Abstract
This study examines the levels of gene flow, the distance and the patterns of pollen and seed dispersal, the intra-population spatial genetic structure (SGS) and the effective population size of a spatially isolated Myracrodruon urundeuva population using five microsatellite loci. The study was carried out in the Paulo de Faria Ecological Station, São Paulo State, Brazil and included the sampling and mapping of 467 adult-trees and 149 juveniles. Open-pollinated seeds (514) from 29 seed-trees were also sampled and genotyped. Significant SGS was detected in both adult (S p = 0.0269) and juveniles trees (S p = 0.0246), indicating short-distance seed dispersal. Using maternity analysis, all juveniles had the mother-tree assigned within the stand. A father-tree within the stand was also assigned for 97.3% of the juveniles and 98.4% of offspring. The average pollen dispersal distance measured in juveniles \( \left( {\hat{\delta } = 1 3 8\pm 1 6 9 {\text{ m}},{\text{ mean}} \pm {\text{SD}}} \right) \) and offspring \( \left( {\hat{\delta } = 2 5 2\pm 20 4 {\text{ m}}} \right) \) were higher than the average seed dispersal distance measured in juveniles \( \left( {\hat{\delta } = 1 2 4\pm 1 50{\text{ m}}} \right) \). About 70% of the pollen from juveniles and 51% from offspring traveled less than 200 m and, 72% of the seeds traveled less than 50 m. The effective population size of the studied sample indicates that the 467 adult-trees and 145 juveniles correspond respectively to 335 and 63 individuals that are neither inbred nor relatives. The results are discussed in relation to their impact on seed collection practices and genetic conservation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The fragmentation of continuous forests into small, isolated stands is a global problem affecting both tropical and temperate forest ecosystems. Spatial isolation of populations by forest fragmentation changes the movement of seeds and dispersal of pollen thus altering the gene flow among populations (Hamilton 1999). Information about distance and abundance of current pollen and seed flow is fundamental to understanding whether remaining populations are functionally connected (Kamm et al. 2009). This knowledge permits the appropriate design of management and conservation strategies (Wang et al. 2007; Bittencourt and Sebbenn 2008). Understanding the levels of gene flow is also important for successful environmental restoration in order to maintain corridors of gene flow and/or avoid reproductive isolation among remnant populations (Wang et al. 2007). Intensive pollen and/or seed flow maintains genetic cohesion among populations, whereas low intensity pollen and/or seed flow is expected to increase the genetic differentiation through genetic drift and natural selection (Young et al. 1996; Young and Boyle 2000; Bittencourt and Sebbenn 2007). Gene flow also affects the genetic diversity and the effective population size of the remaining populations. If the gene flow among populations is high, the genetic diversity and effective population size will increase. In contrast, if populations are reproductively isolated, the genetic diversity and the effective population size within populations will be reduced.
In this study we examine the contemporary pollen and seed flow, intra-population spatial genetic structure, and effective population size of an isolated population (>1,000 km)of the tropical tree Myracrodruon urundeuva (F.F. & M.F. Allemão), occurring in a large forest fragment in Brazil. The study area is located in the Paulo de Faria Ecological Station, in the Northwest of São Paulo State, Brazil. This stand is one of the last remaining forest fragments in the Northwest of São Paulo State where M. urundeuva is found and therefore represents an important population for conservation and regeneration within the State.
Myracrodruon urundeuva is a bee- and wind-pollinated, dioecious species which is characterized by a small flower size. Insects, such as bees and trips have the potential to disperse pollen over long distances (Ghazoul 2005; Dick et al. 2008). This long distance dispersal is supported by empirical data based on highly polymorphic genetic markers such as microsatellite and paternity analysis approaches (White et al. 2002; Dick et al. 2003; Lacerda et al. 2008; Silva et al. 2008; Carneiro et al. 2009). Pollen dispersal has been found to reach distances greater than 800 m in various tropical species, including the insect-pollinated Swietenia humilis (White et al. 2002), bee-pollinated Dinizia excelsa (Dick et al. 2003) and trip-pollinated Bagassa guianesensis (Silva et al. 2008). Long pollen dispersal distance for insect-pollinated tree species have also been found in the dioecious generalist subtropical tree Eurycorymbus cavaleriei (>667 m, Wang et al. 2007) and temperate tree Sorbus domestica (up to 16 km, Kamm et al. 2009). Although these studies show that pollen dispersal can reach long distances, the frequency of these events is low and the majority of the mating occurs between the nearest neighbors (Byrne et al. 2008; Lacerda et al. 2008; Silva et al. 2008; Carneiro et al. 2009; Kamm et al. 2009). Nearest neighbor mating is the result of the foraging behavior of many insects, such as bees, which tend to move from one flowering tree to the nearest flowering tree (Dick et al. 2008). However, forest fragmentation may breakdown this pattern of nearest neighbor mating, as has been found in isolated trees and populations occurring in highly fragmented landscapes, including of Symphonia globulifera (Aldrich and Hamrick 1998), Swietenia macrophylla (White et al. 2002) and Dinizia excelsa (Dick et al. 2003). Asynchronous flowering, pollen carryover and the inability of some small insects to perceive flowering trees beyond a specific distance can also explain the breakdown in nearest neighbor foraging (Dick et al. 2008).
Long distance seed dispersal is also important in connecting isolated populations and colonization (Kamm et al. 2009). In diploid species, seed flow introduces two alleles in each locus in the population and has evolutionary advantages for regeneration over pollen as it is far more likely to contribute to the next generation (Kramer et al. 2008). Wind dispersal of seeds also has the potential to reach long distances. In the tropical species Jacaranda copaia seed dispersal via wind has been shown to occur between 2.7 and 710.5 m (Jones et al. 2005). In the temperate tree Fraxinus excelsior, seeds has been found dispersed by wind at distances greater than pollen dispersal (Bacles et al. 2006). However, the porosity of the landscape may affect the distance of wind dispersed seeds. In open landscapes such as pastures, seeds are likely to be dispersed by wind in longer distance than within dense forests.
As noted above, the aims of this study are to investigate the levels of gene flow, the distance and the patterns of pollen and seed dispersal, the spatial genetic structure (SGS), and the effective population size of the spatially isolated M. urundeuva population using microsatellite loci. In order to address the aims of the study, the following questions were raised: (i) What are the levels of pollen and seed immigration and the distances and patterns of pollen and seed dispersal inside of the population? (ii) Is there SGS in the population? (iii) Are there differences in the levels of genetic diversity between adult-trees, juveniles and offspring? (iv) What is the effective population size of adults, juveniles and open-pollinated progeny?
