Abstract
In most plants, the contributions of pollen and seed flow to their genetic structures are generally difficult to disentangle. For typical wind-pollinated and wind-dispersed species Engelhardia roxburghiana in a 20-ha natural forest plot in lower subtropic China, because the prevailing wind directions change during its pollen release and seed dispersal seasons, we could compare its genetic structures in different directions, which could result primarily from pollen or seed flow. Furthermore, because the plot has undergone from an open to a closed canopy stage historically, we also examined forest canopy effects on gene flow in different generations and different directions. Using 522 E. roxburghiana individuals mapped in the plot, our results revealed that greater pollen flow led to biased gene flow in the pollen dispersal-predominant direction (pollen direction), while greater seed flow generated less spatial genetic structure in the seed dispersal-predominant direction (seed direction). The results predicted from generalized additive models indicated that canopy closure enhanced resistance to gene flow from the old generation to the new generation. Analyses by landscape genetic models for the new generation revealed that gene flow associated with pollen direction was more strongly affected by canopy than with seed direction. Our study is new by proposing an alternative way to separate effects of the pollen and seed flow on spatial variation patterns in E. roxburghiana. To our knowledge, our study is also the first attempt to use landscape genetic models to represent canopy effects for different dispersal vectors in spatial scales only up to a few hundred meters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For plant species that are wind-pollinated and/or that have wind-dispersed seeds, wind is the main effect on their local- and broad-scale genetic structures (Latta and Mitton 1999; Heuertz et al. 2003; Born et al. 2012; Millerón et al. 2012). On a local scale, it is related to the mating success of individuals (Burczyk et al. 2004; Bacles et al. 2005; Robledo-Arnuncio and Gil 2005; Albaladejo et al. 2012; Torimaru et al. 2012), population recruitment patterns (Grivet et al. 2009; Steinitz et al. 2011), and metapopulation dynamics (Bohrer et al. 2005). On a broad scale, it is related to connectivity of individuals in fragmented habitats (Sork and Smouse 2006), speciation (Devey et al. 2009), species conservation (Fatemi and Gross 2009; Saro et al. 2014), and safety of transgenic species (Chandler and Dunwell 2008). In general, for these species, their pollen or seed flow will reflect dominant wind conditions during their pollen or seed dispersal seasons (Latta and Mitton 1999; Steinitz et al. 2011; Born et al. 2012). However, such patterns may be undetectable due to particular features, such as complex landscapes, dense forests, and man-made buildings (Bacles et al. 2005; Sork and Smouse 2006; Austerlitz et al. 2007; Millerón et al. 2012; Torimaru et al. 2012; Viner and Arritt 2012; Sant’Anna et al. 2013; Saro et al. 2014) protruding along the trajectories of the predominant wind, acting as physical barriers to gene flow. In addition, other factors may also cause unclear wind directionality patterns, such as daily or seasonal variations in wind directions, lack of flowering individuals along wind directions, random dispersal of pollen or seeds that are not wind dispersed, and environmental selection (Burczyk and Prat 1997; Dutech et al. 2005; Robledo-Arnuncio and Gil 2005; Albaladejo et al. 2012; Wang et al. 2014). Thus, the spatial genetic structure (SGS) of the wind dispersal species might differ among sites and environmental conditions.
However, the SGS studies at the local scale for many plants including wind dispersal ones have not included local condition elements. They mainly address the local SGSs by spatial autocorrelation methods (Vekemans and Hardy 2004), and the patterns of gene flow within a population are predicted by the shape of the correlogram. Nevertheless, the autocorrelation analyses can detect the isolation by distance processes, but they cannot identify local environmental variation effects during dispersal events on SGS. If local conditions are not uniform and disrupt pollen or seed movements, they cannot provide detailed information about specific factors shaping the effective dispersal, resulting in incomplete interpretations of dispersal patterns.
To overcome this shortcoming, landscape genetic approaches have been used frequently and confirmed to be efficient for identifying the specific environmental elements causing genetic discontinuity (Holderegger et al. 2010; Storfer et al. 2010). However, the majority of landscape genetic studies have dealt with animals and have been conducted on a large spatial scale, and landscape genetic analyses are not fully integrated into local gene flow pattern analyses in plants (but see Rhodes et al. 2014). One of the possible reasons for lack of such integration is that at a small spatial scale, the study site is often assumed to be environmentally homogenous for studied plant species. Landscape genetic approaches require that the study site be systematically divided into numbers of equally sized subareas (cells) where each is then assigned a value for the environmental feature it represents; if the environment is assumed to be homogenous, all cells in the study site will be given the same cost values, and no effective spatial cost map could be generated.
Indeed, heterogeneous environments, even at a small spatial scale, are common in plant populations (Fowler 1988), resulting in some areas that are uninhabitable or unreachable and subsequently leading to discontinuity in gene flow. Even for a generally assumed homogenous environment, plants per se, such as their canopies, are not always uniform in their community. By dividing a 1-ha study plot into 400 5 × 5-m cells, Ueno et al. (2006) defined three types of canopy conditions (with gap, closed canopy, and mixed gap and canopy) in the plot. They then investigated the genetic structures of Camellia japonica saplings and found that gene flow was more extensive in gap areas than in the other areas. This study indicates that the grid-based landscape distance/resistance framework should be valid for local SGS analysis at a small spatial scale if site-specific characteristics are identified properly. Rhodes et al. (2014) used such landscape genetic approach directly to study fine-scale SGS of Oenothera harringtonii, a herb endemic to grasslands of Colorado, USA, in an ∼1100 × 600-m plot and found that local topographic heterogeneity was an important determinant of SGS in the species.
Therefore, we initiated this work to combine local specific environmental conditions and landscape genetic tools to study the influence of wind on the genetic structures of Engelhardia roxburghiana (Juglandaceae), a wind-pollinated and wind-dispersed pioneer plant species. The study took place in a large 20-ha forest plot in Dinghushan (DHS, Dinghu Mountain), a part of lower subtropical China. Two site-specific features in this plot make our study interesting.
One is that in the plot during the E. roxburghiana flowering seasons (May–July), the prevailing winds are southwest, with some high speeds, while during seed dispersal seasons (mainly September and October, and on), the main wind directions are northwest, with lower speeds than those during the flowering seasons (Fig. 1). Therefore, these provide the chance to isolate the effects of wind-assisted dispersal patterns in pollen and seed flow on the genetic variations of E. roxburghiana in the plot. The other is that throughout history, with the exception of the southeast corner, the whole DHS plot was clear-cut and then planted with Pinus massoniana about 60 years ago (Wang et al. 2014). As a consequence, during the early stage of the E. roxburghiana population recovery, the forest canopy was open, but then, it gradually closed. Therefore, if there were differential effects of canopy density over gene flow, we would find different genetic spatial patterns among the various generations of E. roxburghiana in the plot.
