Abstract
NAC transcription factors are plant-specific and play important roles in plant development processes, response to biotic and abiotic cues and hormone signaling. However, to date, little is known about the NAC genes in canola (or oilseed rape, Brassica napus L.). In this study, a total of 60 NAC genes were identified from canola through a systematical analysis and mining of expressed sequence tags. Among these, the cDNA sequences of 41 NAC genes were successfully cloned. The translated protein sequences of canola NAC genes with the NAC genes from representative species were phylogenetically clustered into three major groups and multiple subgroups. The transcriptional activities of these BnaNAC proteins were assayed in yeast. In addition, by quantitative real-time RT-PCR, we further observed that some of these BnaNACs were regulated by different hormone stimuli or abiotic stresses. Interestingly, we successfully identified two novel BnaNACs, BnaNAC19 and BnaNAC82, which could elicit hypersensitive response-like cell death when expressed in Nicotiana benthamiana leaves, which was mediated by accumulation of reactive oxygen species. Overall, our work has laid a solid foundation for further characterization of this important NAC gene family in canola.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abiotic stresses such as high salinity, drought and extreme temperatures are among the major environmental factors that influence the productivity of plants. To survive harsh conditions, plants have developed sophisticated mechanisms to sense environmental cues and transmit these signals to regulate plant development and defense. Transcription factors (TFs) play a pivotal role in the control of downstream gene expression. The Arabidopsis genome encodes a little more than 2,000 TFs, which make up about 8 % of protein-coding genes. The NAC [no apical meristem (NAM), Arabidopsis thaliana transcription activation factor (ATAF1/2) and cup-shaped cotyledon (CUC2)] proteins constitute one of the largest TF families and are plant-specific (Olsen et al. 2005). NAC-type TFs were named after the first characterized members, which are NAM originally identified from petunia (Petunia hybrida) (Souer et al. 1996), ATAF1, ATAF2 and CUC2 in Arabidopsis thaliana (Aida et al. 1997). NAC TFs are characterized by a well-conserved N-terminal NAC domain and highly divergent C-terminus. Based on its motif distribution, the NAC domain, which comprises nearly 160 amino acid residues, can be divided into five subdomains (A–E) (Puranik et al. 2012).
There are 105 putative NAC genes in Arabidopsis (Ooka et al. 2003), and 140–151 in rice (Fang et al. 2008; Nuruzzaman et al. 2010). The NAC transcription factors are multifunctional proteins with various roles in the plant life cycle, such as maintenance of the shoot apical meristem (Takada et al. 2001; Vroemen et al. 2003), regulating cell division and cell expansion in flower organs (Sablowski and Meyerowitz 1998), and mediating auxin signaling to promote lateral root development(Xie et al. 2000). Many other NAC genes have been implicated in diverse cellular processes in various plant species, such as hormone signal pathways (Greve et al. 2003; Jensen et al. 2008, 2010), senescence (Guo and Gan 2006; Kjaersgaard et al. 2011; Yang et al. 2011) and formation of the secondary cell wall (Mitsuda et al. 2005, 2007; Zhong et al. 2006). Moreover, quite a few members of the NAC TF family also function in abiotic and biotic stress signaling and tolerance. For instance, Arabidopsis ANAC072 (RD26), ANAC019 and ANAC055 regulate ABA-dependent drought signaling (Fujita et al. 2004; Tran et al. 2004). Overexpression of rice stress-responsive NAC 1 (SNAC1/OsNAC9) increases drought and salt tolerance (Hu et al. 2006), while SNAC2/OsNAC6-overexpressing rice plants showed enhanced dehydration, cold and salt tolerance and also exhibited increased tolerance to blast disease (Nakashima et al. 2007; Hu et al. 2008). Moreover, overexpression of OsNAC5 in rice improves salt, drought and cold tolerance (Takasaki et al. 2010; Song et al. 2011; Jeong et al. 2012) and root-specific overexpression of OsNAC9 or OsNAC10 enlarges roots and enhances drought tolerance, which increases grain yield significantly under field drought conditions (Jeong et al. 2010; Redillas et al. 2012). In addition, a few NAC TFs positively or negatively regulate plant defense responses. For instance, Arabidopsis ATAF1 and -2 are negative regulators of defense responses against bacterial and fungal pathogens (Delessert et al. 2005; Wang et al. 2009) and, ATAF1 plays a dual role in mediating Arabidopsis response to both abiotic and biotic stresses (Lu et al. 2007; Wu et al. 2009). Arabidopsis NAC019 and NAC055 mediate drought tolerance, but their overexpression also decrease resistance to B. cinerea (Bu et al. 2008). It is thought that such dual modulation of NAC proteins in plants implies their alliance with distinct regulatory complexes. NAC TFs in crop plants have also been identified to mediate pathogen resistance. For example, the barley HvNAC6 positively regulates penetration resistance towards the biotrophic fungus Blumeria graminis f. sp. graminis (Bgh) (Jensen et al. 2007). However, the underlying mechanisms remain to be determined. Recent evidences also indicate that NAC TFs play a role in endoplasmic reticulum (ER) or osmotic stress-induced cell death in Arabidopsis, rice and soybean, possibly through regulating vacuolar processing enzyme (VPE) or caspase-like protein activity (Kaneda et al. 2009; Faria et al. 2011; Mendes et al. 2013; Yang et al. 2014).