Materials and methods
The study species
Myracrodruon urundeuva has a large geographic distribution in South America (3°30′ S to 25° S), occurring in Brazil, Argentina, Bolivia and Paraguay (Carvalho 2003). The species occurs in the Semidecidual Estacional Forest, Decidual Estacional Forest, Savanna, Caatinga, Chaco Sul Matrogrossense and Pantanal Mato-Grossense. The population density of the species varies widely, ranging from 2 to 115 trees per hectare (Carvalho 2003). Trees can reach 30 m in height and 100 cm in diameter at breast height (dbh), but the growth of the species is slow, ranging between 0.45 and 0.62 cm year−1 (Baleroni et al. 2003). The wood has a high economic value due to its quality and durability (Carvalho 2003). In southwest Brazil, especially in São Paulo State, naturally forested areas have been drastically reduced through conversion to agriculture (Ribeiro et al. 2009) and as a result M. urundeuva has become a rare species. The species is now found in only a few remaining small and isolated forest fragments of generally less than 10 ha.
Study site
The study was conducted in a forest fragment at Paulo de Faria Ecological Station (PFES), located in the Northwest of São Paulo State, in Brazil (Fig. 1). The forest fragment has 436 ha and is located on the south side of the Rio Grande (19°58′ S and 49°32′ W). The climate of the region is characterized by two climatic seasons: one dry, from April to September (average rainfall of 167 mm), and the other humid, from October to March (average rainfall of 978 mm) (Barcha and Arid 1971). The altitude ranges between 400 to 495 m and the vegetation is typically of Seasonal Semi-deciduous Forest (one of the sub-categories of the Atlantic Forest), characterized by a high diversity of tree species. A floristic study done in the fragment detected 70 tree species (Stranghetti and Ranga 1998). The fragmentation of this area occurred before 1980 because of land conversion for agricultural activities and in the past the forest stand was exploited using selective logging. The Ecological Station is geographically isolated by the river in the north and pastures and sugar-cane on the south, east and west sides. In this fragment, M. urundeuva occurs mainly grouped in the Northern part of the forest, near to the river (Fig. 1). In this section 470 M. urundeuva adult-trees were found, with a diameter at breast height (dbh) ranging between 11.03 and 94.54 cm (28.62 ± 13.52 cm; mean ± standard deviation; Fig. 2) and ages likely ranging between 20 and 210 years. Many of these trees are remnants of the pre-fragmentation population. This area also features intense M. urundeuva regeneration, with dbh ranging from 0.64 to 10.98 cm (7.76 ± 2.39 cm), suggesting that these individuals are between 1 and 20 years of age.
Sample size
For this study a census was carried out in the PFES in 2008. All adult-trees identified in the census were sampled, mapped and their dbh measured. However, due to poor quality DNA samples, a male and two females were not genotyped, and the analysis was carried out with 467 individuals. The open-pollinated seeds used for parentage analysis in this study were collected 16 year ago (see below). To ensure consistency with previous methodology, only trees in the stand with a dbh of 11 cm or more (age ≥19 years) were considered reproductive for this study; in progeny tests has been observed that M. urundeuva starts flowering at three years of age (trees with dbh of about 2 cm), although the intensity is very low (Moraes et al. 1992). Of the 467 adult trees, 175 were female, 268 were male and 24 adult trees were not sexed due the absence of flowers.
To study the seed and pollen dispersal distance and spatial genetic structure of the regeneration, a sample of 149 juveniles and 29 open-pollinated families established in a progeny test was carried out. Juveniles represent different reproductive events and were sampled within circular subplots with a radius of 30 m around 30 random female-trees. The dbh of the female-trees ranged from 19.7 to 94.5 cm. These 30 female-trees were the source of open-pollinated seeds which were used to establish a progeny test at Selvíria Experimental Station (20°20′ S, 51°23′ W and 370 m of altitude) in Selvíria, Mato Grosso do Sul State in 1992. In the trial, 20 offspring were sampled from each family, totaling 600 offspring. However, from these 600 offspring, 86 were excluded from the analysis because they were genotyped with less than four loci (missing data in more than one locus). The number of sampled families was therefore reduced to 29. The cambium tissue was sampled from 11 of the largest trees, leaves in other adult trees, juveniles and offspring. The cambium tissue was preserved in Eppendorf tubes with a solution of CTAB buffer (1/3) and ethanol (2/3) and stored at −20°C before DNA extraction. Leaf samples from juveniles and offspring were stored in silica gel at room temperature before DNA extraction.
Microsatellite analysis
DNA extraction followed the CTAB protocol (Doyle and Doyle 1990), with the following modifications: the addition of proteinase K (100 ug/ml), PVP-40 (4%), PVP-360 (1%), and β mercaptoetanol (2%) in the extraction lid. The analysis of the microsatellite loci was conducted at the laboratory of Vegetable Genetics (CERNAGEN-EMBRAPA). The amplification reactions were carried out following Caetano et al. (2005), using five Auru initiators (C072, D094, A392, D200 and E062). The amplification reaction of 6 μl was realized using 1 ng genomic DNA, 200 μM of each dNTPs, 2.5 mM of MgCl2, 0.33 μM of each initiator, MilliQ sterile water, and 1× Go Taq Master Mix PROMEGA. The forward component of each primer pair was marked in the 5′ extremity with fluorophores. The amplification reactions were carried out in a thermocicler 9600 and 9700 (Applied Biosystem) programmed for the following conditions: a cycle of 94°C per 3 min., 35 cycles of 94°C per 30 s, primer pareament temperature per 30 s, 72°C per 30 s, and a final cycle of 72°C per 5 min. After the amplification products were diluted (1 μl of reaction to 5 μl of MilliQ water), they were mounted and a multiplex of 2 and 3 distinct amplified loci were prepared, using 1 μl of each amplified, 0.35 ul pattern of molecular size CENARGEN/ROX, 9 μl of formamide Hi Di and 0.65 μl of Milli Q autoclaved water. Subsequently, the samples were denatured at 94°C for 5 min and run on an ABI Prism 3700 capillary sequencer (Applied Biosystem). To detect and estimate the size of the alleles the software ABI Prism GeneScan version 3.7 (Applied Biosystems) was used. To filter the peaks and to interpret the genotypes of the individuals, the estimated size of the alleles were imported using the software ABI Prism Genotyper version 3.7 NT (Applied Biosystems). Finally, the values of the alleles were fitted into discrete categories, using the software AlelloBin (Idury and Cardon 1997).
