Abstract
Extensive realized pollen and seed flow across populations reduces inbreeding and spatial genetic structure (SGS) and increases the genetic diversity and effective size within populations. Inbreeding, SGS and realized patterns of pollen and seed dispersal of the dioecious, wind pollinated Araucaria angustifolia were investigated based on microsatellite loci. The study was conducted in a 7.2 ha plot established within a continuous Araucaria Forest in Southern Brazil. In the plot, all 290 adult trees were mapped, measured (diameter at breast height, dbh), sexed, sampled and genotyped. We also sampled, measured (total height), mapped and genotyped 223 juveniles. A total of 86 alleles were detected in the sample (n = 513). Adults and juveniles showed a positive and significant fixation index (minimum of 0.096), suggesting inbreeding or Wahlund effect. Juveniles presented a significant aggregated distribution which was associated with a genetic aggregation (significant SGS up to 20 m), indicating that near neighbor trees may be related due the limited seed dispersal. However, the intensity of SGS was not significantly higher among juveniles (Sp = 0.0041) than adults (Sp = 0.0026). Realized pollen and seed immigration into the plot was low (pollen = 6 %; seeds = 5 %) and the patterns of pollen and seed dispersal were similar. Pollen was dispersed over long distances (343 m), but 50 % was dispersed up to 124 m. Seeds also reached long distances (318 m), with 50 % being dispersed up to 133 m. Our results are discussed in terms of auto-ecology and the genetic conservation of A. angustifolia populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gene flow together with genetic drift interacts with natural selection to determine the potential for population adaptation (Klein et al. 2011). Extensive gene flow through both pollen and seeds reduces the genetic differentiation among populations, increasing the genetic diversity and effective population size within populations (Sork and Smouse 2006). In contrast, low levels of gene flow increase the genetic differentiation among populations and reduce the genetic diversity and effective population size within demes (Smouse and Sork 2004). Furthermore, in natural populations, gene flow can effectively counter the detrimental effects of genetic drift and may be the source of new alleles (Burczyk et al. 2004).
Determining the influence of pollen and seed flow on the effective population or neighborhood size of a population requires detailed analysis of the mating system and intra-population spatial genetic structure (SGS). Such studies have been conducted using highly polymorphic genetic markers such as microsatellite loci and parentage analysis (Ashley 2010). Analyses based on open pollinated seeds can determine the paternity of seeds and, thus, the pollen dispersal distance immediately after the mating event (effective pollen dispersal), measured by the distance between the maternal plant and pollen donor (Nason et al. 1996; Burczyk et al. 2004). However, the gap between the mating stage and the establishment of seedlings or juveniles (realized pollen dispersal) is substantial and many stochastic and deterministic processes can occur during this period (Burczyk et al. 2004), such as random mortality, predation, long-distance seed dispersal and selection. The realized pollen and seed dispersal distances can be measured by sampling established seedling and juvenile individuals (Burczyk et al. 2004). The realized seed dispersal distance is measured by the spatial distance between the seedling and the mother plant (maternity analysis) and the realized pollen dispersal distance is determined by the distance between the mother plant and pollen donor (paternity analysis).
Here we explore the inbreeding, intra-population SGS and pollen and seed dispersal patterns in the wind pollinated, Neotropical, coniferous tree Araucaria angustifolia (Bertol.) Kuntze (Araucariaceae). This species was one of the most important species for timber production in Brazil and the Araucaria forest was intensely exploited for timber production between the 1950s and 1970s (Guerra et al. 2002). Araucaria angustifolia trees can reach 50 m in height and 2 m in diameter at breast height (dbh), with a cylindrical trunk, that is resinous and rarely branched (Klein 1960). The species is classified as dioecious and is seldom monoecious (Reitz and Klein 1966). Pollen is dispersed by wind and the species’ edible seeds (Reitz and Klein 1966) are dispersed primarily by gravity and secondarily by animals, such as crows, small rodents, and agoutis (Lamberts 2003; Souza et al. 2008). Araucaria angustifolia is a dominant species of the Mixed Ombrophilous Forest (Araucaria Forest) that extends across the southern Brazilian Meridional Plateau, forming pure stands, or stands mixed with hardwoods, sometimes separated by grasslands. Its natural distribution was continuous in the uppermost parts of the Southern Plateau, in the states of Paraná, Santa Catarina and Rio Grande do Sul, and discontinuous in the states of São Paulo, Minas Gerais and Rio de Janeiro (Klein 1960). The extensive loss of habitat that has occurred in the Atlantic Rain Forest threatens the species and it is currently on the International Union for Conservation of Nature’s (IUCN) Red List of endangered species, being classified as critically endangered (Leal and Câmara 2005). The species is also included in Brazil’s endangered species list (MMA 2008).
A previous study exploring the realized pollen and seed flow in A. angustifolia seedlings and juveniles occurring within a small forest fragment (5.4 ha), suggested that both pollen and seed dispersal are restricted (Bittencourt and Sebbenn 2007). In this study, no seed flow was detected and seeds were predominantly dispersed near to mother trees, resulting in an intra-population SGS. Moreover, a low rate of pollen flow was detected in seedling (3 %) and juvenile (7 %) stages and pollen was predominantly dispersed in short distances, although some instances of long-distance dispersal were detected. The absence of seed flow was attributed to the long distances between populations of the species and intensive agriculture throughout the landscape. The low realized pollen flow and the tendency toward short-distance pollen dispersal were attributed to the characteristics of A. angustifolia pollen and the typically high vegetal densities of Araucaria forests. A. angustifolia pollen has a reduced ability to float in the air because it is nonsaccate and the pollen grain is large (Sousa and Hattemer 2003). Furthermore, Araucaria forests generally have a high vegetation density which may create barriers to pollen dispersal, limiting the movement of pollen within the stands (Sousa and Hattemer 2003). However, Bittencourt and Sebbenn’s (2007) study was restricted to a small, spatially isolated population of the species. Further studies of realized pollen and seed dispersal within large, continuous populations of the species are needed to confirm the pollen and seed dispersal patterns observed. Restricted realized pollen and seed flow across populations, as observed by Bittencourt and Sebbenn (2007), may increase SGS and inbreeding and decrease the genetic diversity and effective population size within populations.