Previous reports have indicated that quite a few TFs in Arabidopsis, especially those of bZIP and NAC gene families, are membrane-associated and may have dual localization (Chen et al. 2008; Seo et al. 2008). The number of putative membrane-associated NAC TFs in Arabidopsis and rice is 18 and 5, respectively (Kim et al. 2007, 2010). Upon developmental cues or environmental stress challenges, those membrane associated TFs could be cleaved by proteases and released into nuclei, as a rapid regulatory mechanism (Kim et al. 2010). For example, in Arabidopsis, cold stimulates proteolytic activation of a plasma membrane-tethered NAC transcription factor NTL6/ANAC062. The transcriptionally active NTL6 protein enters the nucleus, where it induces a subset of pathogenesis-related (PR) genes, such as PR1, PR2, and PR5. Transgenic plants overexpressing an active NTL6 form exhibit enhanced disease resistance (Seo et al. 2010). The complex regulation of NAC transcription factors includes microRNA (miRNA)-mediated cleavage of mRNAs and ubiquitin-dependent proteolysis (Xie et al. 2002; Mallory et al. 2004; Guo et al. 2005; Larue et al. 2009).
Though members of NAC gene family in Arabidopsis, rice, and a few other species have been studied or reported (Hegedus et al. 2003; Nuruzzaman et al. 2010; Nakashima et al. 2012; Puranik et al. 2012, 2013; Zhong et al. 2012), so far, there are only two reports conducted with canola (Brassica napus L.), in which 11 NAC genes were cloned and studied (Hegedus et al. 2003; Zhong et al. 2012). Canola is a very important oil crop worldwide and its yield is frequently limited by environmental factors including drought, salinity and cold. However, the identity and role of NAC genes in canola responses to abiotic and biotic stresses are largely unknown. It is therefore necessary to characterize the NAC gene family in canola before stress/disease tolerant canola species can be developed. In our previous transcriptomic profiling studies in canola seedlings treated with a fungal pathogen S. sclerotiorum and drought stress, we identified several NAC genes induced by S. sclerotiorum and/or drought stress (Yang et al. 2007). Hence, to explore the role of NAC genes in canola, we mined the available expressed sequence tags (ESTs) deposited in the public database to identify NAC genes in canola. We then cloned the cDNA sequences of 41 NAC genes from canola. We then analyzed the transcriptional activity of these NAC encoded proteins in yeast. We also examined the responses of selected genes under a range of abiotic stress and hormone treatment conditions. Interestingly, we successfully identified two novel BnaNAC genes that could elicit cell death when transiently expressed in N. benthamiana leaves. Our data presented here will lay a solid foundation for further characterization of this important NAC gene family in canola responses to abiotic and biotic stresses.
Materials and methods
Database search and identification of ESTs for NAC genes in canola
The seed sequences of NAC domain (Pf02365) from Pfam (http://pfam.sanger.ac.uk/family/nac#tabview=tab0) were used to search the Brassica napus ESTs in the NCBI dbEST (http://www.ncbi.nlm.nih.gov/dbEST/index.html, release 01012013), with an e-value cutoff lower than 10−4. After manual curation, these ESTs were clustered and assembled by sequence alignment program of DNASTAR (DNASTAR Inc.). Subsequently, contigs and singletons were run in a reciprocal BLAST search against the Arabidopsis database to assign a putative ortholog based on the best hit (Online Resource 1).
Plant growth and gene cloning
Canola (double haploid DH12075) plants were grown in Pindstrup soil mix (Denmark) in the greenhouse at 22 °C with 16 h light/8 h dark. 18 day-old leaves were harvested for RNA isolation using the Plant RNA kit (Omega bio-tek, USA). First-strand cDNA synthesis and high-fidelity PCR amplification using PrimeSTAR HS DNA polymerase (TaKaRa, Japan) were performed as previously described (Liang et al. 2013). 5′RACE (rapid amplification of cDNA end) and 3′RACE were performed as described earlier (Yang et al. 2009). Primers used were listed in Online Resource 2. PCR products were purified and cloned into pJET1.2 vector supplied in CloneJET PCR cloning kit (Fermentas, USA). At least two clones were sequenced from both ends using T7 promoter primer and a reverse primer for each gene, and sequencing results were analyzed by DNASTAR. The cDNA sequences of genes cloned in this study were deposited in GenBank database under the accession numbers of KC966745-KC966760, KF738267-KF738278, KF974755-KF974766, KJ420439-KJ420441, KJ670121-KJ670124 and KM975667-KM975669.
