Abstract
Background and Aims
Soil microbial communities contribute to organic phosphorus cycling in a variety of ways, including secretion of the PhoD alkaline phosphatase. We sampled a long-term grassland fertilization trial in Switzerland characterized by a natural pH gradient. We examined the effects of phosphate depletion and pH on total and active microbial community structures and on the structure and composition of the total and active phoD-harboring community.
Methods
Archaeal, bacterial and fungal communities were investigated using T-RFLP and phoD-harboring members of these communities were identified by 454-sequencing.
Results
Phosphate depletion decreased total, resin-extractable and organic phosphorus and changed the structure of all active microbial communities, and of the total archaeal and phoD-harboring communities. Organic carbon, nitrogen and phosphorus increased with pH, and the structures of all total and active microbial communities except the total fungal community differed between the two pH levels. phoD-harboring members were affiliated to Actinomycetales, Bacilliales, Gloeobacterales, Planctomycetales and Rhizobiales.
Conclusions
Our results suggest that pH and associated soil factors are important determinants of microbial and phoD-harboring community structures. These associated factors include organic carbon and total nitrogen, and to a lesser degree phosphorus status, and active communities are more responsive than total communities. Key players in organic P mineralization are affiliated to phyla that are known to be important in organic matter decomposition.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soils sustain complex and diverse microbial communities that are intimately associated with their habitat. Soil properties and vegetation fundamentally shape the microbial community by affecting its composition, diversity and functioning (Berg and Smalla 2009; Dequiedt et al. 2011). Within the soil microbial community, archaea, bacteria and fungi are influenced differently by soil properties and by the plants present. Among the soil properties, soil pH has repeatedly been shown to be the primary environmental driver influencing the total bacterial community structure studied at DNA level (Fierer and Jackson 2006; Griffiths et al. 2011; Lauber et al. 2008). The total fungal community has been reported to be less responsive to pH changes than the total bacterial community (Blagodatskaya and Anderson 1998; Wakelin et al. 2008), and only some specific groups of archaea such as ammonia oxidizers have been shown to change with pH (Nicol et al. 2008). Specific taxonomic groups such as the bacterial groups Acidobacteria, Bacteroidetes and Firmicutes, and the fungal Helotiales, Hypocreales and Mitosporic Basidomycetes have been reported to be highly responsive to pH (Lauber et al. 2009; Rousk et al. 2010). However, the effect of pH on the active community of these microorganisms (studied at RNA level) remains unclear.
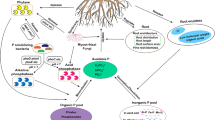
Soil pH also affects the availability of nutrients such as phosphorus (P) by modifying adsorption and desorption reactions (Frossard et al. 1995). This makes it difficult to distinguish the effect of P availability on microbial communities from that of pH. P is often a limiting nutrient for plants and microorganisms in soil (Vitousek et al. 2010) and phosphate fertilizer is therefore commonly applied to agricultural soils to maintain crop productivity. Phosphate addition can lead to a shift in the microbial community composition (Mander et al. 2012; Siciliano et al. 2014). Archaea and bacteria are generally thought to be more responsive than fungi to water-soluble phosphate addition (Bissett et al. 2011; Cruz et al. 2009). Under low P availability, microorganisms up-regulate the genes of the Pho regulon, which includes genes encoding phosphatases, phosphate transporters and other systems for mobilizing soil P (Vershinina and Znamenskaya 2002). Phosphatases can potentially hydrolyze up to 89 % of the total organic P extracted from soil (Jarosch et al. 2015).
The PhoD alkaline phosphatase is found in a range of environments (Luo et al. 2009; Tan et al. 2013) and is mainly produced by bacteria (Ragot et al. 2015). It hydrolyzes phosphomonoesters and –diesters excluding inositol phosphates such as phytate (Kageyama et al. 2011), which represent an important fraction of organic P in soil (Condron et al. 2005). The few studies addressing the diversity, total community structure and composition of the phoD gene report effects of organic and water-soluble phosphate fertilizers, crop management, vegetation and pH (Chhabra et al. 2012; Jorquera et al. 2014; Ragot et al. 2015; Sakurai et al. 2008; Tan et al. 2013; Wang et al. 2012a). Effects of soil pH on the total phoD-harboring community have been reported in studies in which organic C and available P increased together with pH (Cui et al. 2015; Wang et al. 2012b). Using pyro-sequencing, dominant phoD-harboring community members have been shown to be affiliated primarily to Alpha- and Gamma-Proteobacteria but also to Acidobacteria and Cyanobacteria (Tan et al. 2013; Fraser et al. 2015). However, these studies used a set of primers (ALPS primers; Sakurai et al. 2008) that has been shown to have an amplification bias towards Alpha-Proteobacteria and to cover the phoD diversity only partly (Tan et al. 2012; Ragot et al. 2015).
This study aimed to clarify the effect of phosphate depletion and soil pH on microbial communities in a long-term fertilization trial on a permanent grassland characterized by a pH gradient and low P availability (Bünemann et al. 2012). To evaluate the effect of phosphate depletion, we compared microbial communities in plots fertilized with N, P and K, which maintained soil P stocks, with plots fertilized with N and K only, which resulted in phosphate depletion (Liebisch et al. 2013). To evaluate the effect of pH, we sampled plots immediately adjacent to the fertilization trial, which had two different pH values. The total and active archaeal, bacterial, fungal and phoD-harboring community structures were examined, and related to soil and plant properties using multivariate analysis, allowing us to separate the effects of phosphate depletion and pH. In addition, we analyzed the composition of the total and active phoD-harboring communities to identify the key microorganisms that harbor and/or express the phoD gene. It has been shown that the total and active microbial community can differ significantly during decomposition processes, as only some members of the total community (especially Acidobacteria, Actinobacteria and Proteobacteria) are metabolically active (Baldrian et al. 2012). We therefore expected stronger expression of the phoD gene by these taxonomic groups than by other phoD-harboring organisms, and the active community structures to differ more between treatments than the total community structures.