For a continuous population of A. angustifolia only effective pollen dispersal has been studied (Bittencourt and Sebbenn 2008) due to the absence of seedlings and juveniles. This study assessed contemporary pollen dispersal in a 14 ha transect established within a continuous Araucaria forest (Manguerinha Indigenous Reserve), based on open-pollinated seeds (Bittencourt and Sebbenn 2008). The authors found significant intra-population SGS up to 75 m and inbreeding (or Wahlund effect) among both adult trees and seeds. The results also showed a high rate of pollen immigration (54 %) and long distance pollen dispersal within the plot (up to 344 m), but a strong tendency toward short-distance pollen dispersal, with 94 % of pollen being dispersed up to 200 m.
The aims of the present study were to investigate the intra-population SGS, the dynamics of genetic diversity and inbreeding among adults and juveniles, and realized pollen and seed dispersal distances and patterns within a plot established in a continuous forest of A. angustifolia. Specifically we address the following questions: (i) Are there differences in the levels of genetic diversity and inbreeding between adults and juveniles? (ii) Is there SGS among adults and juveniles? (iii) What is the realized pollen and seed immigration rate into the plot? (iv) What are the distance and patterns of pollen and seed dispersal within the plot?
Materials and methods
Study area
The study was carried out at the Caçador Forest Genetic Reserve—CFGR-EPAGRI/EMBRAPA (26º49′ and 26º53′S, 50º59′ and 50º53′W, altitude ranging from 900 to 1,104 m), in the Northern plateau of Santa Catarina state, South Brazil. The CFGR covers an area of 772 ha and is located in the municipality of Caçador. The region has a mesothermal and humid climate, with severe frost in the winter. Predominate soils are nitosols and latosol. The vegetation cover is the Ombrophilous Mixed Forest, a type of Atlantic Forest, dominated by A. angustifolia individuals and associated with the trees Ocotea porosa, Ilex paraguariensis and Dicksonia sellowiana (Klein 1960). The study area is natural with a history of low intensity timber exploitation that occurred 80 years ago; the area is now considered a secondary forest in an advanced stage of succession (Puchalsky et al. 2006).
Sampling and demography
To conduct the genetic and demographic studies, a permanent plot (Fig. 1) of 7.2 ha (300 × 240 m), divided in sub-plots of 100 m2 (10 × 10 m) were established to precisely identify A. angustifolia individuals and quantify density by dbh classes. All A. angustifolia individuals within the plot were mapped (x and y coordinates), measured for dbh (height > 1.5 m) or total height (height ≤ 1.5 m) and a tissue sample was collected (bark from tallest individuals and leaves when was possible). Individuals in the plot present a typical inverted J-shaped distribution for dbh. The majority of adult trees were classified in the first dbh class (20–30 cm), followed by a reduction in the frequency with an increase in the diametric class.
Microsatellite analysis
DNA was extracted from leaves using the procedure developed by Stefenon et al. (2004). Genotypes of all samples were scored at nine microsatellite loci, namely CRCA1 and CRCA2 (Scott et al. 2003), Aang01 (Schmidt et al. 2007), Ag20, Ag23, Ag45, Ag56, Ag62 and Ag94 (Salgueiro et al. 2005). PCR amplifications for locus Aang01 were performed in a 12 μL volume containing 30 ng of genomic DNA, 1× Taq buffer, 1 U Taq polymerase (Biotools), 0.25 μM of each dNTP (Fermentas), 1.5 mM of magnesium chloride (Biotools) and 100 μM of each primer. The amplification for other loci were performed with a tailed-primer technique, comprising 30 ng of genomic DNA, 1× Taq buffer (Biotools), 1 U Taq polymerase (Biotools), 0.25 μM of each dNTP (Fermentas) 1.5 mM of magnesium chloride (Biotools), 100 μM of reverse primer, 100 μM of M-13 tailed forward primer and 100 μM of labeled M-13 (6-FAM, HEX) in a 20 μL reaction. A BioRad C1000 Thermal Cycler was used to amplify the nine SSR loci with the following sequence: 95 °C for 3 min, then 30 cycles of 95 °C for 30 s, annealing temperature (according to the authors) for 45 s, 72 °C for 1 min, and a final extension step of 7 min at 72 °C. Each 5′ forward oligo was labeled with a fluorophore HEX or 6-FAM to enable the automated genotyping. After the reactions, PCR products were diluted 5× in ultrapure water in order to be genotyped by capillary electrophoresis in a MegaBACE 1000 DNA Analysis System (GE Healthcare). Alleles were genotyped by comparison with ET 400-R size standard (GE Healthcare) using Fragment Profiler analysis software version 1.2 (GE Healthcare).
Analysis of spatial aggregation
To verify if adult trees and juveniles present an aggregated distribution within the plot (Fig. 1), the Clark and Evans (1954) aggregation index was used along with SGS software (Degen et al. 2001): \( \hat{R}_{\text{CE}} = \hat{R}_{\text{o}} /\hat{R}_{\text{E}} \), where R o is the average distance of the closest neighbor, and R E is the average expected distance of the closest neighbor. If R CE = 1, then the distribution of individuals in the plot were determined to be random; if R CE < 1, there was an aggregated distribution; or if R CE > 1, then the distribution was uniform.
Analysis of genetic diversity and fixation index
The genetic diversity for adults, juveniles and the total population (all adults and juveniles) was characterized by the total number of alleles across all loci (k), average number of alleles per locus (A), average allele richness per locus (R), observed heterozygosity (H o) and expected heterozygosity in Hardy–Weinberg equilibrium (H e). These analyses were run using FSTAT 2.9.3.2 program (Goudet 1995). Levels of inbreeding were assessed by calculating the fixation index (F). To verify if the estimated F values were significantly different from zero, 10,000 permutations of alleles among individuals were carried out. We also applied a sequential Bonferroni correction for multiple comparisons (95 %, α = 0.05) to avoid false-positives. The fixation index and the permutations were estimated using the Spagedi 1.3 program (Hardy and Vekemans 2002). The fixation index in juveniles was estimated using gene frequencies calculated for the adult trees. For adults, we assumed that the gene frequencies present in this generation represent the parental gene frequencies.