Phylogenetic tree reconstruction, multiple alignment analysis and conserved signature detection
The NAC genes of representative species were identified using a previously constructed domain alignment (from the Pfam database version 27.0) to query the proteome of representative species in Phytozome (http://www.phytozome.com/, v9.0). To investigate the evolutionary relationship among NACs, the predicted amino acid sequences of NACs of canola and other species were aligned using ClustalX1.83 and then phylogenetic tree was reconstructed using the maximum parsimony (MP) algorithm implemented in MEGA6.06 (release 6140226). Motif analysis of BnaNACs was determined by using Prosite program (http://prosite.expasy.org/prosite.html). The respective domains of NAC proteins were aligned using ClutsalX1.83 and illustrated by Boxshade (http://www.ch.embnet.org/software/BOX_form.html). Prediction of membrane-bound BnaNAC proteins was performed in ARAMEMNON protein database (http://aramemnon.botanik.uni-koeln.de) and TMHMM server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The putative ortholog of each BnaNAC was identified by the InParanoid server v. 8.0 (http://inparanoid.sbc.su.se/cgi-bin/blast_search.cgi).
Yeast assay
The coding regions of canola NAC genes were cloned into the pGBKT7 (BD) vector (Clontech, USA), using the primers listed in Online Resource 2. Then the recombinant plasmids were transformed individually into yeast (Saccharomyces cerevisiae) AH109 competent cells according to the Yeast Protocols Handbook (Clontech, USA). The transcriptional activity of BnaNACs was tested by streaking both on nonselective SD-LW (synthetic dropout without leucine and tryptophan) medium, as well as on selective SD-LWH (SD-Leucine–Tryptophan–Histidine + 5 mM 3′AT) and SD-LWHA (SD-Adenine–Histidine–Leucine–Tryptophan) media. Plates were incubated at 30 °C for up to 7 days before photographed. The titration assay was conducted by firstly performing four serial ten-time dilution of overnight cell cultured transformed with plasmids after adjusting the cell density of OD600 to 0.5. Then, two microliters of the original, 1/10, 1/100 and 1/1,000 diluted cell culture were spotted onto SD-LW, SD-LWH + 3′AT and SD-LWHA media and plates were incubated at 30 °C for 7 days before being photographed. The colony-lift filter assay was conducted following the instructions of the Yeast Protocols Handbook.
Quantitative RT-PCR (qRT-PCR) assay
Eighteen-day-old canola grown in a greenhouse with photoperiod of 16 h light/8 h dark with a relative humidity of 60 % were treated with different chemical solutions including 200 mM NaCl, 50 µM abscisic acid (±-ABA, Invitrogen, USA), 10 µM methyl viologen (MV, Sigma-Aldrich) as performed previously (Liang et al. 2013). In addition, 18-day old seedlings were also treated at 4 °C (cold), 37 °C (heat) or dehydration, and mock-treated seedlings were used as controls. Leaves were collected at 6 and 24 h post treatments, flash frozen in liquid nitrogen and stored at −80 °C. Total RNA samples were isolated and the first-strand cDNAs were synthesized from 2.5 μg of total RNA as described previously (Liang et al. 2013). Three independent biological replicates of each sample were prepared at different times.
Quantitative reverse transcriptase PCR (qRT-PCR) was performed using 10-fold diluted cDNA and SYBR Green I kit (CWBIO, China) on the CFX96 real-time PCR machine (Bio-Rad, USA). Primers used for qRT-PCR were designed using PrimerSelect program (DNASTAR Inc. USA), which targeted mainly at 3′UTR with an amplicon size of 75–250 bp (Online Resource 2). The specificity and amplification efficiency of each pair of primers were examined through both BLASTn search in NCBI database and by running standard curves with melting curves. Three independent biological replicates and two technical replicates for each biological replicate were run and the significance was determined through t test of SPSS statistic software (p ≤ 0.05).
Subcellular localization and confocal microscopy
To examine the localization of selected BnaNACs in planta, the coding regions were amplified using Pfu polymerase (Bioer, China) with primers listed in the Online Resource 2. After purification, PCR products were restricted and then fused upstream of green fluorescent protein gene (GFP) in the pYJGFP vector. The cDNA of chalcone synthase (CHS, At5g13930) gene was cloned from Arabidopsis (WT, Col-0) through RT-PCR and inserted upstream of the mCherry reporter gene in the pYJmCherry vector, a binary vector derived from pBS-mCherry (Zhang et al. 2014). These constructs and p19 protein of tomato bushy stunt virus were transformed into Agrobacterium tumefaciens GV3101 individually through a freeze–thaw method and overnight cell cultures were resuspended in infiltration media before infiltration into 5-week-old leaves of N. benthamiana (Liang et al. 2013). Two days later, leaf discs were observed of GFP under confocal microscope LSM510meta (Zeiss, Germany) or A1R (Nikon, Japan).