Mean wind directions and speeds from May to November for 8 years (2003–2012; 2005 and 2008 were excluded due to incomplete records), based on daily records taken at 30-min intervals. Calm periods (no wind) were removed. May–July corresponds to the flowering seasons, and September–November corresponds to seed dispersal seasons for Engelhardia roxburghiana. For each month, mean and max wind speeds were shown on lower right corner, media wind direction were represented by an arrow
Therefore, in this study, we aimed to link the spatial distribution of genetic diversity of E. roxburghiana with existing site-specific conditions to understand the biological processes that governed the dynamics of the population. Specifically, by dividing the gene flow of E. roxburghiana into pollen and seed flow according to their predominant flow directions (Supplementary S1 Fig. S1), we compared pollen and seed flow effects on SGS and assessed the effects of canopy resistance on pollen and seed dispersals. In this study, we used landscape distance/resistance approaches to assess canopy resistance on gene flow in different directions through canopy simulations. To do so, we particularly chose individuals of the new generation with diameter at breast height (DBH) < 30 cm, whose regenerations were most likely influenced by canopies, and built different canopy friction maps for the plot. Then, we used three different kinds of distances (straight line, least cost, and resistance) to test the landscape genetic models. Here, we expected that as dispersal distances of both wind-assisted pollen and seeds were positively related to wind speed (Nathan et al. 2011; Steinitz et al. 2011), the genetic relatedness among individuals would be best explained by the straight line distance for them. However, as other factors, such as turbulent and vertical winds, morphological adaptations for dispersal and landscape features influence the final distance and direction of pollen and seed travel (Nathan and Katul 2005; Wright et al. 2008; Millerón et al. 2012; Viner and Arritt 2012; Maurer et al. 2013; Damschen et al. 2014); the best associated distance may not be straight and may also differ for pollen and seed. Using landscape genetic models, we are able to provide the first comparative test about the effects of local site-specific canopy conditions on pollen and seed flow.
Materials and methods
Study species
E. roxburghiana is an evergreen tree that is widely distributed in Southeast Asia, from eastern Pakistan to southern China and Indonesia (Lu et al. 1999). It is a pioneer canopy tree and grows in various soil types, up to 30 m tall. It is an important medicinal plant in China. Compounds isolated from its leaves, stems, and roots are useful for treating many ailments (Wu et al. 2012; Xin et al. 2012). According to the first stem census data obtained in 2005, it was the third most important tree species with an importance value of 4.8 %; Castanopsis chinensis and Schima superba were first and second with importance values 12.3 and 6.6 %, respectively, in the DHS plot (Ye et al. 2008). Here, the importance value is the sum of relative density, frequency, and dominance of a species in its community.
E. roxburghiana has unisexual flowers and wind-dispersed pollen. Its fruit consists of a single-seeded nut with a relatively large trilobate wing formed by three bracts that facilitate the wind dispersal of seeds. The middle bract is 3–5 cm long, and the other two lateral ones are 0.7–2.7 cm long (Lu et al. 1999; Supplementary S1 Fig. S2).
Although E. roxburghiana is a common species, its reproductive biology is relatively unknown. A phenological study of our group from December 2012 to December 2014 using 11 E. roxburghiana individuals with DBH 8.8–49.0 cm in our plot showed that the one with the smallest DBH flowered, but some individuals with larger DBH did not. During our sample collection, we observed no vegetative reproduction from sprouting and creeping stems in E. roxburghiana, suggesting that this species reproduces sexually. As a light-demanding species, its regeneration in late-successional forests depends on light gap formations.
To compare wind-assisted dispersal patterns among generations, E. roxburghiana trees were divided into three DBH classes (Fig. 2): DBH ≥ 40 cm (old generation, 168 individuals), 30 cm ≤ DBH < 40 cm (young generation, 190 individuals), and DBH < 30 cm (new generation, 164 individuals). The generations were defined mainly to make each contain similar numbers of individuals.
Study site
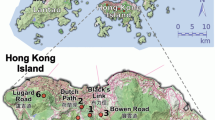
This study was conducted in the 20-ha (400 × 500 m) DHS plot in the 1155-ha DHS National Nature Reserve on the southern verge of the Tropic of Cancer in the subtropical part of South China (Wang et al. 2009). In this plot, the forest at the southeast corner has been well protected, and it is estimated to be more than 400 years old, whereas the rest has frequently been managed (Zhang et al. 1955; Fig. 2b). In the early 1950s, the managed parts were clear cut and reforested with native pines. However, after the whole area of the DHS was chosen as a reserve in 1956, no cutting and forest management activities, such as weeding and fertilization, have been allowed. Therefore, instead of developing into pure, dense pine forest in the managed part, the vegetation in the managed part actually gradually developed into regional evergreen broadleaved forest. At the time of our plot setting, the vegetation of the study area was dominated by Lauraceae, Euphorbiaceae, Rubiaceae, Moraceae, Theaceae, Myrtaceae, and Aquifoliaceae (Ye et al. 2008).
The topography of the plot is complex, including, among other features, elevations ranging from 230.0–476.1 m, high and low ridges, shallow and deep valleys, and hillsides (Wang et al. 2014). On the east side of the plot, there is an eddy flux tower (Supplementary S1 Fig. S3). The tower was established in 2002; it has recorded weather information automatically, including wind, every 30 min each day.
Sample collection and microsatellite analysis
Leaf or cambium tissues of 522 E. roxburghiana individuals in the DHS plot with DBH ≥ 1 cm were collected in 2012 and 2013. The leaves were stored in sealed plastic bags containing silica gel, while cambium tissues were stored in cetyltrimethylammonium bromide (CTAB) solution until DNA extraction. After DNA extraction, nine microsatellites were analyzed, according to procedures of Zhang et al. (2014) (Table 1).
Data analysis
Genetic diversity
We calculated genetic diversity parameters, including allelic richness (A), observed and unbiased expected heterozygosity (H O, H E), and the fixation index (f) using GENETIX 4.05 (Belkhir et al. 1996–2004). Then, we tested deviations from the Hardy–Weinberg equilibrium (HWE) at each locus and genotypic linkage disequilibrium (LD) between all pairs of loci using GENEPOP 4.0.7 (Rousset 2008), adjusting their significance levels by the Bonferroni correction.
Visualization of spatial genetic patterns
We used two methods, genetic landscape shapes (GLS) in Alleles In Space (AIS 1.0; Miller 2005) and spatial principal component analysis (SPCA) in R package ADEGENET (Jombart et al. 2008), to study the general genetic patterns of E. roxburghiana in our plot. In GLS, we calculated genetic distances among neighbors and then interpolated them over the studied area to obtain a graphical representation of the genetic distance pattern of E. roxburghiana. After trying different parameters and values, we chose to use the same ones as previously reported for the same plot (Wang et al. 2014) to display the present results. They included the distance weighting parameter α = 1, a grid size of 100 × 80 m, and raw genetic distances between individuals.
SPCA optimizes genetic variation to find spatial patterns by taking spatial autocorrelation (Moran’s I) into account. Depending on positive or negative spatial autocorrelation, SPCA can identify two types of patterns: global (such as patches and clines) and local (differentiation among neighbors) structuring. We used a Delaunay triangulation network to define the connection of individuals to help estimate spatial autocorrelation. We also used a Monte Carlo approach with 999 permutations to test the significance of the global and local structuring. A bar plot of eigenvalues was used to determine the SPCA eigenvalues.
Anisotropic gene flow
For anisotropic gene flow analysis, we first used Mantel bearing correlograms implemented in PASSAGE 2.0.11.6 (Rosenberg and Anderson 2011) to examine the magnitudes of gene flow in different spatial directions for E. roxburghiana. This procedure calculates a set of directional correlograms for given distance classes and uses the Mantel correlation coefficient (r z ) to measure autocorrelation.
For all individuals and the three DBH classes, we used PASSAGE to automatically choose 20 distance classes, with each containing a similar number of individual pairs (among 6798–6800 for all individuals, 700–702 for individuals with DBH ≥ 40 cm, 897–898 for 30 cm ≤ DBH < 40 cm, and 667–669 for DBH < 30 cm), and observed 36 bearings (every 5° from 0° to 180°) in the correlograms. The angle was measured counterclockwise between the positive x-axis (due east) and the line connecting the two individuals in a pair. Here, we only considered a 0°–180° period for each pair in its geographic angle measurement. We did not use a 0°–360° period because to do so, we must know the order of the two individuals in a pair: the start individual or the mother tree as well as the end individual or the offspring, and we could not determine this. Actually, using methods of Frantz et al. (2010), about 99.984 % of all our 135981 individual pairs were unrelated (Supplementary S2). Considering that the angles were symmetric around the circle, only the bearing in 0°–180° for each individual pair was used in this study.