Analysis of intra-population spatial genetic structure
The analysis of intra-population SGS was carried out using the J. Nason coancestry estimator (θ xy ), described in Loiselle et al. (1995). The coancestry coefficient measures the extent of genetic similarity between individuals x and y, in relation to the average genetic similarity among two random individuals sampled from a population (Hardy and Vekemans 2002). To visualise the SGS, the average values of θ xy were calculated for pairwise individuals within ten pre-determined distance classes of 20 m (20, 40, 60, 80, 100, 120, 140, 160, 180 and 200 m). To verify if the average values of coancestry within each distance class were significantly different from zero, the 95 % confidence interval (95 % CI) was calculated by 10,000 permutations of individuals among different distance classes. The average values of coancestry and the permutations were then used to construct the coancestry correlogram using MS Excel. To compare the extent of SGS between adults and juveniles, we estimated the S p statistic: Sp = −b k/(1 − θ 1) (Vekemans and Hardy 2004), where θ 1 is the average coancestry coefficient calculated in the first distance class (0–20 m), and b k is the slope of the regression curve in relation to the logarithm of the spatial distance (up to 200 m). To test the intensity of SGS, the spatial position of the individual was permutated 10,000 times to obtain the distribution frequency of b k where the null hypothesis states that θ 1 and ln d xy are not correlated (d xy is the spatial distance between individual x and y). These analyses were run using Spagedi 1.3.
Analysis of historic gene dispersal
Spagedi 1.3 was also used to perform the historic gene dispersal analysis in adults and juveniles, assuming equilibrium between migration and genetic drift (Hardy et al. 2006). Gene dispersal in terms of neighborhood size (Nb) was estimated as Nb = −(1 − θ 1)/b k (Vekemans and Hardy 2004), where b k is the slope of the regression curve assumed here within the distance class of σ g < d ij < 200 m for both adults and juveniles. The parameter Nb is an effective estimate of the density of reproductive individuals (D e) (Hardy et al. 2006); we therefore calculated D e = D(N e/N), where D is the population density and the effective density is the ratio between the effective population size (N e) and the total number of individuals (N) in the plot (Vekemans and Hardy 2004). Following Hardy et al. (2006), we used D/10 and D/2 as minimum and maximum estimates of D e. With the D e established, the 95 % CI of Nb was estimated as: Nb (lower) = (θ 1 − 1)/(b k − 2SE b) and Nb (upper) = (θ 1 − 1)/(b k + 2SE b), respectively, where SE b is the standard error of b k, calculated by jack-knifing alleles among individuals (Hardy et al. 2006). The 95 % CI of σ g was estimated as: \( \sigma_{\text{g}} = \sqrt {N_{\text{b}} /4\pi D_{\text{e}} } \), using the upper and lower limit values of Nb (Hardy et al. 2006).
Parentage analysis
Analyses of pollen and seed dispersal in juveniles were carried out using a simple exclusion method of parentage analysis, implemented with the CERVUS 3.0 program (Marshall et al. 1998; Kalinowski et al. 2007). We accepted the paternity and maternity of a juvenile tree only in cases where there was no mismatching among the offspring-mother-father trio. The most probable parent was determined using the gene frequency estimates for adult trees as a reference, as suggested by Meagher and Thompson (1987). The gene frequencies of private alleles in juveniles were added to the reference gene frequencies file of the adults. To do this, the frequency of private alleles was subtracted from the most frequent alleles estimated in the adults. Thus, the sum of the alleles of a locus could reach a maximum value of 1. The paternity and maternity analyses were conducted using the genotypes of 290 adults (dbh ≥ 20 cm) and 223 juveniles. In order to conduct these analyses, the probable age of the juveniles and adults was determined based on the average annual increment (AAI) for dbh: estimated as 1.99 cm at 6 years of age and 0.68 cm at 30 years of age (Sebbenn et al. 2004). The AAI for height was estimated at 1.24 m at 6 years and 0.60 at 30 years (Sebbenn et al. 2004). Thus, the sampled juveniles, with dbh ranging from 0.14 to 6.4 cm and height ranging from 0.2 to 6.4 cm may have ages ranging from <1 year to a maximum of 10 years (6.4/0.68 cm). Based on these estimates, trees with dbh ranging from 20 to 30 cm that were not sexed were excluded as possible parents of the juveniles because these trees likely ranged in age from 29 to 44 years and it is therefore unlikely that they were parents of the sampled juveniles. It is important to note that the dbh class of 20–30 cm was mainly composed of non-reproductive individuals. Of 290 individuals with a dbh ≥ of 20 cm, 60 were identified in the 20–30 cm diameter class and of these 46 were not sexed (77 %). Eight of the 14 remaining were female (dbh ranging 24.6–29 cm) and six were male (dbh ranging 22–27.9 cm). The remaining 28 unsexed individuals with dbh >30 cm were used in the analysis as candidates of both maternal and paternal parents of the juveniles.
The probability of finding a compatible parent candidate within the plot, when the true parent is outside of the plot (cryptic gene flow—CGF), was estimated following Dow and Ashley (1998): \( CGF = 1 - (1 - P_{\text{nExc}}^{n} ) \), where P nExc is the combined non-exclusion probability of the parent pair and n is the number of male and female candidates found as putative parents of the juveniles. Individuals assigned as male and female parents were used to estimate average, median, minimum and maximum pollen and seed dispersal distances. Pollen immigration was estimated as the proportion of juveniles for which a father was not found within the plot, divided by the total number of sampled juveniles. Seed immigration into the plot was estimated as the proportion of juveniles for which the mother and father was not found in the plot, divided by the total number of sampled juveniles. Pollen dispersal distance was measured as the distance between the assigned male and female parents and seed dispersal was measured by the distance between the assigned juvenile and the female parent. The parentage analysis also allowed us to sex some individuals not previously sexed in the field: ten individuals were assigned as male and nine as female. In seven cases an unsexed individual was assigned as both male and female parents of different juveniles; in these cases we assumed the individual was possibly monoecious. In the situation where both parents of a juvenile were not sexed (1 case), the individual located closest to the juvenile was assumed to be the mother and the other was assumed to be the father. As this occurred only once in our study and since seed dispersal in A. angustifolia occurs primarily by gravity and pollen by wind, we believe that such an assumption did not substantially bias our analysis of pollen and seed dispersal distance. To determine if the realized pollen dispersal distance within the plot was a function of the spatial distance between all male trees and female trees, we compared the frequency curve of the pollen dispersal with the frequency curve of the distances between all male trees in relation to the female trees, using the Kolmogorov–Smirnov test (Sokal and Rohlf 1995).