Gene expression in N. benthamiana and physiological assay
The coding regions of respective genes were amplified by high-fidelity Pfu polymerase and primers containing corresponding restriction sites as listed in Online Resource 2. After digestion, the PCR products were inserted downstream of a double CaMV 35S promoter in the binary vector pYJHA, which was modified from the pYJGFP vector. Recombinant plasmids were transformed into A. tumefaciens GV3101 and overnight cell cultures were resuspended in infiltration media containing 10 mM MES-KOH (pH 5.6), 10 mM MgCl2 and 0.15 mM acetosyringon, which was adjusted to an OD600 of 0.5 before a mixture of equal volumes of each cell culture harboring p19 and BnaNAC genes were infiltrated into the lower epidermal side of 5-week-old leaves of N. benthamiana plants. For each construct, 15 independent leaves of five independent plants (three leaves per plant) were used for each time-point tested. After that, infiltrated plants were kept under normal growth condition with the phenotype observed and recorded daily beginning 2 days after infiltration and continuing until day 7. To quantify the degree of cell death, electrolyte leakage was measured according to (Sun et al. 2014). In brief, five leaf discs (10 mm in diameter) were taken from each agro-infiltrated area and kept in deionized water under vacuum for 10 min, followed by incubation for 2 h at 25 °C. Ion conductivity (C1) was measured using a DDS-307 ion conductivity meter (Leici, China). After boiling for 5 min and cooling to room temperature, the ion conductivity (C2) was measured again. Relative conductivity was expressed as C1/C2. Distribution of hydrogen peroxide (H2O2) was detected by 3,3′-diaminobenzidine (DAB) staining according to the previously described protocol (Sun et al. 2014).
Transient expression assay in canola protoplasts
A Sac I-Sph I large fragment (1.9 kb) harboring 35S-GFP-poly(A) was released from pCsGFPBT binary vector and subcloned into pUC19 plasmid using the same restriction sites, with the resultant transient vector called pUC19-GFP. Similarly, the expression cassettes of BnaNAC19 and -82 genes were released through a Sac I-Sph I restriction from the aforementioned pYJHA recombinant plasmids and subcloned into pUC19 plasmid, with the resultant plasmids called pUC19-BnaNAC19 and pUC19-BnaNAC82, respectively. Protoplasts were isolated from leaves of canola plants grown in a 12-h light/12-h dark photoperiod with a relative humidity of 60–70 % according to the protocol for Arabidopsis mesophyll protoplast (Yoo et al. 2007). The protoplast concentration was quantified using a hemocytometer and a light microscope (DM750, Leica, Germany). A PEG4000-mediated transformation of protoplast suspension was performed with 20 μg of the expression plasmids isolated by the E.N.Z.A. Plasmid DNA Midi kit (Omega, USA) and quantified by NanoDrop (Thermo Fisher, USA). After incubated for 22 h at 22 °C, GFP signal was observed under a fluorescent microscope (DM5000B, Leica). For cell death detection in protoplasts, transformed protoplasts were cultivated for 22 h at 22 °C, and then were incubated for 15 min in 0.05 % Evans blue solution (117 mM KCl, 82 mM MgCl2, and 85 mM CaCl2) (Kaneda et al. 2009). After washed, the survival rate of protoplasts were determined by counting under light microscopy and photographed.
Results and discussion
Identification and cloning of NAC genes from canola
Although function of a few NAC genes in Arabidopsis and a few other plant species have been reported, little is known about this gene family in the important oilseed crop, canola (B. napus). Our previous transcriptomic studies of canola responses to a fungal pathogen S. sclerotiorum and drought treatments identified over 10 pathogen or drought responsive NAC genes in canola (Yang et al. 2007), suggesting NAC TFs may play an important role in canola response to pathogen and water deficit. This prompted us to identify, clone and study them in the context of stress conditions and hormone responses in canola.
To identify and characterize NAC genes from canola, we utilized public EST databases, since whole genome sequencing of B. napus is not yet complete. Altogether we obtained 671 unique ESTs representing putative NAC genes in canola (Online Resource 1). To facilitate comparisons between species, we followed the established nomenclature of Arabidopsis NACs when naming the canola NAC genes (Ooka et al. 2003). As a result, we identified 60 NAC genes from canola. We followed the recommended nomenclature of Brassica napus genes and used BnaNAC (for Brassica napus NAC, to differentiate it from Brassica nigra) to represent NAC genes reported in this study (Ostergaard and King 2008). We noted that among all the BnaNAC genes identified, BnaNAC2 had the largest number (72) of ESTs, followed by BnaNAC56 and -81 with a total of 57 and 55 ESTs, respectively; while BnaNAC7, -9, -10,-17, -20, -24, -46, -57, -74, -92, -97, -98 and -103 have only one EST each (Table 1 and Online Resource 1). To facilitate subsequent work, we designed primers based on the identified ESTs to obtain full length cDNA sequences from a double haploid species of canola, at least for the coding regions, employing RT-PCR together with RACE. As a result, we succeeded in cloning the cDNA sequences of 41 BnaNAC genes and deposited them into GenBank, among which we identified two to three slightly different alleles for eight BnaNAC genes (Table 1). Conceptual translation of these cDNA sequences and reciprocal BLAST searches against the Arabidopsis genome indicated that they bore domains and motifs that were typical of NAC proteins (Online Resource 3). A further comparison of BnaNAC genes reported in this study with NAC genes reported in the two previously published work (Hegedus et al. 2003; Zhong et al. 2012) indicate that we cloned the cDNA sequences of 34 novel NAC genes, with sequences of seven out of the 41 BnaNAC genes reported previously (Table 1). The number of amino acids in each BnaNAC protein ranged from 184 to 635, with a pI of 4.59–9.33. Further comparison of these 41 BnaNAC genes with each other demonstrated that they showed an identity of 22.6–80.8 %, with similarity 22.6–82.5 % at the nucleotide level. At the protein level, the maximum identity was 76 % with the minimum 11.6 %, whereas the similarity ranged from 23 to 83.5 %. At this step, we were able to identify putative orthologs of these BnaNAC genes in both Arabidopsis and rice using the program InParanoid (Table 1).
