org/10.1093/nar/25.24.4876
Turner D, Kovacs W, Kuhls K, Lieckfeldt E, Peter B et al (1997) Biogeography and phenotypic variation in Trichoderma sect Longibrachiatum and associated Hypocrea species. Mycol Res 101:449–459. https://doi.org/10.1017/S0953756296002845
Vilgalys R, Hester M (1990) Rapid Genetic Identification and mapping of enzymatically amplified ribosomal DNA from Several Cryptococcus Species. J Bacteriol 172(8):4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
White T, Bruns T, Lee S, Taylor F, White T et al (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: a guide to methods and applications, vol 18. Academic Press, New York, pp 315–322.
Wijayawardene NN, Hyde KD, Lumbsch HT, Jian KL, Maharachchikumbura SSN et al (2018) Outline of Ascomycota: 2017. Fungal Diversity 88(2):167–263. https://doi.org/10.1007/s13225-018-0394-8
Wijayawardene N, Hyde KD, Al-Ani L, Tedersoo L, Haelewaters D et al (2020) Outline of fungi and fungi-like taxa. Mycosphere 11(1):1160–1456. https://doi.org/10.5943/mycosphere/11/1/8
Wu HX, Schoch CL, Boonmee S, Bahkali AH, Chomnunti P et al (2011) A reappraisal of Microthyriaceae. Fungal Diversity 51(1):189–248. https://doi.org/10.1007/s13225-011-0143-8
Wu HX, Li YM, Ariyawansa HA, Li WJ, Hyde KD (2014). A new species of Microthyrium from Yunnan, China. Phytotaxa 176(1):213–218. https://doi.org/10.11646/phytotaxa.176.1.21
Yen LTH, Yamaguchi K, Tsurumi Y, Hop DV, Ando K (2018) Hamatispora, a new genus of aquatic fungi in Microthyriales isolated from fallen leaves in Vietnam. Mycoscience 59(6):467–472. https://doi.org/10.1016/j.myc.2018.04.004
Zhang M, Zhang TY (2004) A new species of Coronospora from China. Mycosystema 23:331–332. https://doi.org/10.3969/j.issn.1672-6472.2004.03.004
Zheng H, Wan Y, Li J, Castañeda-Ruiz RF, Yu ZF (2020a) Phialolunulospora vermispora (Chaetosphaeriaceae, Sordariomycetes), a novel asexual genus and species from freshwater in southern China. Mycokeys 76:17–30. https://doi.org/10.3897/mycokeys.76.57410
Zheng H, Yu Z, Xu J, Castañeda-Ruiz RF, Qiao M (2020b) Ramichloridium endophyticum sp. nov., a novel species of endophytic fungus from Potamogeton pectinatus. Int J System Evolut Microbiol 70(5). https://doi.org/10.1099/ijsem.0.004190
Zheng H, Zhang Z, Liu DZ, Yu ZF (2019) Memnoniella sinensis sp. nov., a new species from China and a key to species of the genus. Int J System Evolut Microbiol 69(10). https://doi.org/10.1099/ijsem.0.003605
Zheng H, Li J, Gou JS, Qiao M, Yu ZF (2021a) Anacraspedodidymum submersum sp. nov. (Chaetosphaeriaceae, Chaetosphaeriales), a new species of freshwater hyphomycetes from southwest China. Int J System Evolut Microbiol 71(2). https://doi.org/10.1099/ijsem.0.004650
Zheng H, Qiao M, Lv Y, Du X, Zhang KQ et al (2021b) New species of Trichoderma isolated as endophytes and saprobes from Southwest China. J Fungi 7(6):467. https://doi.org/10.3390/jof7060467
Zheng H, Qiao M, Xu JP, Yu ZF (2021c) Culture-based and culture-independent assessments of endophytic fungal diversity in aquatic plants in Southwest China. Front Fungal Biol 2:27. https://doi.org/10.3389/ffunb.2021.692549
Acknowledgements
We thank Linlin Fang for her work on this manuscript; we are also grateful for the support of Yunnan University Research and Innovation Fund for Postgraduates (2021Y294).
Funding
This work was financed by the National Natural Science Foundation Program of PR China (31770026, 31760012).
Author information
Authors and Affiliations
Contributions
ZY conceived and designed the study. HZ and MQ wrote the manuscript. JG and JP conducted the experiments. RFC contributed actively in the identification and the taxonomy of the fungal strains.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare to have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zheng, H., Qiao, M., Guo, J. et al. Keqinzhangia aquatica gen. et sp. nov. and Pseudocoronospora hainanense gen. et sp. nov., isolated from freshwater in southern China. Antonie van Leeuwenhoek 115, 203–213 (2022). https://doi.org/10.1007/s10482-021-01688-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-021-01688-3