Variance effective size analysis
The variance effective size of the reproductive population and juveniles was calculated as: \( \hat{N}_{\text{e}} = 0.5/\Uptheta \) (Cockerham 1969); where \( \Uptheta \) is the average group coancestry. For sexed adults and adults assigned by parentage analysis as parents of the juveniles, the average group coancestry was estimated as: \( \hat{\Uptheta } = \sum\nolimits_{x = 1}^{{n_{{f}} }} {\sum\nolimits_{y = 1}^{{n_{{f}} }} {\hat{\theta }_{{f}} /4n_{{f}}^{2} + \sum\nolimits_{x = 1}^{{n_{{m}} }} {\sum\nolimits_{{}}^{{n_{{m}}^{{}} }} {\hat{\theta }_{{m}} /4n_{{m}}^{2} + \sum\nolimits_{x = 1}^{{n_{{f}} }} {\sum\nolimits_{y = 1}^{{n_{{m}} }} {\hat{\theta }_{{fm}} } } /2n_{{f}} n_{{m}} } } } } \) (Lindgren and Mullin 1998); where θ f , θ m , and θ fm are the coancestry coefficient between females, males, and females and males together, respectively; and n f and n m are the number of female and male trees. The pairwise coancestry coefficients were estimated using the J. Nason method described in Loiselle et al. (1995) and the Spagedi program (Hardy and Vekemans 2002). The group coancestry in juveniles was calculated as: \( \hat{\Uptheta } = [0.25(n_{{M(hs)}} + n_{{p(hs)}} ) + 0.5n_{{M(fs)}} ]/(n^{2} - n) \); where n is the sample size; n M(hs) is the number of pairwise maternal half-sibs; n P(hs) is the number of pairwise paternal half-sibs; and n M(fs) is the number of pairwise full-sibs (Gaino et al. 2010). We estimated the relation between the effective size (N e ) and the number of sampled individuals (n) by N e /n.
Results
Population characteristics
All individuals with dbh >20 cm were considered adults. The sex was checked once per year for 3 years. Individuals that did not present male or female strobilus were considered “non-reproductive”. In the plot, the population density of individuals with dbh ≥20 cm was 40.3 trees/ha and the sexual ratio was 1:1.15 (female:male). In the plot, we identified 290 adult trees (dbh ranging from 20 to 147.1 cm, average of 57.2 cm, and median of 51.1 cm), with 100 females (dbh ranging from 24.6 to 147.1 cm, average of 72.2 cm and median of 67 cm), 115 males (dbh ranging from 22 to 125.1 cm, average of 58.1 cm, and median of 53.5 cm), one was monoecious (dbh of 45.2 cm), and 74 were non-reproductive (dbh ranging from 20 to 94.5 cm, average of 35.7 cm, and median of 28.7 cm). Of the 74 non-reproductive trees, 46 (62 %) had a dbh between 20 and 30 cm. We also randomly sampled 223 juveniles (dbh ranging from 0.1 to 6.4 cm, average of 2.6 cm, and median of 2.5 cm; average, median, minimum and maximum height values were 1.7, 1.0, 0.2 and 6.4 m, respectively). The total sampled population included 513 individuals. The spatial distance among pairwise adult trees ranged from 0.7 to 372 m (average of 148 m and median of 143 m) and among juveniles from 0.2 to 364 m (average of 144 m and median of 143 m).
Population spatial structure
The dispersion index for juveniles was significantly lower than unity (R CE = 0.833, P < 0.05), indicating an aggregated distribution (Fig. 1).
Genetic diversity and inbreeding
Considering the total sample of 513 individuals, the number of alleles per locus ranged from 3 to 22, with an average of 9.56 and a total of 86 alleles. The observed heterozygosity ranged from 0.225 to 0.789, with an average of 0.559. The expected heterozygosity was generally higher than the observed, ranging from 0.217 to 0.876, with an average of 0.639. The fixation index was positive and significantly different from zero in seven loci and the average across all loci ranged from −0.038 to 0.343, with an average of 0.125.
Adults presented a greater number of alleles (84) than juveniles (78); nine alleles were private among adults and two among juveniles (Table 1). The observed and expected heterozygosity were significantly higher in adults than in juveniles. The fixation index was significantly different from zero for both adults (\( \hat{F}_{\text{ad}} = 0093 \)) and juveniles (\( \hat{F}_{\text{juv}} = 0.163 \)). However, these values were not significantly different between adult trees and juveniles (Table 1).
Spatial genetic structure (SGS)
A significant decrease in the average coancestry coefficient with an increase in the spatial distance was detected among both adults and juveniles up to 20 m (Fig. 2), indicating that near neighbor individuals may be related. According with 95 % CI estimated by jackknifing loci, the average coancestry coefficient for the ten distance classes are not significantly different between adults and juveniles. The intensity of the SGS (Table 2) was also not significantly higher among juveniles (Sp = 0.0041 ± 0.0014) than adults (Sp = 0.0026 ± 0.0025). These results suggest an average gene dispersal distance in juveniles between 107 and 241 m and in adults between 140 and 312 m, corresponding to an average neighborhood size of 226 juveniles and 381 adults.
Intra-population spatial genetic structure in adults and juveniles of Araucaria angustifolia. The solid line represents the average θ xy value. The dashed lines represent the 95 % (two-tailed) confidence interval of the average θ xy distribution, which was calculated based on 1,000 permutations of the spatial distance among pairs of adults. The vertical lines represent the 95 % confidence interval of the mean coancestry coefficient, calculated by jackknifing loci and using Spagedi 1.3 program (Hardy and Vekemans 2002)
Pollen and seed dispersal
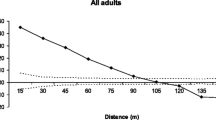
Two of the 223 juveniles were excluded from the parentage analysis because they were genotyped for <4 loci. The combined non-exclusion probability of the parent pair was very low (0.00002), indicating that the loci battery used is suitable to discriminate parents in a parentage analysis. The combined non-exclusion of genetic identity for adults was also very low (0.000846), indicating that genotypes are unique. Of the 221 sampled juveniles, a mother tree was found for 210 individuals and a father tree was found for 208 individuals, indicating a realized pollen and seed immigration into the plot of 6 and 5 %, respectively (Table 3). Juveniles were mothered by 98 (90 %) of the 109 identified female trees (100 visually sexed + 9 sexed by maternity analysis), and of these 98 females, 34 (35 %) mothered 58 % of the assigned juveniles. Of the 125 male trees identified in the plot (115 visually sexed + 10 sexed by paternity analysis), 107 (86 %) fathered the 208 juveniles, and 30 (28 %) of these fathered 50 % of the juveniles. The comparison of the frequency curve of pollen dispersal with the frequency curve measured among all male and female trees was not significantly different from zero (Kolmogorov–Smirnov test (D = 0.08, P < 0.18, Fig. 3), indicating that the spatial distance among males and females explains the observed patterns of pollination.