To measure the genetic differentiation between individuals, we first calculated the Queller and Goodnight (1989) relatedness coefficients between individuals using SPAGEDI 1.2g (Hardy and Vekemans 2002) and subsequently multiplied them by −1. The significance of r z was assessed using 999 permutations. We then performed an anisotropic spatial autocorrelation analysis to compare SGS for all E. roxburghiana individuals and the three DBH classes in different directions. To do so, individual pairs were divided into two groups according to their geographical directions, northeast (NE, 0°–90°) and northwest (NW, 90°–180°), as the SGS of E. roxburghiana could be influenced by different wind speeds in these directions in the plot. Then, in the autocorrelation analysis, we calculated pairwise genetic relatedness coefficients (Queller and Goodnight 1989) and regressed them to different geographic distances with 20-m intervals up to 400 m. The significance of the mean relatedness coefficient in a particular distance class was obtained from 1000 permutations of individual spatial locations.
Generalized additive model and spatial resistance simulation of canopy
To further study the relationship between pairwise genetic relatedness and wind speed/direction, we adapted methods of Carslaw et al. (2006) and Carslaw and Beevers (2013) for modeling varying air pollution with wind speed and direction. To do so, we used geographic distance and direction between individuals to substitute for wind speed and direction as well as genetic relatedness for pollution concentration. To follow Carslaw and Beevers’ (2013) procedures, we applied a generalized additive model (GAM) (Wood 2006) to model r (pairwise genetic relationship) as a smooth function of the pairwise geographic direction and pairwise geographic distance. That was
where r ij is the pairwise genetic relationship between individuals i and j, β 0 is the overall mean of the response, s(dir ij , dis ij ) is the smooth function of covariates dir (pairwise geographic direction, 0°–180° period) and dis (pairwise geographic distance or landscape distance, see below), and ε ij is the residual.
Here, if we consider the pollen and seed of E. roxburghiana released into atmosphere as particles in Carslaw’s model (Carslaw et al. 2006; Carslaw and Beevers 2013), we would expect that if the wind speed was low, they would not be dispersed effectively and thus would be concentrated near their sources. Therefore, a high genetic relatedness between source and sink could only be observed at short distances. Conversely, high wind speed would cause high genetic relatedness even at long distances. In these circumstances, distance-related genetic relatedness patterns match the wind speed patterns well. In fact, the model above is actually a common isolation-by-distance (IBD) model used in SGS analysis except for using direction as a covariate. However, unlike linear regression models, the GAM model used here is a semi- or non-parametric regression model that can fit complex and non-linear relationships between response and predictor variables, and it is more appropriate in spatial genetic pattern analyses (Snäll et al. 2004; Benson et al. 2012) than linear regression models. GAM analysis was performed using the R package MGCV 1.8-3 (Wood 2006, 2011), and model selection was based on generalized cross-validation (GCV) scores, percentage of deviance explained, adjusted R 2, and Akaike information criterion (AIC).
For comparison, we also used surface distance (SD; He et al. 2013) and hypotenuse distance (HD; Supplementary S1 Fig. S4) to replace straight geographic distance (projection distance, PD) to fit the GAM model. If topography was an important factor for influencing the SGS of E. roxburghiana in the DHS plot, we would expect the models with surface or hypotenuse distance to be better compared to the models with traditional geographic distance.
We then simulated forest canopy as an obstacle to gene flow. To do this, we used crown sizes of individuals in the plot to substitute for the forest canopy. Crowns are three-dimensional. Because crown heights were not available, we only considered crown horizontal dimensions. By assuming that a crown was a circular cylinder and its base area represented the linear transform of the actual crown projection area, we used the base area to represent the crown size. The radius of each crown cylinder was estimated from the DBH of individual using log(C (crown radius)) = a + b × log(DBH) reasonable for describing crown radius and the DBH relationship for tree species (O’Brien et al. 1995; Troxela et al. 2013). Then, we calculated the crown sizes (π × C 2 (crown radius)) of all individuals with DBH ≥ 5 cm for all species in the plot recorded with the 2005 stem census. To calculate parameters a and b, we artificially set 10 m as the upper limit for the crown radius of the individuals with the largest DBH of 175 cm and 1.5 m as the lower limit for those with the smallest DBH of 5 cm. According to these, the model became log(C (crown radius)) = −0.1969 + 0.5336 × log(DBH). Then, we assumed that the crowns of the individuals with DBH ≥ 30 cm resulted in resistance to both E. roxburghiana pollen and seed flow during their crown growth. Based on these crowns, the DHS plot was divided into canopy-covered and non-canopy-covered areas. Using these canopy cover models, we subsequently tested the possible canopy influences on gene flow predominated by different wind directions in the new generation (individuals with DBH < 30 cm) of E. roxburghiana.
Following Lander et al. (2013), we used three types of distances to model canopy effects on gene flow: (1) weighted linear distances (WLD), (2) least-cost distances (LCD), and (3) resistance distances (RD). Unlike geographic distance, these three distances take into account the landscape types of which the gene flow encounters so that different cost values can be used for different landscape types, which are canopy types in our case. WLD is based on a straight line path between two individuals, and the lengths of the line segments were weighted by multiplying them with different cost values. LCD and RD are based on the surface cost that we define. LCD is the distance with the lowest cost between two individuals, while RD is the average travel cost during random walk between two individuals by circuit theory (McRae 2006). The models using these distances were also performed in two geographical directions (NE and NW) corresponding to pollen flow or seed flow, respectively.
For WLD, the straight line between two points was subdivided into different sections where canopy or non-canopy areas crossed, and the lines falling in sections of the former were multiplied by 1.5 (low), 2 (medium), or 10 (high) as the additional cost for gene flow. For LCD and RD, we converted the DHS plot to a raster with 0.5 × 0.5-m grid cells. For non-canopy cell types, we assigned a value of 1, meaning no friction. For canopy cell types, we assigned 2, 10, 100, and 1000 to simulate low, medium, high, and very high resistance levels to gene flow, respectively, and tested each level in LCD and RD analyses denoted by LCD2–LCD1000 and RD2–RD1000. In addition, we also made a null model of IBD by assigning 1 to all the cells for comparison. This is equivalent to testing for isolation by linear distance (PD, in our case), but it accounts for the finite size of the study area and, therefore, is more appropriate for comparison with our other models (Lee-Yaw et al. 2009). To examine the influence of grid size, we used a grid size of 1 × 1 m for the analysis and found that the results were not substantially different from those using the 0.5 × 0.5-m grid (Supplementary S1 Tables S1 and S2). Therefore, we only reported results for 0.5 × 0.5-m grid cells.
The partial Mantel test was further used to test the effects of canopy on gene flow while controlling for IBD. If canopy does present a barrier to gene flow, the association between pairwise genetic relatedness and cost distance quantified by the LCD that outperformed in all GAM models (see Results section) should persist when the IBD influence is removed. Because the Mantel test does not support interactive effects, we did not include a pairwise direction matrix in the test. For the controlling distance matrix, we used the results from the null model LCD1. The partial Mantel test was also performed using the PASSAGE software, and the significance of the correlation was evaluated using 999 permutations.