Analysis of genetic diversity and fixation index
The genetic diversity of adults, juveniles and offspring was characterized by the number of alleles (k), allele richness (R), observed heterozygosity (H o), and expected heterozygosity under Hardy–Weinberg equilibrium (H e) for each locus and for all loci together. The allele richness was estimated by rarefaction (El Mousadik and Petit 1996). The levels of inbreeding in the samples were estimated using fixation index (adult-trees: F a; juveniles: F i; offspring: F o). The significance of the F values for each locus and for all loci together was calculated by 10,000 permutations and using a sequential Bonferroni correction for multiple comparisons (95%, α = 0.05). All analyses were run using program Fstat, version 2.9.3.2. (Goudet 1995).
Parentage analysis
The theoretical power to exclude the parent pair \( \left( {P_{Parent - pair} } \right) \) was calculated using the program CERVUS 3.0 (Marshall et al. 1998; Kalinowski et al. 2007). Parentage analysis was carried out by maximum-likelihood maternity and paternity assignment (Meagher 1986) based on multilocus genotype of 149 juveniles, 514 open-pollinated offspring and all 467 adult-trees, also using CERVUS 3.0. The most likely parents and parent pairs were determined by Δ statistic (Marshall et al. 1998) using the allele frequencies calculated in the adult population, as suggested by Meagher and Thompson (1987). To determine the likely mother and father of each juvenile, all 175 females were used as maternal candidates and all 268 males were used as paternal candidates. The significance of Δ was determined through maternity and paternity tests simulated by the software (critical Δ) using a confidence level of 80%. The calculation of critical Δ values used the following parameters: 90% of candidates sampled were located within the population, a genotyping error ratio of 0.01, and 100,000 repetitions. The confirmation of true parenthood (a male or female individually, or a pair) was obtained if an individual (male or female) or a pair of individuals had a calculated Δ index higher than the cryptic Δ derived from simulations. The cryptic gene flow (the probability of finding a compatible mother or father candidate inside of population, when the true parent was outside of the population) was calculated based on the method described by Dow and Ashley (1996). The seed immigration rate \( \left( {m_{Seeds} } \right) \) in juveniles was calculated as the proportion of juveniles that had no determined parents \( \left( {n_{immigrant(seed)} } \right) \) inside of population, relative to the total number of sampled juveniles \( \left( {n_{total} } \right) \): \( \hat{m}_{Seeds} = \hat{n}_{immigrant(seed)} /n_{total} \). The pollen immigration rate \( m_{Pollen} \) in juveniles and offspring was calculated by the proportion of juveniles or offspring that had no pollen parent \( \left( {n_{immigrant(pollen)} } \right) \) inside of the stand in relation to the total number of sampled juveniles or offspring \( \left( {n_{total} } \right) \): \( \hat{m}_{Pollen} = \hat{n}_{immigrant(pollen)} /n_{total} \).
As all sampled individuals were mapped (x and y coordinates) before sample collection, the seed dispersal distance was calculated based on the location of the juveniles relative to their putative mothers. The distance of pollen dispersal was calculated based on the position of the identified mothers in relation to the identified father. In order to investigate if mating success was a function of distance between trees, we compared the frequency distribution of realized pollination with the frequency distribution of the distances among all trees in the fragment using the Kolmogorov–Smirnov test (Sokal and Rohlf 1995). To confirm the parent assignments, the coancestry coefficient between juveniles/offspring and identified parents inside of the population was also calculated. The expected coancestry coefficient value for a parent-sib pair is 0.25 for which the method of J. Nason (Loiselle et al. 1995) and the program Spagedi 1.3 (Hardy and Vekemans 2002) was used.
Spatial genetic structure analysis
Spatial genetic structure (SGS) was examined using the average coancestry coefficient (θ xy) between adult-tree and juvenile pairs. The θ xy values were calculated using the program Spagedi 1.3. The hypothetical coancestry value θ xywas estimated using the allele frequencies of the parent population. This is expected to produce hypothetical values of coancestry between two relatives (half-sibs = 0.125; full-sibs = 0.25; cousins = 0.0625; Hardy and Vekemans 2002). We used the allele frequencies calculated in adult-trees for the analysis of juveniles. For the analysis of adult trees, we assumed that the population has the same allele frequencies as that found in the present adult generation. To visualize the spatial genetic structure, θ xy values were averaged over a set of distance classes, and plotted against the distances. We used 5 m distance intervals for both adults and juveniles with a maximum distance of 70 m. In order to test whether there was significant deviations from SGS, the 95% confidence interval was calculated for each observed value and each distance class, using 1,000 permutations of individuals among locations. We also estimated the coancestry coefficients between juveniles and the fathers and mothers assigned by paternity and maternity analysis (see above) and between juveniles determined as full-sibs, half-sibs and non-siblings.To compare the extent of SGS between adults and juveniles the statistic S p (Vekemans and Hardy 2004) was calculated as follows: \( - b_{k} /(1 - \theta_{1} ) \), where θ 1 is the average coancestry coefficient calculated between all pairwise individuals within the first distance class (0–5 m), and b k is the slope of the regression of coancestry coefficient on the logarithm of spatial distance (0–70 m). To test for SGS, spatial position of the individuals were permuted (10,000 times) to obtain the frequency distribution of b k under null hypothesis that θ 1 and ln(d xy) were uncorrelated.
Coefficient of coancestry and effective population size
The coancestry coefficient and the effective population size were calculated separately for adult-trees, juveniles and offspring. The effective population size (N e) for adult-tree was estimated as (Cockerham 1969):
The parameter Θ is the group coancestry in adult-trees, calculated following the approach for dioecious species proposed by Lindgren and Mullin (1998):
where: θ f, θ m, and θ fm are the coancestry coefficients between females, males, and males and females together, and n f and n m are the number of female and male adult-trees. The coancestry coefficients were calculated using Loiselle′s estimator (described above) and the program Spagedi. The effective population size (N e) for juveniles was calculated using the same expression as for adult-trees, but substituting the coefficient of inbreeding in the juveniles (F i) by F a. The group coancestry in juveniles was calculated by:
where: n is the sample size, \( n_{M(hs)} \) is the number of pairwise maternal half-sibs, \( n_{P(hs)} \) is the number of pairwise paternal half-sibs and, \( n_{M(fs)} \) is the number of pairwise full-sibs. It is important to note that the later expression is used for dioecious or self-incompatible species because the expression does not consider the self-coancestry of individuals (the coancestry between an individual and itself; Lindgren et al. 1996). As dioecious or self-incompatible species are non-selfing, it was excluded from the expression.