The distance of realized pollen dispersal ranged from 5 to 343 m, with an average of 134 m, a median of 124 m, and 95 % of pollen was dispersed within 250 m (Table 3; Fig. 3). There is also a significant negative correlation between the number of juveniles fathered by males (r = −0.64, P < 0.05) and the distance between mother and father tree (Fig. 3), suggesting predominantly short-distance pollen dispersal. The distance of realized seed dispersal ranged from 6 to 318 m, with an average of 131 m, median of 133 m, and 91 % of seeds were dispersed within 250 m (Table 3; Fig. 3). There is a significant negative correlation between distance from the mother tree and number of offspring (r = −0.82, P < 0.01), indicating that seeds were generally established in proximity to the mother tree.
Variance effective size
The average group coancestry coefficient among females, males, and males and females together was 0.000725, −0.000724, and 0.0000434 (total of 0.000769), respectively, indicating that in random mating the expected biparental inbreeding in offspring is very low (0.0000434). The estimated variance effective size of the reproductive population indicates that the 215 sexed adult trees represents 162 unrelated and non-inbred individuals (N e/N = 0.75) and that the 205 reproductive trees assigned by parentage analysis as mother or father of the juveniles represent 145 unrelated and non-inbred individuals (N e/N = 0.71). Among juveniles, the average group coancestry coefficient was 0.00208, resulting in a variance effective size of 106 (N e/N = 0.49).
Discussion
Genetic diversity
The studied population presents high levels of genetic diversity. The estimated levels of expected heterozygosity (\( \hat{H}_{\text{e}} \) = 0.639) were similar to that found using microsatellite markers in other populations of the species (\( \hat{H}_{\text{e}} \) = 0.596, Bittencourt and Sebbenn 2007; \( \hat{H}_{\text{e}} \) = 0.650, Stefenon et al. 2007). Private alleles were found among both adults and juveniles, suggesting firstly that not all adults reproduce, and secondly, the existence of pollen or seed immigration from outside of the plot. Considering the age of some juveniles (probably up to 10 years), an alternative explanation for the occurrence of private alleles in juveniles is that the parents may have died and were therefore not sampled during fieldwork.
The observed and expected heterozygosity were significantly lower in the juveniles than adults (Table 1). Kattle et al. (2007) found a similar pattern in the congener Araucaria nemorosa, in New Caledonia. Considering that allelic richness was not significantly different between the adult and juvenile cohorts in our study, the lower levels of genetic diversity in juveniles may be related to a smaller sample size of juveniles than adults. An alternative explanation is that the adults represent cumulative reproductive events from different generations and thus we can expect higher levels of genetic diversity than that observed in juveniles, as was also observed for A. nemorosa (Kattle et al. 2007).
The relationship between variance effective size and sample size was also lower in juveniles (N e/N = 0.49) than in sexed adults (N e/N = 0.75) and reproductive trees assigned as mother or father of the juveniles (N e/N = 0.71). This is caused by a higher frequency of related individuals and inbreeding in the juvenile stage than adult stages. The relatedness between individuals and inbreeding increases the frequency of identical by descent alleles (IBD) among and within individuals, which consequently reduces the effective population size. However, this pattern is likely dynamic and will change when juveniles reach the adult stage, resulting in a reduction in the difference between effective size and sample size. The difference in the variance effective size of the sexed adults and the trees assigned as mother or father of the juveniles is related to the fact that some sexed individuals were excluded from the analysis because they did not father or mother any of the sampled juveniles; on the other hand some unsexed individuals (previously defined as non-reproductive) were sexed based on the results of parentage analysis.
Inbreeding
Theoretically, high levels of inbreeding are not expected in outcrossing tree species (including dioecious species) due to the strong inbreeding depression that eliminates biparental inbred individuals between seed and adult stages (Hufford and Hamrick 2003; Naito et al. 2008). A positive and significant fixation index was detected in both adult and juvenile cohorts. This suggests inbreeding, the presence of null alleles in the loci or Wahlund effect. To test the segregation of null alleles in the loci, it is preferable to sample progeny arrays, which was beyond the scope of this study. Without control over pedigree, inferences about null alleles are speculative. However, we note that the presence of null alleles in the loci increases the number of false homozygous individuals, resulting in a higher fixation index than expected. While we cannot exclude such a possibility, our estimated fixation index for both adult trees and juveniles were only moderately high and the effect of null alleles on the fixation index is likely not very strong. Moreover, other studies with Araucaria species also found inbreeding in adults and juveniles and higher F values in juveniles than in adults (A. angustifolia, Bittencourt and Sebbenn 2007; A. nemorosa, Kattle et al. 2007). Similar results have also been found in other tropical tree species, such as Hymenaea courbaril (Lacerda et al. 2008), Copaifera langsdorffii (Sebbenn et al. 2010) and Vateriopsis seychellarum (Finger et al. 2012).
Biparental inbreeding, similar to selfing, is expected to result in an increased fixation index across all loci. For adults, F values ranged from negative to significantly positive, suggesting selection favoring heterozygosity in the negative F values and inbreeding in the positive F values. In contrast, F values for juveniles were all positive, suggesting only inbreeding (Table 1). Thus, in juveniles, the high F values may be the result of true inbreeding originating from mating among relatives due to the SGS associated with high intensity, short-distance seed and pollen dispersal as observed here and in other studies of the species (Mantovani et al. 2006; Bittencourt and Sebbenn 2007, 2008; Stefenon et al. 2008; Patreze and Tsai 2010; Ferreira et al. 2012). Furthermore, as mating among relatives produces inbreeding at the same rate as the coancestry coefficient between related parents, the observed inbreeding in juveniles can be explained by mating among relatives ranging from half-sibs (0.125) to full-sibs (0.25). The significant F values for adults and juveniles also indicate that the population is not in Hardy–Weinberg equilibrium. However, it is important to note that the observed positive fixation index in both adults and juveniles may be also due to Wahlund effect as related to the presence of significant SGS, as discussed in Bittencourt and Sebbenn (2007).
Spatial genetic structure and historic gene dispersal
Juveniles showed an aggregated spatial distribution which was associated with genetic aggregation. The population presents significant SGS in both adults and juveniles up to 20 m, suggesting that near neighbor individuals are related. The significant slope of the regression curve for both adults and juveniles confirms the presence of isolation by distance in the plot. The historic gene dispersal suggests average gene dispersal distances between 107 to 241 m in the juveniles and 140 to 312 m in adults. Such patterns are associated with restricted seed dispersal, resulting in related individuals being established near to the mother tree. Aggregation close to the mother tree may result in high mortality rates among juvenile individuals due to predation (Janzen 1970). With high rates of mortality among grouped individuals, the level of aggregation tends to diminish through advanced ontogenetic stages. Significant SGS among A. angustifolia populations has also been reported in other studies at up to 70 m (Mantovani et al. 2006; Bittencourt and Sebbenn 2007); 75 m (Bittencourt and Sebbenn 2008); up to <80 m (Stefenon et al. 2008); and 25 m (Patreze and Tsai 2010).