Material and Methods
Site description and sampling
Soils were sampled in September 2012 from a long-term fertilization trial (1992–2014) at the Agroscope research station near Watt in the Swiss lowland (47°26′44″N, 8°29′32″E, 500 m asl) (Huguenin-Elie et al. 2006). The vegetation consists of a permanent grassland of Arrhenatherion elatioris association with 30 to 40 angiosperm species (Liebisch et al. 2013), which is harvested as hay three times per year with a late first harvest (after 15th June) to preserve plant diversity. The mesic grassland grows on a medium-deep (50–70 cm) Cambisol (WRB 2014) with a medium base saturation. The soil in the upper 10 cm consists of 220, 340 and 440 g kg−1 clay, silt and sand, respectively (Philipp et al. 2004).
The fertilization trial has a randomized block design with four replicates and a plot size of 2 m × 5 m (Fig. 1). The treatments represent different combinations of mineral N, P and K additions. In this study, we selected two treatments: one fertilized with N, P and K (NPK) and one fertilized with N and K only (NK). The amounts added were 45 kg N ha−1 yr.−1 as ammonium nitrate, 17 kg P ha−1 yr.−1 as single superphosphate and 83 kg K ha−1 yr.−1 as potassium chloride. The phosphate input in NPK plots was adequate to reach a zero P balance, while phosphate depletion via the negative P balance of −10 kg P ha−1 yr.−1 in NK decreased P availability (Liebisch et al. 2013).
Previous work indicated the presence of a pH gradient along the field that affects mainly one NPK plot (Southwest corner plot in block IV; Fig. 1), which has a higher pH than the other NK and NPK plots (Liebisch et al. 2013). In order to assess the effect of pH, an additional set of samples was taken in the border strip next to the original field trial at both the lower (pH 5.0) and the higher (pH 6.1) pH end of the trial by extending each block by one plot on each side as indicated in Fig. 1. This border strip has not been fertilized since 1992, but the vegetation has been cut and removed along with harvests in the trial. Five soil cores (0–5 cm) were randomly collected in each plot, and were combined and homogenized by sieving (8 mm sieve) directly after sampling. A subsample of soil was immediately frozen in liquid nitrogen in the field and stored at −80 °C until molecular analysis. Further subsamples were stored at −20 °C for later enzymatic analysis, or dried and finely ground in a ball mill for determination of total nutrient contents. The remaining soil was stored at 4 °C for 2 weeks until analysis. All results are converted to soil dry weight equivalents.
Soil and vegetation analyses
Total C, N and P, and pH
Total organic C (TOC) and total N (TN) were measured on finely ground soil using a CNS analyzer (Thermo-Fisher Flash EA 1112). Total P (TP) in soil was determined by wet digestion with H2O2/H2SO4 (Anderson and Ingram 1993) and measured with malachite green at 610 nm using a UV-1800 Shimadzu Spectrometer (Ohno and Zibilske 1991). Soil pH was measured in a soil suspension in 0.01 M CaCl2 (1 to 2.5 soil mass to volume ratio, shaken for 1 h) using a Benchtop pH 720 A (Orion Research Inc., Jacksonville, FL).
NaOH-EDTA-extractable organic P
Organic P was determined using alkaline extraction after Bowman and Moir (1993). Briefly, 2.5 g of finely ground soil was extracted with 0.25 M NaOH - 0.05 M EDTA in a 1 to 10 (w/v) ratio for 16 h on a horizontal shaker. Soil extracts were then centrifuged and filtered (Whatman No. 1). The concentration of inorganic P in the extract was measured colorimetrically using malachite green (Ohno and Zibilske 1991). The concentration of total P in the extract was measured colorimetrically using malachite green on neutralized extracts after digestion with ammonium persulphate in an autoclave (20 min, 121 °C). Organic P was calculated by subtracting inorganic P from total P in the extract.
Microbial C, N and P and resin-extractable P
Microbial C and N analysis was done using chloroform fumigation-extraction on 25 g of moist soil (Vance et al. 1987). Total organic C and total N in the extracts were measured using a FormacsSERIES TOC/TN analyzer (Skalar, The Netherlands). Concentrations in non-fumigated subsamples were subtracted from those in fumigated ones to calculate chloroform-labile C (MicC) and N (MicN). No conversion factor was applied since this is soil-specific and has not been determined for the soils in this study.
Microbial and resin-extractable P were determined on moist samples by fumigation-extraction using anion-exchange resin membranes (BDH laboratory supplies #55,164 2S, England) charged with CO3 2−and 1-hexanol as fumigant (Kouno et al. (1995) as modified by Bünemann et al. (2007)). Microbial P (MicP) was calculated as the difference between fumigated and non-fumigated subsamples (resin-extractable P, Pres) and corrected for the incomplete recovery of a P spike, which ranged between 88 % and 99 %. No conversion factor was applied, for the same reason as above.
Potential acid and alkaline phosphatase activity
Potential acid (ACP) and alkaline (ALP) phosphatase activity were determined following Marx et al. (2001) as modified by Poll et al. (2006). Briefly, one gram of moist soil was dispersed in 100 mL of autoclaved ddH2O using an ultrasonic probe Labsonic U (Braun Melsungen, Germany) at 50 J s−1 for 120 s. The assay contained 50 μL aliquots of the soil suspension in a microplate with 6 analytical replicates, using 1 mM 4-methylumbelliferylphosphate as substrate and either 0.1 M MES buffer (pH 6.1) or Modified Universal Buffer (pH 11) (Alef et al. 1995) as buffer for ACP and ALP activity, respectively. Microplates were incubated on a horizontal shaker for 3 h at 30 °C. The linear increase in fluorescence over time was measured, and the enzymatic activity (μmol h−1 g soil−1) was calculated by normalization to a 4-methylumbelliferone standard added to the soil suspension.
Vegetation
All vascular plant species were identified and their relative abundance at heading stage of the dominant grass species was visually estimated in May 2013 following Dietl (1995). Plant samples for analysis of P and N content were taken in June 2013, shortly before the first cut of the year. The vegetation was cut at 4 cm above the ground using electric scissors in a randomly selected 50 × 50 cm square with at least 0.5 m distance to the border of the plot. The material was sorted into grasses, legumes and forbs, and the three groups were dried at 60 °C for 3 days and milled. Plant N content was measured by a CNS analyzer. To determine plant P content, 250 mg of plant material was ashed (6 h, 550 °C) and dissolved in 3 mL 10.3 M HNO3 (Westerman 1990). Samples were diluted and neutralized before colorimetric P determination using malachite green.