Results
Genetic diversity
The number of alleles per locus ranged from 4 to 15, and the H E ranged from 0.370to 0.886 among the loci (Table 1). No locus deviated from HWE after the Bonferroni correction. A significant deviation from LD was found for all locus pairs at the 5 % level when all 522 individuals were analyzed. However, no such deviation was found when only individuals with a DBH ≥ 50 cm were analyzed (n = 45).
Visualization of spatial genetic patterns
Generally, the GLS results indicated that the northern portions of the individuals were more genetically similar to each other compared to those in the southern part of the DHS plot, with the highest and lowest similarities near the southern and northern borders (Fig. 3a), respectively.
a The genetic landscape shape of Engelhardia roxburghiana in the DHS plot. Circles represent E. roxburghiana individuals with different DBH sizes in the plot. b Plot for the first global principal component of the spatial principal coordinate analysis (SPCA). Circle sizes represent individual scores on the first component. Bold contour line(s) in both panels delineate the main genetic differentiation and are derived by the interpolation of results for each method
In the SPCA analysis, the global structure was significant (P = 0.001), but the local structure was not (P = 0.661). Based on the eigenvalues (Supplementary S1 Fig. S5), we chose the first positive SPCA component (SPC1) that accounted for 11.6 % of the total variance to present the results graphically, and it revealed a clear separation of the individuals in the plot into two parts: northwest and southeast (Fig. 3b).
Anisotropic gene flow
The Mantel bearing correlogram indicated clear direction-dependent spatial autocorrelation patterns with positive spatial autocorrelation extending to the tenth ring (169.6 m) of distance classes in the NE direction for all individuals analyzed. Furthermore, although significant negative spatial autocorrelation generally occurred in all directions (0 –180°) from the distance class of 198.3 m (the 12th ring) to farther distance classes, the magnitude was greater in the NW direction than in the NE direction, as indicated by the absolute Mantel’s r z values. For different DBH classes, the individuals with DBH ≥ 40 cm showed a similar pattern to all individuals, with the positive spatial autocorrelation extending to the ninth ring (150.3 m) in the NE direction. Those with 30 cm ≤ DBH < 40 cm had a similar trend, while the rest with DBH < 30 cm showed no such directional patterns (Fig. 4).
Mantel bearing correlograms of Engelhardia roxburghiana in the DHS plot for a all individuals, b individuals with DBH ≥ 40 cm, c with 30 cm ≤ DBH < 40 cm, and d with DBH < 30 cm. For each panel, the dots in the lower circular part are absolute Mantel’s r z values (size of the each dot corresponds to its value), and their significance or non-significance is indicated by the colored dots in the diagonal position on the upper circular part of each panel. The red, sky blue, and deep yellow circles represent significant (P < 0.05) positive, negative, and non-significant Mantel’s r z values, respectively. The annuli in a represent distance classes with the upper limits of 38.0, 57.0, 72.5, 86.7, 100.3, 114.3, 128.2, 142.1, 160.0, 169.6, 183.5, 198.3, 214.2, 232.0, 252.2, 275.9, 302.6, 334.1, and 372.0 m; in b with 38.4, 57.2, 72.3, 85.2, 97.3, 109.9, 123.0, 136.5, 150.3, 163.7, 178.6, 195.3, 212.5, 231.1, 251.4, 275.6, 299.3, 327.2, and 364.1 m; in c with 34.6, 52.2, 67.1, 79.3, 91.8, 104.4, 117.2, 130.0, 143.8, 157.4, 171.0, 184.5, 198.5, 214.2, 232.9, 256.0, 282.3, 317.3, and 358.8 m; and in d with 34.7, 56.3, 73.8, 92.0, 109.0, 124.7, 140.0, 156.6, 172.0, 185.1, 199.1, 214.6, 232.7, 250.5, 270.8, 295.0, 319.0, 349.3, and 387.2 m. For each panel, their 20th annulus is not shown in order to make the first inner annuli observable
Significant SGS with positive relatedness coefficients extended up to 140 and 120 m for all of the individual pairs in the NE and NW directions, respectively (Fig. 5a). However, the individual pairs in the NE direction showed higher mean genetic relatedness coefficients in all distance classes than did those in the NW direction. For different DBH classes, individuals in the NE direction also showed higher SGS than did individuals in the NW direction in most distance classes (Fig. 5b–d), especially for individuals with DBH < 30 cm within the distance class of 120 m (Fig. 5d).
Generalized additive model and spatial resistance simulation of the canopy
In the GAM analysis, for all individuals and in the three DBH classes, three geographic distances (PD, HD, and SD) and the IBD null model using LCD1 had comparable results (Table 2). Consistent with the Mantel bearing correlogram, the predicted GAM values (Fig. 6) demonstrated direction-dependent spatial genetic patterns that were clearer for individuals with DBH ≥ 40 cm (extending to about 200 m around 90° and to 100 m around 150°) than for those in the other two DBH classes.
Predicted pairwise genetic relatedness distribution varying with geographic distance (projection distance, PD) and direction, according to generalized additive models for a all individuals, b individuals with DBH ≥ 40 cm, c with 30 cm ≤ DBH < 40 cm, and d with DBH < 30 cm of Engelhardia roxburghiana in the DHS plot. Long and short arrows in a and b indicate wind directionality patterns that roughly represent predominant pollen and seed flow, respectively
For individuals with DBH < 30 cm, LCD10–LCD1000 improved models used for predicting the association between pairwise genetic relatedness and spatial resistance, such as with high adjusted R 2 values of 0.071, 0.074, and 0.074. On the other hand, RD with low friction values performed the worst in all models, with adjusted R 2 values of 0.035 and 0.031 for RD2 and RD10 (Table 2). Models using RD2 and RD10 were even inferior to the IBD null model that used RD1 with adjusted R 2 value of 0.041.
When considering two gene dispersal directions, NE and NW, three geographic distances (PD, HD, and SD) showed similar association values among each other for all individuals and for the three DBH classes (Table 3).
For individuals with DBH < 30 cm, the models using LCD10–LCD1000 were superior, with adjusted R 2 values of 0.075, 0.080, and 0.076, respectively, to those using other distance types with adjusted R 2 values ranging from 0.032 to 0.070 in the NE direction, while no LCD models showed such superiority in the NW direction (Table 3).
Partial Mantel tests revealed that the models using LCD for individuals with DBH < 30 cm remained significant even when controlling for IBD in the NE direction but not in the NW direction (Table 4).
Discussion
Although large wind or weather patterns are often predictable and frequently used in gene flow analyses (e.g., Cox et al. 2011; Kremer et al. 2012), locally, only a few relevant studies have incorporated meteorological data, such as wind (Nathan and Katul 2005; Austerlitz et al. 2007; Wright et al. 2008; Millerón et al. 2012; Viner and Arritt 2012, Maurer et al. 2013), snowmelt timing (Cortés et al. 2014), and the other weather conditions, such as temperature, humidity, and solar radiation intensity (Bohrerova et al. 2009). Results from these studies indicate that the meteorological data are helpful to estimate and simulate gene flow patterns. Generally, setting up meteorological stations is costly and not practical. However, if the topographies of the study area and its surrounding areas are flat, meteorological data from nearby meteorological stations might be useful. For instance, in an area near Montargis in France, a comparison between data from a meteorological station 70 km from the experimental field and the data from another station located inside the field showed little difference (Bensadoun et al. 2014). Therefore, we should try our best to obtain and include possible meteorological data in the analyses of plant gene flow.