Pollen dispersal
Our results also suggest limited pollen immigration into the study plot (5.9 %). This level of pollen flow is low for a continuous forest of a wind pollinated tree species. In fact, the observed seed and pollen immigration can have two origins: (i) it may be attributed to seeds and pollen originating from trees located outside of the plot; or (ii) it may have originated from deceased trees located inside the plot that were not genotyped for this study. Another study examining effective pollen dispersal for A. angustifolia showed high levels of pollen immigration (54 %) in a 14 ha plot in a continuous Araucaria forest (Bittencourt and Sebbenn 2008). However, although the size of the plot was approximately double the plot studied here (7.2 ha), the density of reproductive trees was lower (7.85 trees/ha) than what was observed herein which may encourage pollen dispersal and immigration due to a lower vegetation density. The median distance of realized pollen dispersal (124 m) and average pollen dispersal distance (134 m) were similar to that observed in Bittencourt and Sebbenn (2008) (average of 102 m), with the majority of pollen dispersed over short distances. In contrast to our results from the Kolmogorov–Smirnov test, we found a significant negative correlation between the number of juveniles fathered by males and the distance between mother and father trees, suggesting an effect of short pollen dispersal distance. This result supports the hypothesis of restricted pollen dispersal for this species. Although A. angustifolia pollen is dispersed by wind, it has a reduced ability to float in the air because it is nonsaccate and the pollen grain is large (Sousa and Hattemer 2003). This fact, along with the high vegetal densities typical of Araucaria forests may create barriers to pollen dispersal, limiting the distance of pollen movement within stands (Sousa and Hattemer 2003).
Our results showed a clear deviation from random mating and/or stochastic processes such as random mortality and predation. 90 % of females and 86 % of males produced the juveniles and only a minority of males (28 %) fathered 50 % of the juveniles. These results suggest a high individual variation in male fertility. Mantovani et al. (2004) studying an A. angustifolia population in Campos do Jordão, São Paulo State, Brazil, over two consecutive years, observed flowering synchronization for male and female trees; however, males presented individual variation in both flowering time and intensity. Mantovani et al. (2006) also found that some males produce more strobilus than others, suggesting that some males may produce more pollen than others. This may be explained by the fact that many long-living organisms experience a decline in fecundity due to deterioration of metabolic functions, a phenomenon known as senescence (Ricklefs 2003). A large variance in the number of offspring fathered per individual reduces the effective population size, thus accelerating the process of genetic drift (Hartl and Clark 1997) and evolution. Despite this process being a natural mechanism, it may represent an aggravating factor for species at risk of extinction.
Seed dispersal
Our results suggest a low rate of realized seed immigration into the plot (5 %) and a strong tendency of mating among near neighbor individuals; however, there are some realized instances of long-distance seed dispersal (Table 3; Fig. 3b). We also found a strong negative association between the distance of realized matings and the number of mothered juveniles. This confirms the results found for SGS that offspring are generally established close to the mother tree. These results also suggest that SGS is produced not only because of the short-distance seed dispersal, but also because of the relatively short-distance pollen dispersal. Both short-distance pollen and seed dispersal increase the frequency of local alleles in an area, producing subpopulations where individuals are more related than expected through random pollen and seed dispersal and, consequently, producing SGS. Similar results were also observed in a small forest fragment of A. angustifolia by Bittencourt and Sebbenn (2007) who found seed dispersal distances ranging from 0.35 to 291 m, with 47 % of seeds dispersed effectively within a radius of 60 m around the mother tree. Other studies of barochoric seed dispersal based on genetic markers have also found a high frequency of short-distance seed dispersal. For example, Nakanishi et al. (2009) studying Quercus salicina in Japan found low levels of seed immigration into a plot of 11.56 ha (15 %), indicating short-distance seed dispersal. Asuka et al. (2005), studying Fagus crenata found a seed immigration rate of 21.9 % in two plots and a maximum seed dispersal distance of 20 m for one plot and 30 m for the other. These results, together with the results observed here, suggest a high intensity of short-distance seed dispersal for species with gravity dispersed seeds. However, while the seeds of A. angustifolia are primarily dispersed by gravity, small animals have also been documented as dispersal vectors. Because animals tend to disperse seeds farther away from the mother tree, the instances of long-distance seed dispersal detected in this study can be explained by a secondary seed dispersal stage caused by animal behavior.
Little is known about the connection or distinction between dispersal and seed predation, especially with regards to A. angustifolia which has edible seeds, but no fruit. For this species, when seeds are ingested the embryo is destroyed. Animals are therefore predators of the seeds as well as dispersers, both accidentally through waste, or deliberately through reserves or caches of seeds. Another study examining seed dispersal vectors for A. angustifolia showed that of the total number of seeds analyzed in the study, 13 were consumed in situ and 205 were removed; of the 205 removed seeds, six were lost, 180 were consumed and only 19 were cached (Lamberts 2003). The author identified three groups of disperser animals: crows, small rodents, and agoutis. Crows dispersed seeds the greatest distances and small rodents moved the seeds up to a distance of 26.7 m (with an average of 4.7 ± 0.42 m). However, 99.3 % of seeds removed by small rodents were consumed. The third dispersal vector, the agouti, scattered the seeds in distances up to 50 m (average of 6.50 ± 1.65 m). The percentage of seeds consumed by the agouti was 67.7 % and therefore 32.3 % were stored, thus making the agouti a more efficient dispersal vector than small rodents. Lob (2007) also evaluated the dispersal distance of A. angustifolia seeds and found that of 320 seeds identified, 222 were moved up to a distance of 30 m (average 3.14 ± 5.05 m) and 213 of the total were consumed. These studies, although producing only short-term analysis of seed dispersal via animal vectors, do provide some indication of the spread of A. angustifolia seeds throughout the forest. The Caçador Forest Genetic Reserve currently has a rich fauna (Tortato 2009). However, it is important to note that a decrease in the population of disperser animals may have a serious impact on seed flow among some tree species populations, especially in terms of long-distance seed dispersal, resulting in seed dispersal being confined to the forest fragment and in proximity to the mother tree.