For offspring, the coancestry coefficient was calculated by \( \hat{\Uptheta } = 0.125(1 + \hat{F}_{a} )(1 + \hat{r}_{p} ) \), where F a is the inbreeding coefficient in the parental population, and r p is the paternity correlation (Sousa et al. 2005). The paternity correlation was calculated by \( \hat{r}_{p} = \sum\nolimits_{i = 1}^{{n_{p} }} {\hat{p}_{i}^{2} } \), where n p is the number of analyzed embryos within families and P t is the number of seeds fathered by a male-tree (i) within families. The effective number of pollen donors was calculated by \( \hat{N}_{ep} = 1/\hat{r}_{p} = 1/\sum\nolimits_{i = 1}^{{n_{p} }} {\hat{p}_{i}^{2} } \) (Burczyk et al. 1996). The effective population size for offspring was calculated using the same expression as for adult-trees and juveniles, but substituting the coefficient of inbreeding in the offspring (F o) by F a. The effective pollination neighbor area (A ep) was calculated for each seed-tree from the variance in the distance of pollen dispersal (σ2), assuming a circular area surrounding each seed-tree, \( \hat{A}_{ep} = 2\pi \hat{\sigma }^{2} \) (Levin 1988). The radius of pollen dispersal was calculated from A ep by \( \hat{r}_{ep} = \sqrt {\hat{A}_{ep} /3.1415} \) (Smouse et al. 2001).
Results
Genetic diversity and fixation index
From the DNA analysis of five loci and the total sample of 1130 genotypes (adults + juveniles + offspring), 60 alleles were detected. The number of alleles per locus ranged from 7 (SR62) to 25 (SR72), with an average of 12 alleles per locus. The observed heterozygosity ranged from 0.472 to 0.839, with average of 0.713 and the expected heterozygosity ranged from 0.581 to 0.866, with average of 0.662. The fixation index ranged from −0.348 to 0.215, with average of -0.077 (P < 0.05).
From the five loci, 60 alleles were found in adult-trees, 41 in juveniles, and 50 alleles in offspring (Table 1). Within the population, 19 alleles were found exclusively in adult-trees, suggesting genetic drift during mating. The average number of alleles per locus in adults \( \left( {\hat{A} = 1 2\pm 1. 6 3} \right) \) was significantly higher than in juveniles \( \left( {\hat{A} = 8. 2\pm 0. 7 8} \right) \) and offspring \( \left( {\hat{A} = 10 \pm 0. 5 4} \right) \). However, the average allele richness (R), estimated by rarefaction, and both the observed and expected heterozygosities were not significantly different between the adults, juveniles and offspring. The fixation index was also significantly (P < 0.05) lower in adults \( \left( {\hat{F}_{a} = - 0. 1 5 3\pm 0.0 6 4} \right) \) than juveniles \( \left( {\hat{F}_{j} = - 0.0 1 8\pm 0.0 6 9} \right) \) and offspring \( \left( {\hat{F}_{o} = - 0.0 5 3\pm 0.0 5 3} \right) \). Using a sequential Bonferroni correction (95%, α = 0.05), the fixation index in three of the five loci were significantly different from zero in adults, juveniles and offspring (Table 1). The average fixation index for all loci was also statistically significantly different from zero for adult-trees and offspring.
Parentage analysis
The hypothetical exclusion power of the parent-pair was relatively high, 0.993 (Table 1), and the probability of cryptic gene flow was also high (0.42). Thus, about 42% of assigned maternity and paternity matches could be incorrect. A probable mother-tree was found inside of the stand for each of 149 sampled juveniles (Table 2). Of 175 candidate mother-trees, 74 (42%) mothered at least one juvenile. The number of juveniles mothered by these mother-trees ranged from 1 to 11. No significant association was detected between the number of juveniles mothered by each mother-tree and their respective dbh \( \left( {\hat{r} = - 0. 3 6 8, \, P > 0.0 5,{\text{ df}} = 7 2} \right) \). A probable father-tree was found for 145 (97.3%) of the 149 studied juveniles and for 506 (98.4%) of the 514 offspring. Of the 268 candidate male-trees, 78 (29%) fathered at least one juvenile (ranged from 1 to 7) and 138 (51.5%) fathered at least one offspring (ranged from 1 to 23). No significant association was detected between the number of juveniles \( \left( {\hat{r} = - 0. 20 1, \, P > 0.0 5,{\text{ df}} = 7 6} \right) \) and offspring \( \left( {\hat{r} = - 0.0 6 5, \, P > 0.0 5,{\text{df}} = 1 3 6} \right) \) fathered by each pollen donor and their respective dbh. No mismatching was observed between offspring and their respective seed-tree, indicating that the progeny trial was established with the correctly sampled family. To confirm these mother and father assignments, we estimated the coancestry coefficient between identified mother-trees and juveniles and father-trees with juveniles and offspring. The estimated average coancestry coefficient between assigned mother-trees and juveniles (\( 0.326 \pm 0.037 \), ± CI95%), putative father-trees and juveniles \( \left( {0.288 \pm 0.046} \right) \), and putative father-tree and offspring (0.234 ± 0.012) were significantly different from zero.
Seed and pollen dispersal distance
The distance of seed dispersal measured by paternity analysis ranged between 0.8 and 887 m, with an average of 124 ± 150 m (± SD) and median of 46.3 m (Table 2). The results indicate that about 53% of seeds grow within a 50 m of radius of the mother-tree and about 90% grow within 400 m of the mother-tree (Fig. 3a). The correlation between the number of juveniles mothered by mother-trees and the distance from mother-tree was statistically significant (\( \hat{r} = - 0.83,P < 0.01 \), df = 72).
Pollen was also predominantly dispersed in short-distances, ranging from 2.9–863 m, with an average of 138 ± 169 m (median of 64.4 m), for juveniles and a range of 3.1–890 m, with an average of 252 ± 204 m (median of 191.7 m), for offspring (Table 2, Fig. 3b and 4). These average distances are shorter than the average distance between all male and female-trees (374 ± 235 m) in the stand. For juveniles, about 70% of the pollen was dispersed in distances less that 200 m and 90% of the pollen traveled less than 500 m (Fig. 3b). For offspring, about 87% of the pollen was dispersed in distances less that 500 m (Fig. 4). No juvenile or offspring was fathered by a group of trees further than 4 km (Fig. 1). A significant and strong negative correlation was detected between the number of juveniles \( \left( {\hat{r} = - 0.96,P < 0.01,{\text{ df}} = 7 6} \right) \) and seeds \( \left( {\hat{r} = - 0.92,P < 0.01,{\text{ df}} = 1 3 6} \right) \) fathered by the pollen-donor and the distance between mother and father. The comparison of the curve of frequency of pollen dispersal and the curve of frequency between all male and female-trees rejected the hypothesis of random mating for both juveniles (Kolmogorov–Smirnov test: \( D = 0.713, \) \( P = 0.0001 \)) and offspring (Kolmogorov–Smirnov test: \( D = 0.224, \) \( P = 0.0001 \)).