Theoretically, the utilization of maternally inherited chloroplast DNA (cpDNA) data should enable us to estimate the seed dispersal patterns for E. roxburghiana. However, using eight cpDNA intron or intergenic spacers (Supplementary S3), our results indicated that all 12 individuals we chose shared the same cpDNA haplotype (total length of 7092 bp). Therefore, we gave up using cpDNA markers to detect possible seed dispersal patterns.
Overall, we found significant relationships between site-specific conditions and spatial genetic patterns of E. roxburghiana in the DHS plot, suggesting that these conditions may have played important roles in structuring the genetic variations of this species. Particularly, the nature of the wind system typically resulted in an asymmetrical genetic pattern in E. roxburghiana, with the gene flow clearly biased to the pollen dispersal-predominant NE direction. However, higher SGS also occurred in this direction than that in the seed dispersal-predominant NW direction. The results suggest that canopy closure increased canopy resistance to gene flow, but gene flow predominated by different wind directions displayed different relationships with canopy resistance.
Spatial genetic structure and anisotropic dispersal
We found a clear fine-scale SGS in E. roxburghiana, and it was higher in the NE direction than in the other directions. As there is an apparent prevailing southwest wind during flowering seasons and because we would expect it to homogenize genetic variations by pollen flow in the NE direction within the population, if not in other directions, the observed SGS patterns may most likely be due to restricted dispersal of seeds. Although the seeds of E. roxburghiana are also dispersed by wind, the wind speed in seed dispersal seasons is lower in the NE direction than in the other directions, and thus, seed dispersal by wind might be limited in this direction, causing localized pedigree structures.
Significant SGS was also reported in other tree species with wind-dispersed seeds (Heuertz et al. 2003; Gaino et al. 2010 and the references therein) due to the high frequency of short-distance seed dispersal. In fact, compared to species with animal-dispersed seeds, species with wind-dispersed seeds are generally distributed in clumps, indicating dispersal restriction by the seeds themselves (Li et al. 2009). In diploid plant species, seed flow introduces two alleles at each locus and is far more likely to contribute to the SGS than is pollen flow within the population (Dow and Ashley 1996; Grivet et al. 2009). Therefore, if seed flow is restricted, the overall SGS within the population can be easily established, even under extensive pollen flow (Grivet et al. 2009; Albaladejo et al. 2012; Wang et al. 2014).
Our data suggest that genetic variation patterns vary across generations, as both the Mantel bearing correlograms (Fig. 4) and the GAM analyses (Fig. 6) revealed a clear directional pattern in the old generation (DBH ≥ 40 cm) that gradually diminished in the later generations. The reason for this may be that as the forest recovered after clear-cutting in the plot, the forest density and canopy cover increased and thus hindered the wind-borne gene flow. Therefore, it is possible that as a population develops from open land, conditions for gene flow by wind become less and less favorable. Some old individuals die, and some young regenerate, and, over time, it may become difficult to detect directional genetic variation patterns in the young generations. This suggests that when studying such patterns, we should take population history into consideration.
Landscape genetic models
Locally, unlike previous studies that directly investigated forest canopy effects on pollen or seed dispersal distances (Bacles et al. 2005; Millerón et al. 2012; Sant’Anna et al. 2013; Shohami and Nathan 2014), we used landscape genetic models to assess such effects. To our knowledge, this is the first attempt to associate gene flow processes with cost distances for canopy resistance models at local scales. Our results are discussed according to the following several aspects.
First, GAM results indicate that the association strengths of HD, SD, and simple Euclidean distance (PD distance) with genetic relatedness are similar, implying that topography has little influence on the spatial patterns of E. roxburghiana genetic variations, since HD and SD incorporate topography information and PD does not.
Second, among the three distances used to test the canopy as a barrier to gene flow, both WLD and RD are comparable or inferior to simple PD, indicating that WLD and RD are not suitable for explaining the association between genetic relatedness and canopy resistance. Specifically, our models using RD performed the poorest, and, most of the time, the association results using RD2–RD1000 were even poorer than those of the null models that used RD1. The reasons for these might be that resistance distance using the average resistance over all possible paths between pairwise samples may not be applicable for passive wind-dispersed pollen and seeds that cannot choose their dispersal paths.
Third, we found that least-cost distance with high friction values outperformed the other distances, supporting the idea that canopies can serve as physical barriers to gene flow, such as by pollen (Millerón et al. 2012). However, when we looked at the two different directions that were affected by different wind speeds, we found that the model using LCD was the best for describing the association in the NE direction but not in the NW direction (Tables 3 and 4). Because gene flow in the NE direction was mostly influenced by pollen, the possible reason for this might be that the pollen of E. roxburghiana is very tiny (Manos and Stone 2001), and with strong wind and air turbulence, the pollen could be moved by wind more freely than can seeds bypassing the canopy, increasing the chances of the interception by canopy. In this case, the pollen dispersal route could be partially mirrored by the least-cost path between individuals. On the contrary, the seeds of E. roxburghiana are large. When they are dispersed in the air, they have few chances to be dispersed by anabatic wind. Coupled with moderate wind speed in the NW direction, the pattern of seed flow was more likely to reflect isolation by distance but not by the cost distance used in this study.
Finally, we found that both non-linear (GAM) and linear methods (partial Mantel test) generally gave congruent results for the models using LCD distances with high friction values. For the same direction, the GAM method revealed that the higher the friction values (100 and 1000) used in the LCD, the larger the association values, which means that the canopy only served as a gene flow impediment when there was high spatial resistance. This is understandable because low friction might represent a sparse canopy, while high friction means a dense canopy. However, the highest correlation value detected by the partial Mantel test was using LCD with a low friction value of 2, which is unexplainable. As linear models only account for the linear components of associations, the reasons for the discrepancies between the two methods might be that, at our study scale, the linear method could not fully capture the variations we tested.
A significant strength of the GAM approach adopted here was that splines could be inspected to reveal the true shape of the relationship between genetic relatedness and geographic distance (Fig. 6). Unfortunately, as local wind records were only available since the year 2002, we could not infer the wind dynamics and the relationship with historical gene flow.
Our results indicate that the different genetic patterns obtained for pollen and seed of E. roxburghiana individuals by landscape genetic models are attributable to the differences in canopy resistances to pollen and seed dispersals. This confirms that the grid-based landscape distance/resistance approach used here can gain a much more refined picture of genetic structures and the possible corresponding variables.
However, this study has several limitations in landscape genetic models. First, for each species, we only use the living individuals in the community to simulate previous canopy conditions. The individuals who died in the past may also have contributed to the SGS of E. roxburghiana, but they were not included because of a lack of information about them.
Second, canopy was not directly measured and its heterogeneity, such as canopy height, was not full incorporated in our study. Canopy heterogeneity can modify turbulence leading to different transfer of pollen or seed (Bohrer et al. 2008, 2009; Damschen et al. 2014). Particularly, heterogeneous canopy can create different hot spots for turbulence ejections which can translate to high potential of long distance dispersal (Bohrer et al. 2009). In this study, we used the same model to predict crown sizes of all the species and assumed that their crowns were perfectly round, and this assumption most likely would not adequately reflect the true biological phenomena. Additionally, DBH could explain only part of variation in crown size. Tree heights which characterize the three-dimensional canopy structure were not considered. Individuals with different heights may intercept E. roxburghiana pollen and seed differently. The heights of individuals were also related to the dispersal distances of pollen and seed (Katul et al. 2005; Nathan and Katul 2005; Bohrer et al. 2008). Bohrer et al. (2008) indicated that light particles (corresponding to pollen in our case) released from high individuals would have more advantage in benefiting from canopy hot spots of ejections.