Implications of the results for conservation genetics
Araucaria angustifolia is a key species in the Araucaria forest and our results related to SGS and pollen dispersal have important implications for ex situ conservation of this species. The ideal for ex situ conservation strategies of long-living tree species is to establish germplasm banks through provenance and progeny tests. When the trials are at a reproductive stage, a selection may be carried out among the progenies and seeds can be collected for environmental restoration in order to recombine and replicate the genetic diversity of the conserved population. Our results of SGS indicate that to collect seeds in the studied population for conservation genetics, seed trees must be selected at distances >20 m to avoid collecting seeds from genetically related seed trees. However, the results of parentage analysis suggest that seeds may be dispersed up to 131 m (the average seed dispersal distance). Thus, the ideal is to collect seeds from seed trees located at distances greater than 131 m to decrease the probability of collecting seeds from seed trees that are related. However, such a strategy will not avoid collecting seeds from related trees due to the probability that male trees fertilized several of the selected seed trees, especially if the seed trees and the pollen donors are located in close proximity. In our plot the maximum pollen dispersal distance was estimated at 334 m. While this distance is not the true maximum distance of pollen dispersal as it does not consider distances of pollen immigration into the plot, a distance of 334 m may be used to determine the distance among seed trees which received an overlapping pollen pool. Thus, seeds collected from seed trees at this distance will likely only be related within progenies, thus maximizing the effective size of the collected seed samples in ex situ conservation seed banks.
References
Ashley MV (2010) Plant parentage, pollination, and dispersal: how DNA microsatellites have altered the landscape. Crit Rev Plant Sci 29:148–161
Asuka Y, Tomaru N, Munehara Y, Tani N, Tsumaru Y, Yamamoto S (2005) Half-sib family structure of Fagus crenata samplings in an old-growth beech-dwarf bamboo forest. Mol Ecol 14:2565–2575
Bittencourt JM, Sebbenn AM (2007) Patterns of pollen and seed dispersal in a small fragmented population of a wind pollinated Araucaria angustifolia in southern Brazil. Heredity 99:580–591
Bittencourt JM, Sebbenn AM (2008) Pollen movement in a continuous forest of Araucaria angustifolia, inferred from paternity and TwoGener analysis. Conserv Genet 9:855–868
Burczyk J, DiFazio SP, Adams WT (2004) Gene flow in forest trees: how far do genes really travel. For Genet 11:1–14
Clark DA, Evans FC (1954) Distance to nearest neighbour as a measure of spatial relationship in populations. Ecology 35:445–456
Cockerham CC (1969) Variance of gene frequencies. Evolution 23:72–84
Degen B, Petit R, Kremer A (2001) SGS—Spatial Genetic Software: a computer program for analysis of spatial genetic and phenotypic structure of individuals and populations. J Hered 92:447–448
Dow BD, Ashley MV (1998) High levels of gene flow in bur oak revealed by paternity analysis using microsatellites. J Hered 89:62–70
Ferreira DK, Nazareno AG, Mantovani A, Bittencourt R, Sebbenn AM, Reis MS (2012) Genetic analysis of 50-year old Brazilian pine (Araucaria angustifolia) plantations: implications for conservation planning. Conserv Genet 13:435–442
Finger A, Kettle CJ, Kaiser-Bunbury CN, Valentin T, Mougal J, Ghazoul J (2012) Forest fragmentation genetics in a formally widespread island endemic tree: Vateriopsis seychellarum. Mol Ecol 21:2369–2381
Gaino APSC, Silva AM, Moraes MA, Alves PF, Moraes MLTM, Freitas MLM, Sebbenn AM (2010) Understanding the effects of isolation on seed and pollen flow, spatial genetic structure and effective population size of the dioecious tropical tree Myracrodruon urundeuva. Conserv Genet 11:1631–1643
Goudet J (1995) Fstat. (Version 2.9.3.2.): a computer program to calculate F statistics. J Hered 86:485–486
Guerra MP, Silveira V, Reis MS, Schneider L (2002) Exploração, manejo e conservação da araucária (Araucaria angustifolia). In: Simões, LL; Lino CF (org.) Sustentável Mata Atlântica: a exploração de seus recursos florestais. Editora SENAC, São Paulo, p 85–101
Hardy O, Vekemans X (2002) SPAGeDI: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hardy OJ, Maggia L, Bandou E, Breyne P, Caron J, Chevallier MH, Doligez A, Dutech C, Kramer A, Latouche-Hallé C, Troispoux V, Veron V, Degen B (2006) Fine-scale genetic structure and gene dispersal inferences in 10 neotropical tree species. Mol Ecol 15:559–571
Hartl DL, Clark AG (1997) Principles of population genetics, 3rd edn. Sinauer Associates Inc., Sunderland
Hufford KM, Hamrick JL (2003) Viability selection at three early life stages of the tropical tree, Platypodium elegans (Fabaceae, Papilionoideae). Evolution 57:518–526
Janzen DH (1970) Herbivores and the number of tree species in tropical forests. Am Nat 104:501–528
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Kattle JC, Hollingsworth M, Jaffré T, Moran B, Ennos A (2007) Identifying the early genetic consequences of habitat degradation in highly threatened tropical conifer, Araucaria nemorosa Laubenfels. Mol Ecol 16:3581–3591
Klein RM (1960) Aspectos dinâmicos do pinheiro brasileiro. Sellowia 12:17–44
Klein EK, Carpentier FH, Oddou-Muratorio S (2011) Estimating the variance of male fecundity from genotypes of progeny arrays: evaluation of the Bayesian forward approach. Method Ecol Evol 2:349–361
Lacerda EBL, Kanashiro M, Sebbenn AM (2008) Effects of selective logging on genetic diversity and spatial genetic structure of a Hymenaea courbaril population in the Brazilian Amazon Forest. For Ecol Manag 255:1034–1043
Lamberts AVDH (2003) Predação e sobrevivência de sementes de Araucaria angustifolia (Bert.) Kuntze em áreas de mata nativa e plantação de Pinus eliotti na Floresta Nacional de São Francisco de Paula, RS. Dissertação (Mestrado em ecologia), Unicamp
Leal CG, Câmara IG (2005) Status do hotspot Mata Atlântica: uma síntese In: Leal, C.G, Câmara I.G. Mata Atlântica: Biodiversidade, ameaças e perspectivas. Fundação SOS Mata Atlântica, São Paulo, p 3–11