Molecular analysis
Nucleic acid extraction and reverse transcription
Nucleic acids were extracted in analytical duplicates from 2 g of snap-frozen soil using the RNA Powersoil® Total RNA isolation kit (MO BIO, Carlsbad, CA, USA) according to the manufacturer’s instructions, with an additional homogenizing step (2.8 mm zirconium beads for 1 min at 5 m s−1) using the Omni Bead Ruptor Homogenizer (Omni International, Kennesaw, GA) prior to isolation. RNA was eluted from the RNA/DNA Capture Column, precipitated and resuspended following the manufacturer’s instructions. Subsequently. DNA was eluted using 4 mL of DNA elution solution (1 M NaCl, 50 mM MOPS, 15 % (v/v) isopropanol, pH 7). DNA was precipitated with isopropanol, washed, dried, and redissolved in DEPC-treated H2O. RNA samples were treated with Turbo DNase (Thermo Fisher Scientific Inc., Waltham, MA) to remove DNA contamination. RNA concentrations were measured using a Qubit® 2.0 Fluorometer (Life Technologies, Inc., Grand Islands, NY). Complementary DNA (cDNA) was synthesized from 200 ng of total RNA, using 500 ng random hexamers (Thermo Fisher Scientific Inc.) and 160 Units GoScript™ Reverse Transcriptase (Promega, Madison, WI) according to the manufacturer’s recommendations.
Analysis of the archaeal, bacterial and fungal community structures using T-RFLP
Archaeal, bacterial and fungal communities were studied using regions of the 16S rRNA gene for archaea and bacteria, and the ITS region for fungi, as detailed below. All PCRs (25 μL) contained 0.6 Units GoTaq (Promega), 1X reaction GoTaq Buffer, 2 mM MgCl2, 0.2 mM dNTPs, 0.5 μM of each primer, and 1–2 ng template DNA. Amplification of archaeal 16S DNA and cDNA was carried out with Ar109F/FAM-Ar915R primers (Lueders and Friedrich 2000). Bacterial 16S DNA and cDNA were amplified using 1064R/FAM-356F primers (Winsley et al. 2012). The fungal community was investigated with FAM-ITS1-F (Gardes and Bruns 1993)/ITS4-R (White et al. 1990) primers. After initial denaturation for 5 min at 95 °C, amplification was done for 35 cycles comprising a denaturation step of 30 s at 95 °C, an annealing step of 30 s at 53 °C (archaea), 60 s at 52 °C (bacteria) or 30 s at 60 °C (fungi), and an extension step of 60 s at 72 °C. A final extension step of 5 min at 72 °C was carried out to complete the reaction. Results from DNA- and cDNA-analysis are referred to as measurements of the total and active communities, respectively.
Restriction enzyme digestion reaction contained 10 μl PCR product, 5 U of MspI (Promega), 2 μg acetylated bovine serum albumin and water to a final volume of 20 μL, and was carried out for 3 h at 37 °C. Digested products were precipitated by adding 3 μL of 3 M CH3COONa (pH 5.2), 3 μL of 100 mM Na2EDTA pH 8.0), 400 μg of glycogen and 60 μL 95 % (v/v) ice-cold ethanol. Products were centrifuged, washed with 70 % (v/v) ice-cold ethanol and redissolved in 15 μL DEPC-treated H2O. The purified products (3 μL) were analyzed on an ABI 3130XL sequencer using Liz500 as standard (Life Technologies, Inc.). The T-RFLP electropherograms of the archaeal, bacterial and fungal communities were analyzed in GeneMapper v3.7 (Applied Biosystems, Foster City, CA) with a fragment length range of 50–500 base pairs. T-RFLP profiles were processed and analyzed with the T-REX program (Culman et al. 2009), using the peak area to identify operational taxonomic units (OTUs) and a clustering threshold of 0.5 bp.
Analysis of total and active phoD-harboring community structure and composition using 454-sequencing
The phoD alkaline phosphatase gene was amplified by PCR from DNA and cDNA using phoD-F733/R1083 primers (Ragot et al. 2015) in a reaction mixture (25 μL) containing 0.6 Units GoTaq (Promega) 1X GoTaq buffer, 2 mM MgCl2, 0.2 mM dNTPs, 0.5 μM of each primer, and 1–2 ng DNA or cDNA as template. Amplification included an initial denaturation step of 5 min at 95 °C, followed by 35 cycles of a denaturation step of 30 s at 95 °C, an annealing step of 30 s at 58 °C and an extension step of 60 s at 72 °C. The final extension step was carried out for 10 min at 72 °C. Field replicates of DNA and cDNA extracts of each plot were diluted to 20 ng μL−1, pooled and analyzed by 454-sequencing using a GS-FLX+ platform (Roche 454 Life Sciences, Branford, CT), at Research and Testing Laboratory (Lubbock, TX). The Standard Flowgram Format (SFF) files were submitted to the European Nucleotide Archive (ENA) under the accession number ERP010357.
Reads resulting from the 454-sequencing were analyzed using MOTHUR (Schloss et al. 2009) as described in Ragot et al. (2015). Briefly, resulting reads were trimmed at 150 bp and 450 bp as minimum and maximum length, respectively, and aligned using the Needleman-Wunsch global alignment algorithm as implemented in MOTHUR, using 6-mers searching and aligned reference sequences as template as in Ragot et al. (2015). The pairwise distance matrix was calculated from the alignment and sequences clustered using the k-furthest method as implemented in MOTHUR with a similarity cutoff at 75 % to define the operational taxonomic units (OTUs) as calculated by Tan et al. (2013). OTU matrices were then normalized to the smallest library size using the normalized.shared command as implemented in MOTHUR to allow comparison between plots (Supplementary Table S4). Taxonomy was assigned using BLAST+ (Camacho et al. 2009) with a minimum e-value of 1e−8 to retrieve NCBI sequence identifiers (gi accession number), and in-house Perl scripts were then used to obtain the taxonomy for each read (scripts written by Stefan Zoller, Genetic Diversity Centre, ETH Zurich) (Supplementary Table S5).
Statistical analysis
Since the plots with different pH levels were not part of the randomized block design of the fertilization trial, soil properties and plant data from the fertilization trial and from the two pH levels were subjected to independent statistical analysis. Redundancy analysis (RDA) was then used to evaluate the effect of soil and plant properties on microbial community structures across the whole dataset.