Third, in the models, we used all the species with their DBH larger than 30 cm including E. roxburghiana. For these E. roxburghiana individuals, we ignored their fecundity and pollen production. In fact, they might not be physical barriers but rather competitive seed sources in the plot, especially for the larger ones.
Four, although our association analysis indicates that topography has little influence on the spatial patterns of E. roxburghiana genetic variations, it has been reported that landscape heterogeneity plays an important role on dispersal (Damschen et al. 2014; Trakhtenbrot et al. 2014) that subsequently influences long-term genetic patterns in the species. To explore the effects of topography on the seed dispersal by wind, Trakhtenbrot et al. (2014) utilized a topography simulation and showed that even gentle hill could generate large differences in seed-mediated gene flow, such as hill top vs. upwind or downwind slopes. Damschen et al. (2014) found that open gaps and corridors affected the wind speed and direction and thus influenced both short and long distance dispersals. In our study, we only consider the undulating terrain of the plot but neglect its interactions with wind and canopy, which may have resulted in our failure to find the relationship of landscape and genetic patterns. In addition, wind data was only obtained at a single point (Supplementary S1 Fig. S3) and a single elevation for the complex landscape of our plot, and such data cannot reflect the characteristics of the wind conditions over the entire plot.
Finally, our models are limited by the fact that the relatedness coefficient we used is symmetric, and they could not be used to disentangle true pollen and seed flows in the 0°–180° period we used. Consequently, the genetic structure in different directions was mostly due to the intensity of pollen or seed dispersal, and future parentage analysis using seeds and seedlings would allow for more robust insight into the impact of anisotropic winds on genetic variation in this species.
Therefore, our landscape genetic model is simple and needs further improvements in future studies by incorporating these factors into analyses.
Conclusions
Seasonal changes in wind directions occur in many locations throughout the world (Komdeur and Daan 2005; Abe et al. 2008; Nazemosadat and Ghaedamini 2010; Cheung et al. 2011). For example, regionally, they are common in India and Southeast Asia, known as monsoon winds. Therefore, the seasonal wind direction-dependent pollen and seed dispersal systems in E. roxburghiana should not be considered unique features in our study plot but should be considered common elsewhere.
Our results demonstrate site-specific conditions resulting in local-scale genetic variations in E. roxburghiana, suggesting the importance of integrating these conditions into our future estimation and simulation of pollen and seed flow. Regarding the analytical approach, our study shows that landscape genetic approaches offer great potential in revealing factors of local-scale gene flow patterns, although the model still needs further improvements.
Finally, considering that rapid climate change causes wind conditions to vary unpredictably, further long-term realistic assessments of wind-aided gene dispersal using parentage analysis of pollen and seeds are required in E. roxburghiana and other similar species for better biodiversity conservation and resource management in the future.
References
Abe T, Sakamoto T, Nobuhiro T, Kabeya N, Hagino H, Tanaka H (2008) Wind data recorded at Ogawa Forest Reserve, Japan, from November 2003 to April 2006. Bull For Forest Prod Res Inst 7(4):245–266
Albaladejo R, Guzmán B, Gonzalez-Martinez SC, Aparicio A (2012) Extensive pollen flow but few pollen donors and high reproductive variance in an extremely fragmented landscape. PLoS One 7(11):e49012. doi:10.1371/journal.pone.0049012
Austerlitz F, Dutech C, Smouse PE, Davis F, Sork VL (2007) Estimating anisotropic pollen dispersal: a case study in Quercus lobata. Heredity 99:193–204. doi:10.1038/sj.hdy.6800983
Bacles CFE, Burczyk J, Lowe AJ, Ennos RA (2005) Historical and contemporary mating patterns in remnant populations of the forest tree Fraxinus excelsior L. Evolution 59:979–990. doi:10.1111/j.0014-3820.2005.tb01037.x
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996-2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. – Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier (France)
Bensadoun A, Monod H, Angevin F, Makowski D, Messéan A (2014) Modelling of gene flow by a Bayesian approach: a new perspective for decision support. AgBioForum 17(2):213–220
Benson JF, Patterson BR, Wheeldon TJ (2012) Spatial genetic and morphologic structure of wolves and coyotes in relation to environmental heterogeneity in a Canis hybrid zone. Mol Ecol 21:5934–5954. doi:10.1111/mec.12045
Bohrer G, Nathan R, Volis S (2005) Effects of long-distance dispersal for metapopulation survival and genetic structure at ecological time and spatial scales. J Ecol 93:1029–1040. doi:10.1111/j.1365-2745.2005.01048.x
Bohrer G, Katul GG, Nathan R, Walko RL, Avissar R (2008) Effects of canopy heterogeneity, seed abscission, and inertia on wind-driven dispersal kernels of tree seeds. J Ecol 96:569–580. doi:10.1111/j.1365-2745.2008.01368.x
Bohrer G, Katul GG, Walko RL, Avissar R (2009) Exploring the effects of microscale structural heterogeneity of forest canopies using large-eddy simulations. Bound-Lay Meteorol 132(3):351–382
Bohrerova Z, Bohrer G, Cho KD, Bolch MA, Linden KG (2009) Determining the viability response of pine pollen to atmospheric conditions during long-distance dispersal. Ecol Appl 119(3):656–667. doi:10.1890/07-2088.1
Born C, Le Roux P, Spohr C, McGeoch MA, Van Vuuren BJ (2012) Plant dispersal in the sub-Antarctic inferred from anisotropic genetic structure. Mol Ecol 21:184–194. doi:10.1111/j.1365-294X.2011.05372.x
Burczyk J, Prat D (1997) Male reproductive success in Pseudotsuga menziesii (Mirb.) Franco: the effects of spatial structure and flowering characteristics. Heredity 79:638–647
Burczyk J, DiFazio SP, Adams WT (2004) Gene flow in forest trees: how far do genes really travel? For Genet 11(3-4):179–192
Carslaw DC, Beevers SD (2013) Characterising and understanding emission sources using bivariate polar plots and k-means clustering. Environ Model Softw 40:325–329. doi:10.1016/j.envsoft.2012.09.005
Carslaw DC, Beevers SD, Ropkins K, Bell MC (2006) Detecting and quantifying aircraft and other on-airport contributions to ambient nitrogen oxides in the vicinity of a large international airport. Atmos Environ 40:5424–5434. doi:10.1016/j.atmosenv.2006.04.062
Chandler S, Dunwell JM (2008) Gene flow, risk assessment and the environmental release of transgenic plants. Crit Rev Plant Sci 27(1):25–49. doi:10.1080/07352680802053916