Domain analysis of BnaNAC proteins and phylogenetic tree reconstruction
We further examined conserved domains and motifs within BnaNAC proteins. From this alignment (Online Resource. 3), it was clear that each BnaNAC protein possessed a ca. 160 amino acid amino-terminal NAC domain, which is further divided into five highly conserved subdomains (A–E) (Ooka et al. 2003; Puranik et al. 2012). These five subdomains were interspersed by short and variable sequences. In subdomain D, there is a short sequence containing conserved basic amino acids, which is the nuclear localization signal (NLS). Moreover, the carboxyl-terminal regions of a few BnaNAC proteins were predicted to include a possible PEST motif, which is rich in proline (P), glutamic acid (E), serine (S), and threonine (T) (Online Resource. S1). This PEST motif could be targeted by proteasome-mediated degradation, which negatively affect the stability or half-life of NAC in cells (Rechsteiner and Rogers 1996). Previous studies with Arabidopsis NAC021(NAC1), NAC030(VND7) and NAC083(VNI2) have indicated that turnover could be mediated by ubiquitin-mediated proteolysis (Xie et al. 2002; Yamaguchi et al. 2010).
To examine the evolutionary relationships of canola NACs to other representative crops and models and trace the origins of NAC genes, we searched putative NAC genes in representative, lower species using Pfam seed sequences in Phytozome. The species queried included the bryophyte Physcomitrella patens (Pp), lycophyte Selaginella moellendorffii (Sm), Chlamydomonas reinhardtii (Cr), Ostreococcus tauri (Ol) and Ostreococcus lucimarinus (Ot). As a result, 32 and 20 NAC genes were identified from P. patens and S. moellendorffii, respectively (Online Resource 4). However, no NAC genes could be identified from the marine green alga O. tauri, which is the smallest free-living eukaryote (Derelle et al. 2006) or from a pico-eukaryotic (bacterial-sized) prasinophytic green alga Ostreococcus lucimarinus, which has one of the highest gene densities known in eukaryotes (Lanier et al. 2008). This suggests that NAC genes may only exist in the land plants (embryophytes). In addition, the presence of a smaller set of NAC members in primitive land plants including P. patens and S. moellendorffii compared to eudicots (Arabidopsis) and monocots (rice) indicated significant expansion of the NAC gene family during the evolution of the angiosperms.
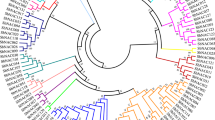
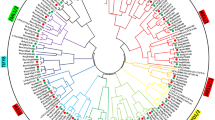
Next, we constructed a phylogenetic tree using a maximum parsimony (MP) algorithm after performing alignment of full-length amino acid sequences of NAC proteins from Arabidopsis, canola, rice, P. patens and S. moellendorffii, as well as a few from published references (Fig. 1). It can be seen that the NAC proteins from representative species could be clustered into three major groups, A to C. For canola NACs, BnaNAC2, -13, -14, -18, -19, -25, -29, -32, -36, -41, -42, -47, -50, -51, -55, -56, -57, -60, -69, -72, -81, -82, -83, -89, -91, -102, -103, -104 belong to group A, while BnaNAC7, -12, -22, -54, -59, -74, -79, -84, -92, -100 and -105 belong to group B. In contrast, group C only contains three BnaNAC proteins (BnaNAC10, -97 and -104). Moreover, the NAC proteins with group A could be clustered into six subgroups (I to VI), while those of group B into two subgroups (I and II). According to a recent report, a group of stress-responsive NAC (SNAC) genes exists in both higher and lower plants and, they could be clustered into multiple subgroups (Nakashima et al. 2012). In the present phylogenetic analysis, we identified that subgroup A-I is SNAC-A, while subgroup A-I and subgroup A-II are SNAC-B1 and SNAC-B2, respectively (Fig. 1). This observation could be a guideline to further study the function of these SNAC genes in canola in the context of abiotic and biotic stresses.