Two-tailed t-test was used to examine differences differed between fertilization treatments, and between pH levels, respectively. Analysis of dissimilarity (anosim) in the vegan package (vegan: Community Ecology Package) in R v.2.15.0 (R Core Team, http://www.R-project.org/, 2014) was used to test whether the vegetation composition was significantly different between fertilization treatments and between pH levels, respectively. Linear correlations between soil properties were tested in a Pearson’s correlation matrix (Supplementary Table S1).
T-RFs of the archaeal, bacteria and fungal communities were normalized using Hellinger distance (Legendre and Gallagher 2001) and T-RFs were further converted into a binary matrix. Species richness was represented by the number of OTUs found in the T-RF profiles in each community (Supplementary Table S3). Analysis of dissimilarity was used to evaluate whether the T-RF profiles of the total and active microbial communities were significantly different between fertilization treatments and between pH levels.
Rarefaction curves based on phoD reads were calculated and extrapolated to an average library size of 5099 reads to standardize the samples using EstimateS (Version 9, http://purl.oclc.org/estimates). Species richness of the phoD-harboring community was calculated using the Chao1 index (Chao and Shen 2003) (Supplementary Table S3). Structures of the phoD-harboring community within the fertilization treatments and within the pH levels were compared using pairwise Libshuff analysis with 1000 iterations as implemented in MOTHUR (Schloss et al. 2004).
Relationships between the microbial and phoD-harboring community structures and soil and plant properties standardized by the z-score method (Ramette 2007) were analyzed using RDA on T-RFLP data for the archaeal, bacterial and fungal community and on 454-sequencing data for the phoD-harboring community. Plant species were only included in the analysis if they had an abundance of more than 2 % and differed significantly in abundance (p < 0.05) in either fertilization treatments or pH levels (Supplementary Table S2).
Results
General soil physicochemical and biological properties
Soil pH was similar in the two fertilization treatments, although one NPK replicate had a pH of 5.9, while pH in all other NK and NPK plots varied between 4.9 and 5.3 (Table 1). Phosphate depletion in NK decreased TP, Porg, Pres, MicP and ACP activity compared to NPK. The pH levels differed significantly by one pH unit. At higher pH values, concentrations of TOC, TN and TP were greater than at lower pH.
Potential ACP and ALP activities were strongly correlated with pH, with ALP activity being five times greater at pH 6.1 than at pH 5.0, whereas ACP activity showed the opposite trend (Supplementary Table S1). Among the other soil properties, pH was linearly correlated with TOC, TN, TP, MicC, MicN and MicP (Supplementary Table S1).
Plant productivity and composition
Phosphate depletion led to a 1.8-fold decrease in total plant yield, though different plant groups were differently affected, with a 2.7-fold decrease in grass biomass contrasting with a 2-fold increase in forb biomass (Table 2). Phosphate depletion also decreased plant N and P content by factors of two and four, respectively. In total, 40 plant species were observed in the fertilization treatments (Supplementary Table S2). The vegetation composition was significantly different between the fertilization treatments, while the plant richness remained similar (p-value =0.03). Among the eight plant species differing in abundance between NK and NPK treatments, the legumes Trifolium pratense and Lotus corniculatus were more abundant in NK than NPK, while the grasses Arrhenatherum elatius and Holcus lanatus were more abundant in NPK than NK.
Grass, legume and forb yields were similar at both pH levels (Table 2). Lower pH led to a higher plant N content, associated with a slightly reduced species richness. A total of 42 plant species was observed at the two pH levels (Supplementary Table S2). Vegetation composition was significantly different between the pH levels (p-value =0.03). Acidomesophilic plant species such as Anthoxanthum odoratum, Cynosurus cristatus and Rumex acetosa were more abundant at pH 5.0, while alkalomesophilic plant species such as Knautia arvensis and Galium album were more abundant at pH 6.1 (Hill et al. 1999). This shows that one unit difference in pH can significantly influence the vegetation composition.
Effect of phosphate depletion and soil pH on total and active archaeal, bacterial, fungal and phoD-harboring community structures
Phosphate depletion led to a significant shift in both total and active archaeal and phoD-harboring community structures (Table 3). For bacteria and fungi, an effect of phosphate depletion on active but not on total community structures was observed.
The difference in pH of about one unit between pH levels affected the structure of both total and active communities of archaea, bacteria and phoD (Table 3). The total fungal community structure was similar at both pH values, whereas the active fungal community structure differed.
Correlations between the total and active archaeal, bacterial, fungal and phoD-harboring community structures and environmental factors
Based on the first and second component of the RDA, the total archaeal community structure in the NPK treatment was positively correlated to Pres, plant P content, grass yield and total plant yield, and negatively correlated to forb yield (Fig. 2a and b, Table 4). In addition to pH, the total archaeal community structure at pH 6.1 was also positively correlated to TOC, TN and the abundances of Galium album and Knautia arvensis, while the community structure in NK and at pH 5.0 was negatively related to these variables. The abundance of Anthoxanthum odoratum decreased with pH and was thus negatively related to the communities at pH 6.1. The active archaeal community structure was correlated to the same plant species as the total archaeal community, and was mainly affected by pH, TOC and TN.
Redundancy analysis (RDA) of the community structures of archaea (a, b), bacteria (c, d) and fungi (e, f) based on the T-RF profiles in NK (●) and NPK (▲) and at pH 5.0 (Δ) and 6.0 (○) (means of four field replicates with standard deviation). Displayed vectors represent the soil and plant properties that are significantly correlated to the community structures (p ≤ 0.05)
Both total and active bacterial community structures showed a strong separation between the two pH levels and to a lesser degree between the fertilization treatments, which was mostly correlated with pH, TOC, TN and the abundances of Galium album and Knautia arvensis (Fig. 2c and d, Table 4). In contrast, the total fungal community structure was not correlated to any of the soil and plant properties (Fig. 2e, Table 4). The active fungal community structure, however, was related to soil pH as well as to plant N and P content and grass yield, which were all significantly higher in NPK than in NK treatment (Fig. 2f, Table 4).