Cheung K, Daher N, Kama W, Shafer MM, Ning Z, Schauer JJ, Sioutas C (2011) Spatial and temporal variation of chemical composition and mass closure of ambient coarse particulate matter (PM10e2.5) in the Los Angeles area. Atmos Environ 45:2651–2662. doi:10.1016/j.atmosenv.2011.02.066
Cortés AJ, Waeber S, Lexer C, Sedlacek J, Wheeler JA, Van Kleunen M, Bossdorf O, Hoch G, Rixen C, Wipf S, Karrenberg S (2014) Small-scale patterns in snowmelt timing affect gene flow and the distribution of genetic diversity in the alpine dwarf shrub Salix herbacea. Heredity 113:233–239. doi:10.1038/hdy.2014.19
Cox K, Vanden Broeck A, Van Calster H, Mergeay J (2011) Temperature-related natural selection in a wind-pollinated tree across regional and continental scales. Mol Ecol 20:2724–2738. doi:10.1111/j.1365-294X.2011.05137.x
Damschen EI, Baker DV, Bohrer G, Nathan R, Orrocka JL, Turner JR, Brudvig LA, Haddadf NM, Levey DJ, Tewksburyh JJ (2014) How fragmentation and corridors affect wind dynamics and seed dispersal in open habitats. P Natl Acad Sci USA 111:3484–3489. doi:10.1073/pnas.1308968111
Devey DS, Bateman RM, Fay MF, Hawkins JA (2009) Genetic structure and systematic relationships within the Ophrys fuciflora aggregate (Orchidaceae: Orchidinae): high diversity in Kent and a wind-induced discontinuity bisecting the Adriatic. Ann Bot 104:483–495. doi:10.1093/aob/mcp039
Dow BD, Ashley MV (1996) Microsatellite analysis of seed dispersal and parentage of saplings in bur oak, Quercus macrocarpa. Mol Ecol 5:615–627. doi:10.1111/j.1365-294X.1996.tb00357.x
Dutech C, Sork VL, Irwin AJ, Smouse PE, Davis FW (2005) Gene flow and fine-scale genetic structure in a wind-pollinated tree species, Quercus lobata (Fagaceaee). Am J Bot 92(2):252–261. doi:10.3732/ajb.92.2.252
Fatemi M, Gross CL (2009) Life on the edge—high levels of genetic diversity in a cliff population of Bertya ingramii are attributed to B. rosmarinifolia (Euphorbiaceae). Biol Conserv 142:1461–1468. doi:10.1016/j.biocon.2009.02.014
Fowler NL (1988) The effects of environmental heterogeneity in space and time on the regulation of populations and communities. In: Davy AJ, Hutchings MJ, Watkinson AR (eds) Plant population ecology. Blackwell, Oxford, pp 249–269
Frantz AC, Pope LC, Etherington TR, Wilson GJ, Burke T (2010) Using isolation-by-distance-based approaches to assess the barrier effect of linear landscape elements on badger (Meles meles) dispersal. Mol Ecol 19:1663–1674. doi:10.1111/j.1365-294X.2010.04605.x
Gaino APSC, Silva AM, Moraes MA, Alves PF, Moraes MLT, Freitas MLM, Sebbenn AM (2010) Understanding the effects of isolation on seed and pollen flow, spatial genetic structure and effective population size of the dioecious tropical tree species Myracrodruon urundeuva. Conserv Genet 11:1631–1643. doi:10.1007/s10592-010-0046-3
Grivet D, Robledo-Arnuncio JJ, Smouse PE, Sork VL (2009) Relative contribution of contemporary pollen and seed dispersal to the effective parental size of seedling population of California valley oak (Quercus lobata, Nee). Mol Ecol 18:3967–3979. doi:10.1111/j.1365-294X.2009.04326.x
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620. doi:10.1046/j.1471-8286.2002.00305.x
He J, Li X, Gao D, Zhu P, Wang Z, Wang Z, Ye W, Cao H (2013) Topographic effects on fine-scale spatial genetic structure in Castanopsis chinensis Hance (Fagaceae). Plant Spec Biol 28:87–93. doi:10.1111/j.1442-1984.2011.00365.x
Heuertz M, Vekemans X, Hausman J-F, Palada M, Hardy OJ (2003) Estimating seed vs. pollen dispersal from spatial genetic structure in the common ash. Mol Ecol 12:2483–2495. doi:10.1046/j.1365-294X.2003.01923.x
Holderegger R, Buehler D, Gugerli F, Manel S (2010) Landscape genetics of plants. Trends Plant Sci 15:675–683. doi:10.1016/j.tplants.2010.09.002
Jombart T, Devillard S, Dufour A-B, Pontier D (2008) Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101:92–103. doi:10.1038/hdy.2008.34
Katul GG, Por-porato A, Nathan R, Siqueira M, Soons MB, Poggi D, Horn HS, Levin SA (2005) Mechanistic analytical models for long-distance seed dispersal by wind. Amer Natur 166:368–381. doi:10.1086/432589
Komdeur J, Daan S (2005) Breeding in the monsoon: semi-annual reproduction in the Seychelles warbler (Acrocephalus sechellensis). J Ornithol 146:305–313. doi:10.1007/s10336-005-0008-6
Kremer A, Ronce O, Robledo-Arnuncio JJ, Guillaume F, Bohrer G, Nathan R, Bridle JR, Gomulkiewicz R, Klein EK, Ritland K (2012) Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecol Lett 15:378–392. doi:10.1111/j.1461-0248.2012.01746.x
Lander TA, Klein EK, Stoeckel S, Mariette S, Musch B, Oddou-Muratorio S (2013) Interpreting realized pollen flow in terms of pollinator travel paths and land-use resistance in heterogeneous landscapes. Landscape Ecol 28:1769–1783. doi:10.1007/s10980-013-9920-y
Latta RG, Mitton JB (1999) Historical separation and present gene flow through a zone of secondary contact in ponderosa pine. Evolution 53:769–776. doi:10.2307/2640717
Lee-Yaw JA, Davidson A, McRae BH, Green DM (2009) Do landscape processes predict phylogeographic patterns in the wood frog? Mol Ecol 18:1863–1874. doi:10.1111/j.1365-294X.2009.04152.x
Li L, Huang Z, Ye W, Cao H, Wei S, Wang Z, Lian J, Sun I-F, Ma K, He F (2009) Spatial distributions of tree species in a subtropical forest of China. Oikos 118:495–502. doi:10.1111/j.1600-0706.2009.16753.x
Lu AM, Stone DE, Grauke LJ (1999) Juglandaceae. In: Wu ZY, Raven PH (eds) Flora of China, vol 4. Science Press, Beijing, China and Missouri Botanical Garden Press, St. Louis. USA, p 278
Manos PS, Stone DE (2001) Evolution, phylogeny, and systematics of the Juglandaceae. Ann Mo Bot Gard 88:231–269
Maurer KD, Bohrer G, Medvigy D, Wright SJ (2013) The timing of abscission affects dispersal distance in a wind-dispersed tropical tree. Funct Ecol 27:208–218. doi:10.1111/1365-2435.12028
McRae BH (2006) Isolation by resistance. Evolution 60:1551–1561. doi:10.1111/j.0014-3820.2006.tb00500.x
Miller MP (2005) Alleles in space (AIS): computer software for the joint analysis of interindividual spatial and genetic information. J Hered 96:722–724. doi:10.1093/jhered/esi119
Millerón M, de Heredia UL, Lorenzo Z, Perea R, Dounavi A, Alonso J, Gil L, Nanos N (2012) Effect of canopy closure on pollen dispersal in a wind-pollinated species (Fagus sylvatica L.). Plant Ecol 213:1715–1728. doi:10.1007/s11258-012-0125-2
Nathan R, Katul GG (2005) Foliage shedding in deciduous lifts up long-distance seed dispersal by wind. P Natl Acad Sci USA 102:8251–8256. doi:10.1073/pnas.0503048102
Nathan R, Katul GG, Bohrer G, Kuparinen A, Soons MB, Thompson SE, Trakhtenbrot A, Horn HS (2011) Mechanistic models of seed dispersal by wind. Thero Ecol 4:113–132. doi:10.1007/s12080-011-0115-3