Reproductive effective neighbor area
The reproductive neighbor area was estimated for each family (Table 3). The result varied substantially among seed-trees, indicating that seed-trees received pollen from male-trees within circles of 3.8–56.1 ha, with an average of 22.1 ha. This produces a radius of effective pollen dispersal ranging from 110 to 422 m, with average of 251 m.
Spatial genetic structure (SGS)
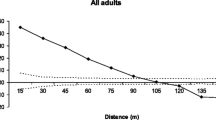
Significant SGS was detected in both adult and juvenile stages (Fig. 5a, b). In adults, the estimated coancestry coefficient (θ xy) was significantly positive in the distance class 0 to 42 m and in juveniles in the distance class 0–32 m, suggesting isolation by distance. In adult trees, the θ xy value in the first distance class \( \hat{\theta }_{xy} = 0.0 5 9 \) was similar to that expected between first-cousins \( \left( {\hat{\theta }_{xy} = 0.0 6 2 5} \right) \) and in juveniles \( \left( {\hat{\theta }_{xy} = 0. 1 2 1} \right) \) it was similar to the expected value between half-sibs \( \left( {\hat{\theta }_{xy} = 0. 1 2 5} \right) \). The regression slope b k of pairwise coancestry coefficient on the logarithm of spatial distance (0–70 m) was significantly negative for both adults (b k = −0.0253, P < 0.001) and juveniles (b k = −0.0217, P < 0.001), thus confirming the presence of isolation by distance. The intensity of SGS as measured by S p statistic, was similar in adults (S p = 0.0269) and juveniles (S p = 0.0246). These results indicate a strong short distance SGS for both adults and juveniles.
Correlograms of average coancestry coefficient (θ xy) for (a) 14 distance classes of adults and (b) 14 distance classes of juveniles of M. urundeuva, each with intervals of 5 m. The solid line represents the average θ xy values. The dashed lines represent the 95% (two-tailed) confidence interval of the average θ xy distribution calculated by 1000 permutation of spatial distance among pairs of adults and juveniles
Coancestry coefficient and effective population size
Both the coancestry coefficient between sexes and the effective population size were estimated for the 443 adult-trees whose sex was determined (Table 3). The average coancestry coefficient between females (θ xyf), males and females (θ mf), and male trees (θ m) were 0.000359, 0.00000409, and −0.00003869 (assumed as zero), respectively. Thus, in random mating a low level of biparental inbreeding is expected in this population (F << 1%). From the effective population size it was determined that of the 443 adult-trees whose sex was identified in this study, 335 individuals were neither inbred, nor related \( \left( {\hat{N}_{e} /N = 0.76} \right) \).
In juveniles the sex was not determined; therefore, the coancestry coefficient was estimated for 145 juveniles that had both identified parents within the stand (Table 3). From all paired juveniles (10,440 pairs), 2% were maternal half-sibs, 0.29% full-sibs, 1.05% paternal half-sibs and 96.66% were not related. The average coancestry coefficient was 0.00454 and the effective population size was 62.9. Thus, the 145 juveniles consist of 63 individuals that are neither inbred nor relatives \( \hat{N}_{e} /N = 0.43 \).
For open-pollinated seeds, the paternity correlation (r p) ranged among families from 0.061 to 0.167, with an average of 0.100, indicating that the offspring are comprised of mixtures of half- and full-sibs (Table 3). The effective number of pollen donors (N ep) ranged among families from 6 to 16.61, with an average of 10.83. The average coancestry coefficient and the effective population size within families was lower than expected in open-pollinated seeds collected from panmictic populations (Θ = 0.125; N e = 4).
Discussion
Stand isolation
The results of parentage analysis in this study show a total absence of seed immigration in established juveniles and low levels of pollen immigration in both juveniles (2.7%) and offspring (1.6%). Myracrodruon urundeuva seeds are dispersed by anemochory and trees are pollinated mainly by bees (Nunes et al. 2008). Theoretically, anemochory and bees have the potential to disperse seeds and pollen over long distances; however, many factors can limit the distance of such dispersal methods such as high vegetal density and the absence of other seed and pollen sources surrounding the stand. The high vegetal density of the studied stand (personal observation) can limit both seed and pollen dispersal due to the low porosity inside of the stand. With regards to pollen, some studies have shown that vegetal density can limit the distance of pollen dispersal of both wind (Dyer and Sork 2001; Robledo-Arnuncio et al. 2004) and insect-pollinated tree species (Sork et al. 2005). Thus, the high vegetal density of the stand likely contributes to the observed absence of seed immigration and low pollen immigration into the stand. However, we believe that a more probable cause of the low levels of immigration is the absence of others seed and pollen sources in the vicinity of the stand. We observed some small, isolated forest fragments in the region, but M. urundeuva was not represented in these fragments. Moreover, it is important to replicate the analysis of seed and pollen immigration using more loci in order to decrease the observed rate of cryptic gene flow and to confirm the observed patterns and high levels of isolation. Nevertheless, the detected high levels of genetic isolation suggest an increase in the genetic differentiation of this isolated population in relation to other populations of the species. The genetic isolation of the population will likely result in a reduction of genetic diversity and effective population size in future generations.