The total phoD-harboring community structure was correlated with pH, TOC and TN, whereas the active phoD-harboring community structure was only correlated with pH and TOC. In both cases, these soil properties were mainly associated with the first component, which explained 19.9 and 17.9 % of the variability in the total and active phoD-harboring community structures, respectively (Fig. 3a and b, Table 4). Additionally, the active phoD-harboring community structure of the NK and NPK treatments showed tighter clustering than for the total phoD-harboring community structure, suggesting a more similar community structure of the active than of the total phoD-harboring members (Fig. 3a and b). The distribution of the total and active phoD-harboring community structures was similar at both pH levels (Fig. 3a and b).
Taxonomic composition of the total and active phoD-harboring community as related to soil and plant properties
The taxonomic composition of the total and active phoD-harboring community was investigated using 454-sequencing and BLAST+ to elucidate the key phoD-harboring members in the studied soils. Taxonomy was assigned to the majority of OTUs, which were all affiliated with the bacterial kingdom. A remainder of 692 reads could not be identified and represented 0.8–3.1 % of the reads before normalization (Supplementary Table S4 and S5). The phoD gene was found in 28 bacterial orders (Fig. 4). The dominant bacterial orders across all samples were Actinomycetales (15–32 %), Bacilliales (5–15 %), Gloeobacterales (6–23 %), Planctomycetales (17 %–30 %) and Rhizobiales (5 %–26 %).
The dominant bacterial orders that harbor and express the phoD gene were similar in all plots. Nonetheless, some differences between treatments and/or between the total and active community were observed. For example, Rhizobiales had a low relative abundance in the NK treatment, while they were dominant in the NPK treatment (Fig. 4). Xanthomonadales had a low relative abundance in the total community, but a high relative abundance in the active community of the NK treatment. Finally, the relative abundance of some groups such as Gloeobacterales was not affected by the treatments.
Furthermore, the relative abundance of several phoD-harboring orders was strongly correlated to individual soil properties (Fig. 4). The abundance of Caulobacteriales and Pseudomonadales decreased with increasing Porg and Pres, while that of Planctomycetales increased with Porg and Pres. Additionally, the abundance of Actinomycetales and Gemmatimonadales increased together with pH, TN, TOC and TP, while the abundance of Burkholderiales decreased with increasing pH, TN and TP, and the abundance of Gloeobacterales decreased with increasing pH, TOC and TN. All correlated soil properties were in turn affected by either phosphate depletion or pH.
Discussion
Effect of phosphate depletion on the total and active archaeal, bacterial, fungal and phoD-harboring community structures
In this work, we studied the effects of phosphate depletion and soil pH on the archaeal, bacterial, fungal and phoD-harboring community structures in a long-term fertilization trial characterized by a pH gradient on site. Our results show that the active archaeal, bacterial and fungal community structures are more responsive to phosphate depletion than their corresponding total community structures, except for the total archaeal community structure, which was affected by phosphate depletion and correlated to phosphate availability (Pres). Indicators of phosphate depletion (TP, Porg and Pres) were otherwise not significant drivers of the communities in the first and second components of the RDA (Fig. 2, Table 4). The active community structures of archaea, bacteria and fungi were all correlated with pH and grass yield, which is an indicator of productivity (Fig. 2, Table 4).
Our results are in agreement with those of He et al. (2007) who showed a significant shift in total archaeal community structure between NK and NPK treatments in a wheat-maize rotation cropping system. In their study, phosphate addition specifically promoted ammonia-oxidizing Crenarchaeota. Our results on the total bacterial community, however, contrast with several studies that reported a significant effect of phosphate addition on the total bacterial community structure in grassland, cropped and forest soils (Beauregard et al. 2009; Liu et al. 2012; Tan et al. 2013; Zhong and Cai 2007). The total fungal community has also been reported to be shaped primarily by phosphate availability (Lauber et al. 2008). The fact that in our study only the active bacterial and fungal community responded to phosphate depletion may be related to the level of phosphate fertilization. Previous studies in the same long-term trial revealed that microorganisms were P limited in the NK treatment (Bünemann et al. 2012) and that the P balance equaled zero in NPK treatment, suggesting that phosphate addition was adequate to maintain plant productivity but did not lead to a net increase in available and total P (Liebisch et al. 2013). Our results suggest that phosphate depletion over time impacts only the active bacteria and fungi, but does not change the structure of the greater total community. Our data also suggest that archaea are more sensitive to phosphate depletion than bacteria and fungi.
Since phoD is part of the Pho regulon (Vershinina and Znamenskaya 2002), we expected available P (Pres) to be correlated with the active phoD-harboring community structure. Moreover, low available P (Pres) was expected to result in an increase of ACP/ALP activity in NK compared to NPK. Our results showed that phosphate depletion led to a significant shift in the total and active phoD-harboring community structure and increased ACP activity (Fig. 3, Table 3). However, this shift in the total and active phoD-harboring community structure was not correlated to any of the phosphate depletion indicators (TP, Porg and Pres) (Fig. 3, Table 4). Based on the first and second RDA components, our results show that the total and active phoD-harboring community structures were most strongly correlated to pH and TOC, suggesting that the effect of phosphate depletion may be masked by the strong effect of pH and TOC in the RDA. Previous DNA-based studies on phoD have shown contrasting results of phosphate addition. Diversity of phoD has been reported to increase, decrease and not to be affected by phosphatase addition (Chhabra et al. 2012; Jorquera et al. 2014; Tan et al. 2013). In a long-term trial including non-fertilized soils, soil fertilized with composted manure or water-soluble phosphate, and prairie soils, Fraser et al. (2015) observed a general positive correlation between phoD gene abundance and potential ALP activity in soil, suggesting that PhoD contributes significantly to the total ALP activity. However, comparing the non-fertilized and the water-soluble fertilized soils, they did not observe a consistent response to phosphate addition in ALP activity and phoD gene abundance over 2 years of the experiment, suggesting that water-soluble phosphate addition does not always decrease potential ALP activity. Our results are in agreement with the outcome of their study and showed that phosphate depletion did not increase ALP activity nor change the total phoD-harboring community structure as hypothesized. However, our results suggest that phosphate depletion affects significantly the active phoD-harboring community composition, even though phosphate depletion indicators (TP, Porg and Pres) were not significant drivers in the first and second components of the RDA (Fig. 3, Table 4).