Phylogenetic analysis of NAC proteins. Reconstruction of the phylogenetic tree through maximum parsimony method. Members of canola (B. napus), Arabidopsis (A. thaliana), rice (O. sativa), lycophyte (S. moellendorffii), and moss (P. patens) were denoted by diamond, empty triangle, circle, square, filled triangle, respectively. The numbers on the nodes are percentages from a bootstrap analysis of 500 replicates. Zm, Zea mays; Ta, Triticum aestivum; Sl, Solanum lycopersicum; Ph, Petunia × hybrida
Transcriptional activity of BnaNAC TFs
To explore the function of NAC genes in canola, it is necessary to identify their ability to activate transcription. To this end, we analyzed the ability of 33 selected BnaNAC TFs to activate reporter genes LacZ,TRP1, LEU1 and ADE2 in budding yeast. Firstly, the coding regions of BnaNAC genes were fused to the GAL4 DNA-binding domain to examine their ability to activate transcription from the GAL4 upstream activation sequence (UAS) and thereby promote yeast growth. The yeast cells containing individual BnaNAC plasmids and control plasmid pGBKT7 all grew well on SD-LT medium, while on SD medium without histidine, tryptophan and adenine hemisulfate, only yeast cells containing pGBKT7-BnaNAC2, -12, -13, -14, -19, -29, -42, -54, -56, -59, -60, -69, -72, -79, -82, -89, -91, -92, -100, -102 and -104 plasmids could grow (Fig. 2). In the β-galactosidase activity assay, when the yeast cells containing a specific construct turned blue, it indicates the activation of another reporter gene LacZ. The results from both titration and X-gal staining assays demonstrated that BnaNAC2, -12, -13, -19, -29, -42, -54, -59, -60, -69, -72, -79, -82, -89, -91, -92, -100, -102 and -104 proteins showed evident transcriptional activation activity, while BnaNAC14 and -56 had only weak transcriptional activation activity. In contrast, BnaNAC10, -32, -41, -47, -57, -74, -83, -84, -97, -103 and -105 did not show any transcriptional activation activity, suggesting that they are transcriptional repressors (Fig. 2). The pink to red color of colonies transformed with BnaNAC10, -32, -41, -47, -57, -74, -83, -84, -97, -103 or -105 plasmids also indicated that ADE2 reporter gene was not expressed in these colonies. Previous studies with Arabidopsis NAC TFs using a similar approach have demonstrated that ATAF1(ANAC002), ANAC059(AtNAC3), ANAC089 and ANAC092(AtNAC2), ANAC105(VND3) have transcriptional activation activity (He et al. 2005; Lu et al. 2007; Jensen et al. 2010; Li et al. 2010), which is consistent with the behavior of their orthologs in canola. However, while ANAC103 and ANAC105(VND3) shows transcriptional activation activity in yeast-based reporter assays (Jensen et al. 2010; Sun et al. 2013), BnaNAC103 and -105 expression did not activate yeast reporter genes, which suggests that they have either no transactivational activity in yeast or could be transcriptional repressors. Besides, in accordance with two previous reports, full-length ANAC019 did not show any transactivation potential, while its C-terminal TRD region has transactivation activity in yeast (Bu et al. 2008; Jensen et al. 2010). In contrast, BnaNAC19 showed strong transactivation activity in the present study, which suggests difference in regulatory mechanism also exists between canola and Arabidopsis orthologous genes.
Analysis of transactivational activity of BnaNAC in budding yeast. The yeast cells of strain AH109 harboring the indicated plasmid were grown on either the nonselective (SD-LW) or selective (SD-LWH + 3-AT and SD-LWHA) media, followed by β-galactosidase assay (X-Gal staining). Decreasing cell densities in the dilution series are illustrated by narrowing triangles. BD at the last row represents empty GAL4-BD vector
Expression analysis of BnaNAC genes in response to stress treatments
To elucidate of the functions of BnaNAC genes in the context of abiotic and hormone stimuli, the expression patterns of 17 selected BnaNAC genes, representing all three groups in the phylogenetic tree, were studied in response to a variety of stress conditions using quantitative real-time PCR (qRT-PCR). The stress conditions applied to 18-day old canola seedlings included high salinity (200 mM NaCl), cold (4 °C), drought, heat (37 °C), abscisic acid (ABA) and Paraquat (methyl viologen, MV). Among these stress treatments, ABA is a well-known abiotic stress hormone and MV is a herbicide triggering ROS burst. We subjected canola seedlings to moderate stress treatments and measured the response at two time-points to better monitor the transcript changes of the investigated BnaNAC genes. Data of three independent biological replicates were analyzed to identify BnaNAC genes responsive to one or a combination of stress conditions (Fig. 3). As a result, we found that salt increased expression of BnaNAC2, -10, -19, -29, -51, -84 and -92 at either 6 h or 24 h or both time-points. Cold stress up-regulated the transcript abundance of BnaNAC2, -22, -36, -51, -57, -59 and -92, while it down-regulated BnaNAC84 and -100. Drought or water deficit stress induced transcript accumulation of BnaNAC2, -22, -29 and -57, whereas it repressed BnaNAC81 transcription at either early or late time-point. Heat treatment up-regulated the transcript levels of BnaNAC29, -51, -59 and -104, while down-regulating those of BnaNAC19 and -92. ABA induced the expression of BnaNAC51 and -81 at the 24 h time-point; however, it repressed BnaNAC84 expression 6 h after treatment. Lastly, Paraquat-triggered ROS accumulation repressed both BnaNAC57 and -84 expression at the 6 h time-point. Taken together, these data indicate that on one hand, some BnaNAC genes participate in transduction of multiple stresses; and on the other hand, a specific stress activates transcription of more than one BnaNAC, providing evidence that some BnaNACs may functionally redundant. Moreover, since expression of BnaNAC2, -19, -22, -29 and so on were induced by multiple stresses, it suggests that they could play a role in the cross-talk of multiple stresses, including both abiotic and hormone stresses.