Seed dispersal distance
The results of this study show a high frequency of seeds dispersed over short distances in relation to the mother-tree (Fig. 3a). The results also show a strong association between the number of juveniles mothered by mother-trees and their respective distance from the mother-tree, suggesting isolation by distance. This short-distance seed dispersal can explain the observed SGS in both adult trees and juveniles (Figs. 4 and 5). A high frequency of short-distance seed dispersal has been found in other wind seed dispersed tropical tree species. Examining two yearly events of seed dispersal in the tropical tree Jacaranda copaia, Jones et al. (2005) found seed dispersal distances ranging from 2.7 to 421.8 m, with average of 40.1 m, in 2000, and from 2.9 to 710.5 m, with average of 58.8 m, in 2002. This range of seed dispersal is similar to that observed here (between 0.8 and 887 m), although the average was smaller than detected in M. urundeuva (124 ± 150 m). However, the average seed dispersal distance in Jacaranda copaia may be underestimated due the fact that the authors were able to locate the mother tree of only 23% of the sampled seeds in 2000 and 11% in 2002. A high frequency of short-distance seed dispersal has been also found in the wind seed dispersed temperate tree species Fraxinus excelsior (38–100 m), although the majority occurred over long distances (53% > 2.7 km, Bacles et al. 2006). The authors concluded that the deforested landscape in their case study favored long-distance seed and pollen dispersal. Short-distance seed dispersal has also been found in the animal seed dispersed subtopical/tropical tree Araucaria angustifolia (~ 80% under 160 m, Bittencourt and Sebbenn 2007) and animal seed dispersed tropical tree Copaifera langsdorffii (82% under 100 m, Sebbenn et al. 2009). In contrast, long distance seed dispersal has been found in vertebrate seed dispersed species, such as the tropical tree Simarouba amara (Hardesty et al. 2006) and the tropical palm Iriartea deltoidae (mean 391.6 m, Sezen et al. 2005).
Pollen dispersal distance
The observed pattern of pollen dispersal was the tipical near-neighbor with a high frequency of short-distance dispersal, with mating probabilities rapidly decreasing with distance (Table 2, Figs. 3 and 4). The results also indicate that the reproductive neighbor area of individual seed-trees is smaller than the total area of the fragment (maximum 56.1 ha) because nearer male trees contribute to a higher fraction of pollination events than more distant male trees. Bees are able to travel long distances during foraging; however, many bee species show local site fidelity or are territorial (Ghazoul 2005). For example, forest honeybees forage in very small patches (a few tens of meters wide) and other bees tend to transport pollen between only one or a few adjacent individuals (Wilmer et al. 1994). This pattern of foraging can explain the observed short-distance pollen dispersal. Another factor could be the high vegetal density of the stand which can restrict the movement of bees among more distant conspecifics, as already discussed.
Intra-population spatial genetic structure
Short-distance seed dispersal resulted in family structures inside of the stand. We found SGS in both adults and juveniles and a higher level of SGS in juveniles compared to adult-trees. The higher level of SGS in juveniles than adult-trees indicates that the relatedness inside of the stand is increasing. This is likely due to the isolation of the population and short seed and pollen dispersal distances. Since the population does not receive new immigrants and the levels of pollen immigration is also low, mating predominantly occurs only between individuals within the population thus increasing relatedness in subsequent generations. In reproductive populations with SGS and a high frequency of short-distance pollen dispersal, mating among relatives is inevitable. Mating among relatives produces biparental inbreeding. However, using the mixed mating model and MLTR program (Ritland 2002), we did not find significant mating among relatives (t m − t s = 0.007, P > 0.05) and inbreeding in offspring (Table 1). This discrepancy can be explained by inbreeding depression which eliminates inbred individuals between the fertilization event and the moment of genotype analysis by microsatellite markers. Inbreeding depression changes the genetic structure between different ontogenetic stages and has been reported in some tree species, such as Pseudobombax munguba (Gribel and Gibbs 2002), Platypodium elegans (Hufford and Hamrick 2003), and Neobalanocarpus heimii (Naito et al. 2005). Within the present M. urundeuva population, inbreeding depression is possible considering that the juveniles are individuals established in the population and the studied offspring were established in progeny tests.
Genetic diversity of different ontogenetic stages
The results of this study show that the effects of stand isolation on genetic diversity in different ontogenetic stages are unclear as the allele richness and heterozygosities were not significantly different between generations (Table 1). The main explanation for this lack of evidence is the long-life of the species and the small number of generations which have developed after isolation, as has been discussed in other studies (Lowe et al. 2005; Kramer et al. 2008; Aguilar et al. 2008). Another issue affecting the results is the slow depletion of expected heterozygosity by genetic drift in large populations (Savolainen and Kärkkäinen 1992).
Effective population size
The effective population size in both adults and juveniles was lower than the number of individuals in the population and this difference was greater among the juvenile population than the adult (Table 3). This reflects the relatedness between trees inside of the stand; as the relatedness was higher among juveniles than adults, the relationship \( N_{e(adults)} /N \) was lower in juveniles. These results suggest that the frequency of related individuals is increasing in new generations of the population, a phenomenon expected in isolated populations. Regarding the open-pollinated families, the effective population size (N e) was lower than expected in open-pollinated seeds collected from panmictic populations. This result is due to non-random mating (correlated mating) thus indicating that seeds for conservation genetics must be collected from a large number of seed-trees. Assuming that (i) seeds will be collected from non related seed-trees, and (ii) the seed-trees sampled are from different pollen pools, the number of seed-trees needed for seed collection can be determined by dividing the effective population size intended for conservation by the average effective population size within families, \( \hat{m} = N_{e(reference)} /\hat{N}_{e(within\_familes)} \) (Sebbenn 2002). Thus, for example, to conserve an effective population size of 150 (3 × 50, Nunney and Campbell 1993), the necessary number of seed-trees for seed collection in this population would be 48.
Conclusion
In conclusion, due to the large distances between the stand and other conspecifics or populations of the species, no seed immigration was detected in the stand and only low levels of pollen immigration were observed. Inside of the stand, a high frequency of seeds and pollen were dispersed in short-distances, thus explaining the observed patters of SGS. Consequently, the population was organized in family structures, resulting in a reduction in the effective population size of both adult and juvenile generations in relation to the total number of individuals in the population. This study has also shown that it is necessary to collect seeds from a large number of seed-trees in order to compensate for the increase in the relatedness within sampled families due to non-random mating. Such results indicate the need to consider pollen and seed dispersal and SGS when designing sampling strategies for seed collection in conservation, breeding and environmental restoration projects.