Effect of soil pH on the total and active archaeal, bacterial, fungal and phoD-harboring community structures
One unit difference in pH value affected all investigated total and active microbial community structures except for the total fungal community (Fig. 2, Fig. 3, Table 4). Soil pH also changed some soil and plant properties such as TOC and TN, which in turn were important drivers of the microbial community structures. While bacterial community structure is widely known to be strongly correlated with soil pH (Fierer and Jackson 2006; Griffiths et al. 2011; Lauber et al. 2008; Nacke et al. 2011), less information is available for archaea and fungi. Studies of Crenarchaeota (Lehtovirta et al. 2009) and crenarchaeal ammonium-oxidizing archaea (Nicol et al. 2008) showed that pH can select for particular archaeal groups. Lauber et al. (2008) reported that the total fungal community structure is stable over pH values ranging from 3.6 to 4.7, though it is possible that only specific fungal taxa such as Helotiales and Hypocreales are responsive to pH changes (Rousk et al. 2010). Our results suggest that the total fungal community is also stable over pH values ranging from 5.0 to 6.0, while one unit difference in pH value significantly affected the active fungal community. This suggests that the active fungal community is more sensitive to pH differences than the greater total fungal community. More importantly, we could not distinguish the effect of pH from that of TOC and total N, which are both commonly correlated with pH in soil (Drenovsky et al. 2004; Lauber et al. 2008; Siciliano et al. 2014). Previous studies have reported pH as the main driver of the total phoD-harboring community in arable and grassland soils (Cui et al. 2015; Ragot et al. 2015; Wang et al. 2012b). This suggests that pH rather than TOC and TN led to a shift in the structure of the total and active phoD-harboring community.
Key phoD-harboring microorganisms
The phoD gene was only found in bacteria in this study, although it is also present in archaea (e.g. Euryarcheota) and fungi (e.g. Ascomycetes) (see Integrated Microbial Genomes database, https://img.jgi.doe.gov/). Our results showed that despite differences in soil and plant properties, the dominant bacterial orders that harbor and express the phoD gene were similar in all plots, suggesting that the composition of the total and active phoD-harboring community is rather stable. Nonetheless, differences between treatments in relative abundances of both the total and active communities were also observed. For example, phoD-harboring Gemmatimonadales were dominant at pH 5.0 in both the total and active communities, whereas they were almost absent at pH 6.1 (Fig. 4). Furthermore, differences between total and active phoD-harboring community compositions within one treatment indicate that several orders are more active than others under certain soil conditions. For example, the relative abundance of phoD-harboring Xanthomonadales was higher in the active than in the total community of the NK treatment, while it was lower in the active than in the total community of the NPK treatment. Similarly, the active phoD-harboring Planctomycetes and Pseudomonadales had a lower relative abundance in the NPK than in the NK treatment, while they had a similar relative abundance in the total community. Additionally, the relative abundance of the total and the active phoD-harboring Bacilliales at pH 5.0 and 6.1 suggests that higher pH strongly up-regulates the expression of the phoD gene in this taxonomic group. Additionally, strong correlations were observed between soil pH, TOC, TP and TN and the relative abundance of some bacterial orders such as Actinomycetales (Fig. 4).
In contrast to our results, Tan et al. (2013) observed that water-soluble phosphate addition decreased the relative abundance of phoD-harboring Acidobacteria and Pseudomonas fluorescens. They reported that the dominant phoD-harboring phyla were the same at all studied phosphate fertilization regimes (0, 15 and 30 kg P ha−1) and included mainly Proteobacteria (predominantly Alphaproteobacteria), Actinobacteria and Cyanobacteria. In our study, the key phoD-harboring phyla were Actinobacteria, Cyanobacteria, Firmicutes and Planctomycetes. The differences in the phoD-harboring community composition between our results and those of Tan et al. (2013) may be attributed to the fact that Tan et al. (2013) used primers designed by Sakurai et al. (2008), which have been shown to have an amplification bias, amplifying only a limited number of microbial groups and resulting in an over-representation of Alphaproteobacteria in the phoD-harboring community (Ragot et al. 2015; Tan et al. 2013). Nonetheless, our findings show that the regulation of phoD expression in Bacilliales, Burkholderiales, Chroococcales, Gloeobacterales, Gemmatimonadales, Planctomycetales, Rhizobiales and Xanthomonadales may be linked to phosphate depletion and pH.
In support of our findings, some of the key phoD-harboring taxonomic groups identified in our study are known to play an important role in other nutrient cycles. For example, Proteobacteria (e.g. Rhizobiales) are known to be important players in C cycling as decomposers (Štursová et al. 2012) and in N cycling as N2-fixing bacteria (Hayatsu et al. 2008). Actinobacteria (e.g. Actinomycetales) and Cyanobacteria (e.g. Gloeobacterales) also play a relevant role in N cycling as N2-fixing and nitrifiers (Fierer et al. 2007; Hayatsu et al. 2008). Moreover, although their ecological role is not clear, Deinococcus-Thermus and Planctomycetales have been reported to grow well in high organic matter soils (Buckley et al. 2007; Štursová et al. 2012), suggesting that they also play a role as decomposers. Our results show that these taxonomic groups play an additional ecological function as organic P mineralizers in soil.
Conclusions
Our results showed that the active communities are generally more responsive to pH and phosphate depletion and, thus, may reflect better the response of microbial communities to their environment. Additionally, our results suggest that the fungal community is less responsive to pH and phosphate depletion than the archaeal, bacterial and phoD-harboring communities. Soil pH, Corg and TN were more important drivers than P status (TP, Porg and Pres) which did not appear in the first and second components of the RDA. Despite differences in soil and plant properties, the dominant phoD-harboring members were overall the same in all plots (Actinomycetales, Rhizobiales, Gloeobacterales and Planctomycetales). This suggests that the potential key player in organic P mineralization remain similar despite differences in certain soil conditions.