Nazemosadat MJ, Ghaedamini H (2010) On the relationships between the Madden–Julian oscillation and precipitation variability in Southern Iran and the Arabian Peninsula: atmospheric circulation analysis. J Climate 23:887–904. doi:10.1175/2009JCLI2141
O’Brien ST, Hubbell SP, Spiro P, Condit R, Foster RB (1995) Diameter, height, crown, and age relationship in eight neotropical tree species. Ecology 76:1926–1939
Queller DC, Goodnight KF (1989) Estimating relatedness using genetic markers. Evolution 43:258–275
Rhodes M, Fant JB, Skogen KA (2014) Local topography shapes fine-scale spatial genetic structure in the Arkansas Valley evening primrose, Oenothera harringtonii (Onagraceae). J Hered 105:806–815. doi:10.1093/jhered/esu051
Robledo-Arnuncio JJ, Gil L (2005) Patterns of pollen dispersal in a small population of Pinus sylvestris L. revealed by total-exclusion paternity analysis. Heredity 94:13–22. doi:10.1038/sj.hdy.6800542
Rosenberg MS, Anderson CD (2011) PASSaGE: Pattern Analysis, Spatial Statistics, and Geographic Exegesis. Version 2. Methods Ecol Evol 2(3):229–232. doi:10.1111/j.2041-210X.2010.00081.x
Rousset F (2008) GENEPOP ‘007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106. doi:10.1111/j.1471-8286.2007.01931.x
Sant’Anna CS, Sebbenn AM, Klabunde GHF, Bittencourt R, Nodari RO, Mantovani A, dos Reis MS (2013) Realized pollen and seed dispersal within a continuous population of the dioecious coniferous Brazilian pine [Araucaria angustifolia (Bertol.) Kuntze]. Conser Genet 14:601–613. doi:10.1007/s10592-013-0451-5
Saro I, Robledo-Arnuncio JJ, González-Pérez MA, Sosa PA (2014) Patterns of pollen dispersal in a small population of the Canarian endemic palm (Phoenix canariensis). Heredity 113:215–223. doi:10.1038/hdy.2014.16
Shohami D, Nathan R (2014) Fire-induced population reduction and landscape opening increases gene flow via pollen dispersal in Pinus halepensis. Mol Ecol 23:70–81. doi:10.1111/mec.12506
Snäll T, Fogelqcist J, Ribeiro PJ Jr, Lascoux M (2004) Spatial genetic structure in two congeneric epiphytes with different dispersal strategies analysed by three different methods. Mol Ecol 13:2109–2119. doi:10.1111/j.1365-294X.2004.02217.x
Sork VL, Smouse PE (2006) Genetic analysis of landscape connectivity in tree populations. Landscape Ecol 21:821–836. doi:10.1007/s10980-005-5415-9
Steinitz O, Troupin D, Vendramin GG, Nathan R (2011) Genetic evidence for a Janzen–Connell recruitment pattern in reproductive offspring of Pinus halepensis trees. Mol Ecol 20:4152–4164. doi:10.1111/j.1365-294X.2011.05203.x
Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP (2010) Landscape genetics: where are we now? Mol Ecol 19:3496–3514. doi:10.1111/j.1365-294X.2010.04691.x
Torimaru T, Wennström U, Lindgren D, Wang X-R (2012) Effects of male fecundity, interindividual distance and anisotropic pollen dispersal on mating success in a Scots pine (Pinus sylvestris) seed orchard. Heredity 108:312–321. doi:10.1038/hdy.2011.76
Trakhtenbrot A, Katul GG, Nathan R (2014) Mechanistic modeling of seed dispersal by wind over hilly terrain. Ecol Model 274:29–40. doi:10.1016/j.ecolmodel.2013.11.029
Troxela B, Pianaa M, Ashtona MS, Murphy-Dunning C (2013) Relationships between bole and crown size for young urban trees in the northeastern USA. Urban For Urban Gree 12(2):144–153. doi:10.1016/j.ufug.2013.02.006
Ueno S, Tomaru N, Yoshimaru H, Manabe T, Yamamoto S (2006) Effects of canopy gaps on the genetic structure of Camellia japonica saplings in a Japanese old-growth evergreen forest. Heredity 96:304–310. doi:10.1038/sj.hdy.6800804
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935. doi:10.1046/j.1365-294X.2004.02076.x
Viner BJ, Arritt RW (2012) Small-scale circulations caused by complex terrain affect pollen deposition. Crop Sci 52:904–913
Wang ZG, Ye WH, Cao HL, Huang ZL, Lian JY, Li L, Wei SG, Sun IF (2009) Species-topography association in a species-rich subtropical forest of China. Basic Appl Ecol 10:648–655. doi:10.1016/j.baae.2009.03.002
Wang ZF, Liang JY, Ye WH, Cao HL, Wang ZM (2014) The spatial genetic pattern of Castanopsis chinensis in a large forest plot with complex topography. Forest Ecol Manag 318:318–325. doi:10.1016/j.foreco.2014.01.042
Wood SN (2006) Generalized Additive Models: An Introduction with R. Chapman and Hall/CRC
Wood SN (2011) Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J R Stat Soc B 73(1):3–36. doi:10.1111/j.1467-9868.2010.00749.x
Wright SJ, Trakhtenbrot A, Bohrer G, Detto M, Katule GG, Horvitz N, Muller-Landau HC, Jones FA, Nathan R (2008) Understanding strategies for seed dispersal by wind under contrasting atmospheric conditions. P Natl Acad Sci USA 105:19084–19089. doi:10.1073/pnas.0802697105
Wu H-C, Cheng M-J, Peng C-F, Yang S-C, Chang H-S, Lin C-H, Wang C-J, Chen I-S (2012) Secondary metabolites from the stems of Engelhardia roxburghiana and their antitubercular activities. Phytochemistry 82:118–127. doi:10.1016/j.phytochem.2012.06.014
Xin W, Huang H, Yua L, Shi H, Sheng Y, Wang TTY, Yu L (2012) Three new flavanonol glycosides from leaves of Engelhardtia roxburghiana, and their anti-inflammation, antiproliferative and antioxidant properties. Food Chem 132:788–798. doi:10.1016/j.foodchem.2011.11.038
Ye W-H, Cao H-L, Huang Z-L, Lian J-Y, Wang Z-G, Li L, Wei S-G, Wang Z-M (2008) Community structure of a hm2 lower subtropical evergreen broadleaved forest plot in dinghushan, China. J Plant Ecol (Chinese Version) 32(2):274–286
Zhang HD, Wang BS, Zhang CC, Qiu HX (1955) Forest community of Dinghushan in Gaoyao of Guangdong Province. Acta Scientiarum Naturalium Universitatis Sunyatseni 3:159–225
Zhang DD, Luo P, Chen Y, Wang ZF, Ye WH, Cao HL (2014) Isolation and characterization of 12 polymorphic microsatellite markers in Engelhardia roxburghiana (Juglandaceae). Silvae Genet 63:109–112
Acknowledgments
We thank Lin-Fang Wu and Ying Chen for their assistance in collecting samples and performing laboratory analyses. We are grateful to the two anonymous reviewers and editors for their valuable suggestions on how to improve the manuscript. The National Natural Science Foundation of China (31170352, 41371078, 31100312) and the Chinese Forest Biodiversity Monitoring Network funded this study.
Data archiving statement
Sampling locations, microsatellite genotypes file: DRYAD entry https://www.researchgate.net/publication/295712804_Engelhardia_roxburghiana-sample_locations_and_genotypes_data
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Kremer
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary S1
(DOC 9.51 mb)
Supplementary S2
(PDF 9 kb)
Supplementary S3
(PDF 67 kb)
Rights and permissions
About this article
Cite this article
Wang, ZF., Lian, JY., Ye, WH. et al. Pollen and seed flow under different predominant winds in wind-pollinated and wind-dispersed species Engelhardia roxburghiana . Tree Genetics & Genomes 12, 19 (2016). https://doi.org/10.1007/s11295-016-0973-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-0973-3