Lindgren D, Mullin TJ (1998) Relatedness and status number in seed orchard crops. Can J For Res 28:276–283
Lob G (2007) Influência de frutos e sementes na abundância de pequenos mamíferos e a relação com a predação e a dispersão de sementes da Araucária (Araucaria angustifolia). Universidade Federal do Rio Grande do Sul, Dissertação (mestrado em ecologia)
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Mantovani A, Morellato LPC, Reis MS (2004) Fenologia reprodutiva e produção de sementes em Araucaria angustifolia (Bert.) O. Kuntze Rev Bras Bot 27:787–796
Mantovani A, Morellato LPC, Reis MS (2006) Internal genetic structure and outcrossing rate in a natural population of Araucaria angustifolia (Bert.) O. Kuntze. J Hered 97:466–472
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655
Meagher TR, Thompson E (1987) Analysis of parentage for naturally established seedlings of Chamaelirim luteum (Liliaceae). Ecology 68:803–812
MMA (Ministério do Meio Ambiente BR) (2008) Lista das Espécies da Flora Brasileira Ameaçadas de Extinção. Instrução Normativa n 6, de 23 de Setembro de 2008
Naito Y, Kanzaki M, Iwata H, Obayashi K, Lee SL, Muhammad N, Okudo T, Tsumura Y (2008) Density-dependent selfing and its effects on seed performance in a tropical canopy tree species, Shorea acuminate (Dipterocarpaceae). For Ecol Manag 256:375–383
Nakanishi A, Tomaru N, Yoshimaru H, Manabe T, Yamamoto S (2009) Effects of seeds- and pollen-mediated gene dispersal on genetic structure among Quercus salicina samplings. Heredity 96:79–84
Nason JD, Herre EA, Hamrick JL (1996) Paternity analysis of the breeding structure of strangler fig populations: evidence for substantial long-distance wasp dispersal. J Biogeogr 23:501–512
Patreze CM, Tsai SM (2010) Intrapopulational genetic diversity of Araucaria angustifolia (Bertol.) Kuntze is different when assessed on the basis of chloroplast or nuclear markers. Plant Syst Evol 284:111–122
Puchalsky A, Mantovani M, Reis MS (2006) Variação em população naturais de Araucaria angustifolia (Bert.) O. Kuntze associada a condições edafo-climáticas. Sci For 70:137–148
Reitz R, Klein RM (1966) Araucariáceas. Flora Ilustrada Catarinense. 1–62
Ricklefs RE (2003) A Economia da Natureza. Quinta edição. Editora Guanabara Koogan
Salgueiro F, Caron H, de Sousa MIF, Kremer A, Margis R (2005) Characterization of nuclear microsatellite loci in South American Araucariaceae species. Mol Ecol Notes 5:256–258
Schmidt AB, Ciampi AY, Guerra MP, Nodari RO (2007) Desenvolvimento e caracterização de marcadores microssatélites (SSRs) para Araucaria angustifolia (Bert.), O. Kuntze. Mol Ecol Notes 7:340–342
Scott LJ, Shepherd R, Henry RJ (2003) Utility and evolution of microsatellites in the Araucariaceae. Plant Syst Evol 236:115–123
Sebbenn AM, Pontinha AAS, Freitas SA, Freitas JA (2004) Variação genética em cinco procedências de Araucaria angustifolia (Bert.) O. Ktze. no Sul do estado de São Paulo. Rev Inst Flor 16:91–99
Sebbenn AM, Carvalho ACM, Freitas MLM, Moraes SMB, Gaino APSC, da Silva JM, Jolivet C, Moraes MLT (2010) Low levels of realized seed and pollen gene flow and strong spatial genetic structure in a small, isolated and fragmented population of the tropical tree Copaifera langsdorffii Desf. Heredity 106:134–145
Smouse PE, Sork VL (2004) Measuring pollen flow in forest trees: an exposition of alternative approaches. For Ecol Manag 197:21–38
Sokal RR, Rohlf FJ (1995) Biometry: principles and practices of statistics in biological research, 3rd edn. W.H Freeman and Company, New York
Sork VL, Smouse PE (2006) Genetic analysis of landscape connectivity in tree populations. Lands Ecol 21:821–836
Sousa VA, Hattemer HH (2003) Pollen dispersal and gene flow by pollen in Araucaria angustifolia. Aust J Bot 51:309–317
Souza AF, Forgiarini C, Longhi SJ, Brena DA (2008) Regeneration of a long-lived dominant conifer and the effects of logging in southern South America. Acta Oecol 34:221–232
Stefenon VM, Nodari RO, Guerra MP (2004) Genética e conservação de Araucaria angustifolia: III. Protocolo de extração de DNA e capacidade informativa de marcadores RAPD para análise da diversidade genética em populações naturais. Biotemas 17:47–63
Stefenon VM, Gailing O, Finkeldey R (2007) Genetic structure of Araucaria angustifolia (Araucariacea) populations in Brazil: implications for the in situ conservation of genetic resources. Plant Biol 9:516–525
Stefenon VM, Gailing O, Finkeldey R (2008) The role of gene flow in shaping genetic structures of the subtropical conifer species Araucaria angustifolia. Plant Biol 10:356–364
Tortato MA (2009) Mamíferos da Reserva Florestal da Embrapa/Epagri Caçador—Santa Catarina. Relatório técnico de atividades de campo, Embrapa
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analysis in plant populations. Mol Ecol 13:921–935
Acknowledgments
This study was supported financially by the Fundação de Amparo a Pesquisa e Inovação do Estado de Santa Catarina (FAPESC). The article is part of the master’s thesis of Cristina S. Sant’Anna who would like to thank CAPES for the MSc scholarship and EPAGRI for accommodation during fieldwork. We acknowledge the Universidade Federal de Santa Catarina and the staff of the Nucleo de Florestas Tropicais who cooperated in the data collection in the field and in the lab. We also thank staff of the laboratory DNA/UDESC Lages. We thank Valdir Marcos Stefenon and Andrea Schmidt for their support throughout this project and Dr. Evelyn Nimmo for her editing of the English manuscript. Scholarships were provided by the National Research Council (CNPq) to Rubens Onofre Nodari, Alexandre Magno Sebbenn and Maurício Sedrez dos Reis. Finally, we thank two reviewers whose suggestions and corrections improved a previous version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sant’Anna, C.S., Sebbenn, A.M., Klabunde, G.H.F. et al. Realized pollen and seed dispersal within a continuous population of the dioecious coniferous Brazilian pine [Araucaria angustifolia (Bertol.) Kuntze]. Conserv Genet 14, 601–613 (2013). https://doi.org/10.1007/s10592-013-0451-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-013-0451-5