Abbreviations
- ACP:
-
Potential acid phosphatase activity
- ALP:
-
Potential alkaline phosphatase activity
- MicC:
-
Microbial C
- MicN:
-
Microbial N
- MicP:
-
Microbial P
- P:
-
Phosphorus
- Porg :
-
Organic P
- Pres :
-
Resin-extractable P
- TN:
-
Total N
- TOC:
-
Total organic C
- TP:
-
Total P
References
Alef K, Nannipieri P, Trasar-Cepeda C (1995) Phosphatase activity. In: Alef K, Nannipieri P (eds) Methods in Applied Soil Microbiology and Biochemistry. Academic Press, London
Anderson JM, Ingram JSI (1993) Tropical soil biology and fertility. A handbook of methods. CAB International, Wallingford
Baldrian P, Kolařík M, Štursová M, Kopecký J, Valášková V, Větrovský T, Žifčáková L, Šnajdr J, Rídl J, Vlček Č (2012) Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J 6:248–258
Beauregard MS, Hamel C, Nayyar A, St-Arnaud M (2009) Long-term phosphorus fertilization impacts soil fungal and bacterial diversity but not AM fungal community in Alfalfa. Microb Ecol 59:379–389
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Bissett A, Richardson AE, Baker G, Thrall PH (2011) Long-term land use effects on soil microbial community structure and function. Appl Soil Ecol 51:66–78
Blagodatskaya EV, Anderson TH (1998) Interactive effects of pH and substrate quality on the fungal-to-bacteria ratio and qCO2 of microbial communities in forest soils. Soil Biol Biochem 20:1269–1274
Bowman RA, Moir JO (1993) Basic EDTA as an extractant for soil organic phosphorus. Soil Sci Soc Am J 57:1516–1518
Buckley DH, Huangyutitham V, Hsu S-F, Nelson TA (2007) Stable isotope probing with 15N2 reveals novel noncultivated diazotrophs in soil. Appl Environ Microbiol 73:3196–3204
Bünemann EK, Marschner P, McNeill AM, McLaughlin MJ (2007) Measuring rates of gross and net mineralisation of organic phosphorus in soils. Soil Biol Biochem 39:900–913
Bünemann EK, Oberson A, Liebisch F, Keller F, Annaheim KE, Huguenin-Elie O, Frossard E (2012) Rapid microbial phosphorus immobilization dominates gross phosphorus fluxes in a grassland soil with low inorganic phosphorus availability. Soil Biol Biochem 51:84–95
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC bioinformatics 10:421
Chao AC, Shen T-J (2003) Nonparametric estimation of Shannon's index of diversity when there are unseen species in sample. Environ Ecol Stat 10:429–443
Chhabra S, Brazil D, Morrissey J, Burke J, O’Gara F, Dowling DN (2012) Fertilization management affects the alkaline phosphatase bacterial community in barley rhizosphere soil. Biol Fertil Soils 49:31–39
Condron LM, Turner BL, Cade-Menun BJ (2005) Chemistry and dynamics of soil organic phosphorus. In: Sims JT, Sharpley AN (eds) Phosphorus: agriculture and the environment. ASA, CSSA and SSSA, Madison
Cruz AF, Hamel C, Hanson K, Selles F, Zentner RP (2009) Thirty-seven years of soil nitrogen and phosphorus fertility management shapes the structure and function of the soil microbial community in a Brown Chernozem. Plant Soil 315:173–184
Cui H, Zhou Y, Gu Z, Zhu H, Fu S, Yao Q (2015) The combined effects of cover crops and symbiotic microbes on phosphatase gene and organic phosphorus hydrolysis in subtropical orchard soils. Soil Biol Biochem 82:119–126
Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH (2009) T-REX: software for the processing and analysis of T-RFLP data. BMC bioinformatics 10:171–181
Dequiedt S, Saby NPA, Lelievre M, Jolivet C, Thioulouse J, Toutain B, Arrouays D, Bispo A, Lemanceau P, Ranjard L (2011) Biogeographical patterns of soil molecular microbial biomass as influenced by soil characteristics and management. Glob Ecol Biogeogr 20:641–652
Dietl W (1995) Wandel der Wiesenvegetation im Schweizer Mittelland. Zeitschrift für Ökologie und Naturschutz 4:239–249
Drenovsky R, Vo D, Graham K, Scow K (2004) Soil water content and organic carbon availability are major determinants of soil microbial community composition. Microb Ecol 48:424–430
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Fraser T, Lynch DH, Entz MH, Dunfield KE (2015) Linking alkaline phosphatase activity with bacterial phoD gene abundance in soil from a long-term management trial. Geoderma 257:115–122
Frossard E, Brossard M, Hedley MJ, Metherell A (1995) Reactions controlling the cycling of P in soils. In: Tiessen H (ed) Phosphorus cycling in terrestrial and aquatic ecosystems: a global perspective. SCOPE/Wiley, New York
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS (2011) The bacterial biogeography of British soils. Environ Microbiol 13:1642–1654
Hayatsu M, Tago K, Saito M (2008) Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci Plant Nutr 54:33–45
He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, Xu MG, Di H (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:2364–2374
Hill MO, Mountford J, Roy D, Bunce RGH (1999) Ellenberg's indicator values for British plants. ECOFACT Volume 2 Technical Annex. Institute of Terrestrial Ecology
Huguenin-Elie O, Gago R, Stutz C, Lüscher A, Kessler W (2006) Long-term effects of fertilisation on herbage composition, yield and quality of an Arrhenatherion-type meadow. Grass Sci Eur 11:550–552
Jarosch KA, Doolette AL, Smernik RJ, Tamburini F, Frossard E, Bünemann EK (2015) Characterisation of soil organic phosphorus in NaOH-EDTA extracts: A comparison of 31 P NMR spectroscopy and enzyme addition assays. Soil Biol Biochem 91:298–309
Jorquera MA, Martinez OA, Marileo LG, Acuna JJ, Saggar S, Mora ML (2014) Effect of nitrogen and phosphorus fertilization on the composition of rhizobacterial communities of two Chilean Andisol pastures. World J Microbiol Biotechnol 30:99–107
Kageyama H, Tripathi K, Rai AK, Cha-um S, Waditee-Sirisattha R, Takabe T (2011) An alkaline phosphatase/phosphodiesterase, PhoD, induced by salt stress and secreted out of the cells of Aphanothece halophytica, a halotolerant cyanobacterium. Appl Environ Microbiol 77:5178–5183
Kouno K, Tuchiya Y, Ando T (1995) Measurement of soil microbial biomass phosphorus by an anion exchange membrane method. Soil Biol Biochem 27:1353–1357
Lauber CL, Strickland MS, Bradford MA, Fierer N (2008) The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40:2407–2415