References
Aguilar R, Quesada M, Ashworth L, Herrerias-Diego Y, Loco J (2008) Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol Ecol 17:5177–5188. doi:j.1365-294X.2008.03971.x
Aldrich PR, Hamrick JL (1998) Reproductive dominance of pasture trees in a fragmented tropical forest mosaic. Science 281:103–105. doi:10.1126/science.281.5373.103
Bacles CFE, Lowe A, Ennos R (2006) Effective seed dispersal across a fragmented landscape. Science 311:628. doi:10.1126/science.1121543
Baleroni CRS, Alves PF, Santos EBR, Cambuim J, Andrade JAC, Moraes MLT (2003) Variação genética em populações naturais de aroeira em dois sistemas de plantio. Rev Inst Flor 15:125–136
Barcha SF, Arid FM (1971) Estudo de evapotranspiração na região norte-ocidental do Estado de São Paulo. Rev Ciências 1:99–122
Bawa KS (1994) Pollinators of tropical dioecious angiosperms: a reassessment? No, not yet. Am J Bot 81:456–460
Bittencourt JM, Sebbenn AM (2007) Patterns of pollen and seed dispersal in a small fragmented population of a wind pollinated Araucaria angustifolia in southern Brazil. Heredity 99:580–591. doi:10.1038/sj.hdy.6801019
Bittencourt JM, Sebbenn AM (2008) Pollen movement in a continuous forest of Araucaria angustifolia, inferred from paternity and TwoGener analysis. Conserv Genet 9:855–868. doi:10.1007/s10592-007-9411-2
Burczyk J, Adams WT, Shimizu JY (1996) Mating patterns and pollen dispersal in a natural knobcone pine (Pinus attenuata Lemmon) stand. Heredity 77:251–260. doi:10.1038/hdy.1996.139
Byrne M, Elliott CP, Yates CJ, Coates DJ (2008) Maintenance of high pollen dispersal in Eucalyptus wandoo a dominant tree of the fragmented agricultural region in Western Australia. Conserv Genet 9:97–105. doi:10.1007/s10592-007-9311-5
Caetano S, Silveira P, Spichiger R, Naciri-Graven Y (2005) Identification of microsatellite markers in a neotropical seasonally dry forest tree, Astronium urundeuva (Anacardiaceae). Mol Ecol Notes 5:21–23. doi:10.1111/j.1471-8286.2004.00814.x
Carneiro FS, Degen B, Kanashiro M, Lacerda AEB, Sebbenn AM (2009) High levels of pollen dispersal in Symphonia globulifera in a dense Brazilian Amazon forest revealed by paternity analysis. Forest Ecol Manag 258:1260–1266. doi:10.1016/j.foreco.2009.06.019
Carvalho PER (2003) Espécies arbóreas brasileiras. Embrapa Informação Tecnológica, Brasília
Cockerham CC (1969) Variance of gene frequencies. Evolution 23:72–84
Dick CW, Etchelecu G, Austerlitz F (2003) Pollen dispersal of Neotropical trees (Dinizia excelsa: Fabaceae) by native insects and Africa honeybees in pristine and fragmented Amazonian rainforest. Mol Ecol 12:753–764. doi:10.1046/j.1365-294X.2003.01760.x
Dick CW, Hardy OJ, Jones FA, Petit RJ (2008) Spatial scales of pollen and seed-mediated gene flow in tropical rain forest trees. Tropic Plant Biol 1:20–33. doi:10.1007/s12042-007-9006-6
Dow BD, Ashley MV (1996) Microsatellite analysis of seed dispersal and parentage of sampling in bur oak, Quercus macrocarpa. Mol Ecol 5:615–627. doi:10.1111/j.1365-294X.1996.tb00357.x
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dyer RJ, Sork LJ (2001) Pollen pool heterogeneity in shortleaf pine, Pinus schinata Mill. Mol Ecol 10:859–866. doi:10.1046/j.1365-294X.2001.01251.x
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839. doi:10.1007/BF00221895
Ghazoul J (2005) Pollen and seed dispersal among dispersed plants. Biol Rev 80:413–443. doi:10.1017/S1464793105006731
Goudet J (1995) Fstat. (Version 2.9.3.2.): a computer program to calculate F-statistics. J Heredity 86:485–486
Gribel R, Gibbs PE (2002) High outbreeding as a consequence of selfed ovule mortality and single vector bat pollination in the Amazonian tree Pseudobombax munguba (Bombacaceae). Int J Plant Sci 163:1035–1043. doi:10.1086/342518
Hamilton MB (1999) Tropical tree gene flow and seed dispersal. Nature 401:129–130. doi:10.1038/43597
Hardesty BD, Hubbell S, Bermingham E (2006) Genetic evidence of frequent long-distance recruitment in a vertebrate-dispersed tree. Ecol Lett 9:516–525. doi:10.1111/j.1461-0248.2006.00897.x
Hardy O, Vekemans X (2002) SPAGeDI: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620. doi:10.1046/j.1471-8286.2002.00305.x
Hufford KM, Hamrick JL (2003) Variability selection at three early life stages of the tropical tree Platypodium elegans (Fabaceae, Papilonoidae). Evolution 57:518–526
Idury RM, Cardon LR (1997) A simple method for automated allele binning in microsatellite markers. Genome Res 11:104–1109. doi:10.1101/gr.7.11.1104
Jones FA, Chen J, Weng G-J, Hubbell SP (2005) A genetic evaluation of seed dispersal in the Neotropical tree, Jacaranda copaia (Bignonianaceae). Am Nat 166:543–555. doi:10.1086/491661
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how computer program Cervus accommodates genotyping error increase success in paternity assignment. Mol Ecol 16:1099–1106. doi:10.1111/j.1365-294X.2007.03089.x
Kamm U, Rotach P, Gugerli F, Siraky M, Edwards P, Holderegger R (2009) Frequent long-distance gene flow in a rare temperate forest tree (Sorbus domestica) at the landscape scale. Heredity. doi:10.1038/hdy.2009.70
Kramer AT, Ison JL, Ashley MV, Howe HF (2008) The paradox of forest fragmentation genetics. Conserv Biol 22:878–885. doi:10.1111/j.1523-1739.2008.00944.x
Lacerda EBL, Sebbenn AM, Kanashiro M (2008) Long-pollen movement and deviation of random mating in a low-density continuous population of Hymenaea courbaril in the Brazilian Amazon. Biotropica 40:462–470. doi:10.1111/j.1744-7429.2008.00.402.x
Levin DA (1988) The paternity pool plants. Am Nat 132:309–317
Lindgren D, Mullin TJ (1998) Relatedness and status number in seed orchard crops. Can J For Res 28:276–283. doi:10.1139/cjfr-28-2-276
Lindgren D, Gea L, Jefferson P (1996) Loss of genetic diversity by status number. Silva Genet 45:52–59
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Lowe AJ, Boshier D, Ward M, Bacles CFE, Navarro C (2005) Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for Neotropical trees. Heredity 95:255–273. doi:10.1038/sj.hdy.6800725
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655. doi:10.1046/j.1365-294x.1998.00374.x
Meagher TR (1986) Analysis of paternity within a natural population of Chamaelirium luteum. 1. Identification of most-likely male parents. Am Nat 128:199–215