Expression analyses of BnaNAC genes in response to various treatments, including 10 μM Paraquat (MV), 50 μM ABA, 200 mM NaCl, heat (37 °C), cold (4 °C). Data is the mean of three biological replicates ± SE. Asterisks denote significant differences (compared to 1) by Student’s t test analysis (p ≤ 0.05)
Among the 17 BnaNAC genes profiled, six genes (BnaNAC2, -19, -29, -32, -72 and -81) belong to the SNAC subgroup (Fig. 1). Indeed, we observed that under abiotic stress and hormone treatments, BnaNAC2, -19, -29 and -81 were responsive to most treatments; however, BnaNAC32 and -72 were induced only under heat treatment and no significant changes to other stress conditions were observed. In addition, BnaNAC82 expression was slightly induced by ROS-generation MV treatment at both time-points, whereas BnaNAC19 was not induced by MV and instead was significantly repressed by MV at the 24 h time-point. However, since both BnaNAC19 and -82 expression induced evident ROS accumulation in leaf tissues (see below text) and exogenous application of MV had slightly different effect on their expression, which implies that underlining mechanisms of ROS accumulation and signaling modulated by these two BnaNAC genes may be different. However this hypothesis needs be experimentally tested.
Subcellular localization of canola NAC proteins
To investigate the subcellular localization of canola NAC proteins, selected genes representing the three groups of BnaNACs were chosen to express as fusion proteins with GFP in leaves of N. benthamiana. The coding regions of eight BnaNAC genes (BnaNAC2, -10, -32, -54, -79, -82, -83 and -92) were fused in frame to the N-terminus of GFP reporter gene with a Gly-Ala rich peptide linker in between. The recombinant plasmids were transformed into agrobacteria and then infiltrated into the lower epidermal leaves of N. benthamiana, with the GFP signal observed 2 days later. We found that in leaf cells harboring the fusion proteins of BnaNAC2, -10, -32, -54, -79, -82, -83 and -92, the GFP signals were present in the nucleus (Fig. 4a–h), which is in agreement with their role of transcription factors. As a control, we also examined the subcellular localization of the GFP protein in leaf cells, and green signals were present obviously both in cytosol and nuclei (Fig. 4i).
Subcellular localization of BnaNAC proteins in N. benthamiana cells using green fluorescence protein (GFP). a–k Represented BnaNAC2, -10, -32, -54, -79, -82, -83, -92, GFP alone, BnaNAC69 and -89, respectively. In each panel, the extreme left is GFP fluorescence, the middle bright or mCherry field and the right an overlay of the two images as indicated on the top of the picture. Bar 50 μm
Previous reports and predictions have indicated that quite a few NAC TFs in Arabidopsis and rice are associated with the membrane, which serves as an adaptive strategy in response to unfavorable growth conditions or inner developmental cues (Kim et al. 2010). To predict whether the proteins encoded by the 38 cloned BnaNAC genes contained transmembrane helices (TMH), we used TMHMM (http://www.cbs.dtu.dk/ser-vices/TMHMM-2.0/) program. As a result, a TMH was identified for six of the 38 BnaNAC proteins, and they are BnaNAC13, -14, -60, -69, -89 and -91 with the TMH located at the C-terminus (Online Resource. 5), suggesting that some of these NAC TFs are putatively transmembrane proteins. Phylogenetic analysis of putative membrane-associated NAC proteins from canola, Arabidopsis and rice divided them into five subgroups, I–V (Online Resource. 5).
We examined the subcellular localization of two putative membrane-associated BnaNAC69 and -89, using GFP as the reporter. An ER marker, chalcone synthase (CHS), which is reported to be located in ER (Hrazdina et al. 1987; Tzfira et al. 2005), was co-expressed as a fusion protein with another reporter mCherry. The results from confocal laser scanning microscopy showed that both BnaNAC69 and -89 were able to be localized in ER (Fig. 4j, k). The subcellular localization of BnaNAC89 was similar to that of its ortholog in Arabidopsis, ANAC089 (Li et al. 2010).