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120
Legendre P, Gallagher ED (2001) Ecologically meaningful transformations for ordination of species data. Oecologia 129:271–280
Lehtovirta LE, Prosser JI, Nicol GW (2009) Soil pH regulates the abundance and diversity of Group 1.1c Crenarchaeota. FEMS Microbiol Ecol 70:367–376
Liebisch F, Bünemann E, Huguenin-Elie O, Jeangros B, Frossard E, Oberson A (2013) Plant phosphorus nutrition indicators evaluated in agricultural grasslands managed at different intensities. Eur J Agron 44:67–77
Liu L, Gundersen P, Zhang TQ, Mo J (2012) Effects of phosphorus addition on soil microbial biomass and community composition in three forest types in tropical China. Soil Biol Biochem 44:31–38
Lueders T, Friedrich M (2000) Archaeal population dynamics during sequential reduction processes in rice field soil. Appl Environ Microbiol 66:2732–2742
Luo H, Benner R, Long RA, Hu J (2009) Subcellular localization of marine bacterial alkaline phosphatases. Proc Natl Acad Sci U S A 106:21219–21223
Mander C, Wakelin S, Young S, Condron L, O’Callaghan M (2012) Incidence and diversity of phosphate-solubilising bacteria are linked to phosphorus status in grassland soils. Soil Biol Biochem 44:93–101
Marx MC, Wood M, Jarvis SC (2001) A microplate fluorimetric assay for the study of enzyme diversity in soils. Soil Biol Biochem 33:1633–1640
Nacke H, Thürmer A, Wolher A, Will C, Hodac L, Herold N, Schöning I, Schrumpf M, Rolf D (2011) Pyrosequencing-based assessment of bacterial community structure alon different management types in German forest and grassland soils. PLoS One 6:e17000
Nicol GW, Leininger S, Schleper C, Prosser JI (2008) The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978
Ohno R, Zibilske LM (1991) Determination of low concentrations of phosphorus in soil extracts using malachite green. Soil Sci Soc Am J 55:892–895
Philipp A, Huguenin-Elie O, Flisch R, Gago R, Stutz C, Kessler W (2004) Effects of phosphate input on a low input meadow. Agrarforschung Schweiz 11:86–91
Poll C, Ingwersen J, Stemmer M, Gerzabek MH, Kandeler E (2006) Mechanisms of solute transport affect small-scale abundance and function of soil microorganisms in the detritusphere. Eur J Soil Sci 57:583–595
Ragot SA, Kertesz MA, Bünemann EK (2015) phoD alkaline phosphatase gene diversity in soil. Appl Environ Microbiol 81:7281–7289
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62:142–160
Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351
Sakurai M, Wasaki J, Tomizawa Y, Shinano T, Osaki M (2008) Analysis of bacterial communities on alkaline phosphatase genes in soil supplied with organic matter. Soil Sci Plant Nutr 54:62–71
Schloss PD, Larget BR, Handelsman J (2004) Integration of microbial ecology and statistics: a test to compare gene libraries. Appl Environ Microbiol 70:5485–5492
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Siciliano SD, Palmer AS, Winsley T, Lamb E, Bissett A, Brown MV, van Dorst J, Ji M, Ferrari BC, Grogan P (2014) Soil fertility is associated with fungal and bacterial richness, whereas pH is associated with community composition in polar soil microbial communities. Soil Biol Biochem 78:10–20
Štursová M, Žifčáková L, Leigh MB, Burgess R, Baldrian P (2012) Cellulose utilization in forest litter and soil: identification of bacterial and fungal decomposers. FEMS Microbiol Ecol 80:735–746
Tan H, Barret M, Mooij MJ, Rice O, Morrissey JP, Dobson A, Griffiths B, O’Gara F (2013) Long-term phosphorus fertilisation increased the diversity of the total bacterial community and the phoD phosphorus mineraliser group in pasture soils. Biol Fertil Soils 49:661–672
Vance ED, Brookes PC, Jenkinson DS (1987) An extraction method for measuring soil microbial biomass C. Soil Biol Biochem 19:703–707
Vershinina OA, Znamenskaya LV (2002) The Pho regulons of bacteria. Microbiology 71:497–511
Vitousek PM, Porder S, Houlton BZ, Chadwick OA (2010) Terrestrial phosphorus limitation: mechanisms, implications, and nitrogen-phosphorus interactions. Ecol Appl 20:5–15
Wakelin SA, Macdonald LM, Rogers SL, Gregg AL, Bolger TP, Baldock JA (2008) Habitat selective factors influencing the structural composition and functional capacity of microbial communities in agricultural soils. Soil Biol Biochem 40:803–813
Wang Y, Marschner P, Zhang F (2012a) Phosphorus pools and other soil properties in the rhizosphere of wheat and legumes growing in three soils in monoculture or as a mixture of wheat and legume. Plant Soil 354:283–298
Wang Y, Zhang F, Marschner P (2012b) Soil pH is the main factor influencing growth and rhizosphere properties of wheat following different pre-crops. Plant Soil 360:271–286
Westerman RL (1990) Soil testing and plant analysis. SSSA, Madison
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18:315–322
Winsley T, van Dorst JM, Brown MV, Ferrari BC (2012) Capturing greater 16S rRNA gene sequence diversity within the domain Bacteria. Appl Environ Microbiol 78:5938–5941
WRB (2014) World reference base for soil resources. World Soil Ressources Reports, FAO, Rome
Zhong W, Cai Z (2007) Long-term effects of inorganic fertilizers on microbial biomass and community functional diversity in a paddy soil derived from quaternary red clay. Appl Soil Ecol 36:84–91
Acknowledgments
The authors would like to thank Dr. Stefan Zoller for the Perl scripts for taxonomy analysis and the Genetic Diversity Center (Zürich, Switzerland) for technical assistance. We also thank Monika Mascai for vegetation analyses, and Laurie Schönholzer for total C and N and TOC/N analyses. This work was supported by the Swiss National Science Foundation (SNF, project number 140900).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Paul Bodelier.
Electronic supplementary material
ESM 1
(PDF 905 kb)
Rights and permissions
About this article
Cite this article
Ragot, S.A., Huguenin-Elie, O., Kertesz, M.A. et al. Total and active microbial communities and phoD as affected by phosphate depletion and pH in soil. Plant Soil 408, 15–30 (2016). https://doi.org/10.1007/s11104-016-2902-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-016-2902-5