Meagher TR, Thompson E (1987) Analysis of parentage for naturally established seedlings of Chamaelirim luteum (Liliaceae). Ecology 68:803–812. doi:10.2307/1938351
Moraes MLT, Kageyama PY, Siqueira ACMF, Kano NK, Cambuim J (1992) Variação genética em duas populações de aroeira (Astronium urundeuva—Fr. All.—Engl.—Anacardiaceae). Rev Inst Flor 4:1241–1245
Naito Y, Konuma A, Iwata H, Suyama Y, Seiwa K, Okudo T, Lee SL, Muhammad N, Tsumura Y (2005) Selfing and inbreeding depression in seeds and seedlings of Neobalanocarpus heimii (Dipterocarpaceae). J Plant Res 118:423–430. doi:10.1007/s10265-005-0245-z
Nunes YRF, Fagundes M, Almeida HS, Veloso MDM (2008) Aspectos ecológicos da aroeira (Myracrodruon urundeuva Allemão- Anacardiaceae): fenologia e germinação de sementes. Rev Árv 32:233–243. doi:10.1590/S0100-67622008000200006
Nunney L, Campbell KA (1993) Assessing minimum viable population size: demography meets population genetics. Tree 8:234–239. doi:10.1016/0169-5347(93)90197-W
Ribeiro MC, Metzger JP, Martensen AC, Ponzoni FJ, Hirato MM (2009) The Brazilian Atlantic Forest: how much is left, and how is the remaining forest distributed? Implictions for conservation. Biol Conserv 142:1141–1153. doi:10.1016/j.biocon.2009.02.021
Ritland K (2002) Extensions of models for the estimation of mating systems using n independent loci. Heredity 88:221–228. doi:10.1038/sj.hdy.6800029
Robledo-Arnuncio JJ, Smouse PE, Gil L, Alía R (2004) Pollen movement under alternative silvicultural practices in native population of Scots pine (Pinus sylvestris L.) in central Spain. Forest Ecol Manag 197:245–255. doi:10.1016/j.foreco.2004.05.016
Savolainen O, Kärkkäinen K (1992) Effect of forest management on gene pools. New Forest 6:329–345
Sebbenn AM (2002) Número de árvores matrizes e conceitos genéticos na coleta de sementes para reflorestamentos com espécies nativas. Rev Inst Flor 14:115–132
Sebbenn AM, Carvalho ACM, Freitas MLM, Moraes SMB, Gaiano APSC, Silva JM, Jolivet C, Moraes MLT (2009) Low level of realized seed and pollen gene flow and strong spatial genetic structure in a small, isolated and fragmented population of the tropical tree Copaifera langsdorffii Desf. Heredity (in press)
Sezen UU, Chazdon RL, Holsinger KE (2005) Genetic consequences of tropical second-growth forest regeneration. Science 307:891. doi:10.1126/science.1105034
Silva MB, Kanashiro M, Ciampi AY, Tompson I, Sebbenn AM (2008) Genetic effects of selective logging and pollen gene flow in a low-density population of the dioecious tropical tree Bagassa guianensis in the Brazilian Amazon. Forest Ecol Manag 255:1548–1558. doi:10.1016/j.foreco.2007.11.012
Smouse PE, Dyer RJ, Westfall RD, Sork VL (2001) Two-generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evolution 55:260–271
Sokal RR, Rohlf FJ (1995) Biometry: principles and practices of statistics in biological research, 3rd edn. W.H. Freeman and Company, New York
Sork VL, Smouse PE, Apsit VJ, Dyer RJ, Westfall RD (2005) A Two-generation analysis of pollen pool genetic structure in flowering dogwood, Cornus florida (Cornaceae), in the Missouri Ozarks. Am J Bot 92:261–271
Sousa VA, Sebbenn AM, Hattemer H, Ziehe M (2005) Correlated mating in populations of a dioecious Brazilian conifer Araucaria angustifolia (Bert.) O. Ktze. For Genet 12:107–119
Stranghetti V, Ranga NT (1998) Levantamento florístico das espécies vasculares da floresta estacional mesófila semidecídua da Estação Ecológica de Paulo de Faria–SP. Rev Bras Bot 21:295–304. doi:10.1590/S0100-84041998000300008
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analysis in plant populations. Mol Ecol 13:921–935. doi:10.1046/j.1365-294X.2004.02076.x
Wang J, Ye Q, Kang M, Huang H (2007) Novel polymorphic microsatellite loci and patterns of pollen-mediated gene flow in an ex situ population of Eurycorymbus cavaleriei (Sampindaceae) as revealed by categorical paternity analysis. Conserv Genet 9:559–567. doi:10.1007/s10592-007-9369-0
White GM, Boshier DH, Powell W (2002) Increased pollen flow counteracts fragmentation in tropical dry forest: an example from Swietenia humilis Zuccarini. Proc Natl Acad Sci USA 99:2038–2042. doi:10.1073/pnas.042649999
Wilmer P, Gilbert F, Ghazoul J, Zalat S, Semida F (1994) A novel form to territoriality—daily paternal investment in an anthophorid bee. Anim Behav 48:535–549
Young AG, Boyle TJ (2000) Effects of logging and other forms of harvesting on genetic diversity in humid tropical forest. For Conserv Genet, pp 115–123
Young AG, Boyle T, Brown ADH (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418. doi:10.1016/0169.5347(96)10045-8
Acknowledgements
This study was supported financially by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP contract number: 06/53357-0). The article was part of the doctoral thesis of A.P.S.C. Gaino in the Faculdade de Engenharia de Ilha Solteira/UNESP, Ilha Solteira, São Paulo, Brazil. We would like to thank to José Cambuim for his assistance in sample collection and Selma M.B. Moraes, Juliana P. Moreira, and Laila T. Cardin for their lab work. A.P.S.C. Gaino would like to thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for the support of a doctoral scholarship at Faculdade de Engenharia de Ilha Solteira/UNESP (Brazil). The author M.A. Moraes and P.F. Alves thank FAPESP for the support of a Graduate scholarship. The author A.M. Sebbenn and M.L.T. Moraes thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for providing each with a Research Fellowship. We are also grateful to Dr. André E.B. Lacerda and Dr. Evelyn Nimmo for their suggestions and English correction in a previous version of this manuscript. The authors also are very grateful to an anonymous reviewer for corrections, suggestions and constructive criticism of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gaino, A.P.S.C., Silva, A.M., Moraes, M.A. et al. Understanding the effects of isolation on seed and pollen flow, spatial genetic structure and effective population size of the dioecious tropical tree species Myracrodruon urundeuva . Conserv Genet 11, 1631–1643 (2010). https://doi.org/10.1007/s10592-010-0046-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-010-0046-3