Identification of two novel BnaNAC genes mediating cell death
In the aforementioned GFP subcellular assay using N. benthamiana, we observed that expression of BnaNAC19 and BnaNAC82 in leaves led to hypersensitive response (HR) symptoms within 2 days after infiltration. HR is a common feature of plant immune responses and a type of programmed cell death (PCD). Activation of cell death is one of the aspects of plant defense responses where ROS play a crucial role (Coll et al. 2011). Hence, we performed 3, 3′-diaminobenzidine (DAB) staining of the agroinfiltrated leaves and identified evident staining, indicative of ROS accumulation (data not shown). To further explore the roles of BnaNAC genes in cell death, we constructed the coding regions of BnaNAC19 and -82 into another binary vector pYJHA (GFP deleted) individually under the driving of CaMV 35S promoter. To examine which domain is essential for inducing cell death, we also constructed two truncated versions of each gene, representing the DNA-binding NAC domain and transcriptional regulatory domain into the same vector (Fig. 5b). Then we performed agroinfiltration of them individually into N. benthamiana leaves and evaluated the phenotype daily for a total of 7 days. Interestingly, expression of either BnaNAC19 or -82 indeed caused pathogen-independent cell death compared with GFP vector control, beginning 48 h post-infiltration (hpi) and proceeded as expected (Fig. 5a). We observed that the water-soaking symptom appeared a little earlier and was more severe in BnaNAC82-expressing leaves than that of BnaNAC19. However, expression of any of the two truncated versions of either BnaNAC19 or BnaNAC82 did not induce evident cell death as observed with the full-length coding region, indicating that both the DNA-binding and transcriptional regulatory domains are required for full function of these two NAC genes. To explore the role of ROS during cell death, we also performed DAB staining and the results showed there was staining in sites expressing BnaNAC19 or -82 beginning at 48 hpi and continued till 144 h, but not or very weak in sites expressing the truncated fragments or GFP gene alone (Fig. 5a). Moreover, we examined the electrolyte leakage of leaf discs taken from leaves expressing full-length BnaNAC19, BnaNAC82 or the truncated versions as well as the GFP alone. The results showed that a significant increase in ion leakage was visible 3 days (72 hpi) after agroinfiltration of BnaNAC19 or -82 in contrast to that of leaves expressing the GFP gene alone (Fig. 5c). This increase in ion leakage was even more pronounced for leaf discs expressing BnaNAC82 gene than those expressing BnaNAC19. However, in leaf discs expressing either the N- or C- terminal of BnaNAC19 or -82, there was only slight or non-significant increase compared to the GFP control.
Overexpression of BnaNAC19 and -82 induced pathogen-independent cell death in N. benthamiana leaves. Leaves were infiltrated with agrobacteria carrying individual plasmids. All experiments were performed three times with similar results obtained. a Symptoms of N. benthamiana leaf areas expressing BnaNAC19, -82 and their deletion versions 0, 24, 48, 72, 96, 120 and 144 h post-infiltration (hpi). The left, middle and right panels represent the front, back sides and DAB staining, respectively. b Schematic representation of domain structures of BnaNAC19 and -82 proteins. The DNA-binding NAC domain (open boxes) is located at their N-terminal regions and the transcriptional regulatory domain is located at their C-terminal regions. The number of total amino acid residues of each protein or truncated fragments is shown at the right side of each structure. Numbers at the bottom row indicate residue positions. c Comparison of relative conductivity in leaf discs expressing BnaNAC19, -82 or their deletion versions at 72, 96,120 and 144 hpi. Values represent the means of three independent assays for each time-point ± SE. Identical and different letters represent non- and significant differences (p ≤ 0.05)
To determine the role of BnaNAC19 and -82 genes in the induction of HR-like cell death in canola, we used a transient assay system in protoplasts of canola based on the Arabidopsis transient assay system (Yoo et al. 2007). Firstly, we constructed three transient expression plasmids based on the pUC19 backbone, which harbored the expression cassettes of GFP (control), BnaNAC19 and BnaNAC82 genes under the control of constitutive CaMV35S promoter. Secondly, we introduced the three expression vectors into protoplasts prepared from canola leaves, and then examined cell death using Evans blue dye, a marker of plasma membrane integrity (Kaneda et al. 2009). As expected, we identified that in protoplasts transformed with either BnaNAC19 or BnaNAC82 plasmids, an average of 20.5 and 17.2 % of protoplasts displayed plasma membrane (PM) shrinkage and loss of PM integrity, respectively (Fig. 6a). As a result, the dead protoplasts accumulated the dye, whereas in protoplasts expressing the GFP gene alone, almost all protoplasts excluded the dye (Fig. 6a). Therefore, 20.5 and 17.2 % of the protoplasts were scored as dead or broken after transient overexpression of BnaNAC19 and -82 genes in canola protoplasts, respectively, whereas the death rate in GFP control was not detectable (Fig. 6b).
Expression of BnaNAC19 and -82 genes in canola protoplasts induced cell death. a Detection of cell death through Evans blue staining. Protoplasts were transfected with the pUC19-GFP (control), pUC19-BnaNAC19 or pUC19-BnaNAC82 plasmids, respectively. After 22 h, viability of protoplasts was assayed with Evans blue staining. Two representative protoplasts for each gene were shown. Bar 10 μm. b Percentages of dead protoplasts transfected with pUC19-GFP (control), pUC19-BnaNAC19 or pUC19-BnaNAC82 plasmids. 22 h after transfection, dead protoplasts were scored using a light microscope. The values with error bars were derived from three to four independent experiments
Taken together, our data presented here demonstrates that hypersensitive response-like cell death associated with hydrogen peroxide production is triggered by high expression of BnaNAC19 and -82. Since orthologs of BnaNAC19 and -82 in Arabidopsis and rice or any other plant species have not been reported to modulate cell death and/or ROS accumulation, they are two novel members. However, the inner mechanisms and signaling pathways of ROS accumulation and cell death mediated by BnaNAC19 and -82 need to be studied both in canola and N.benthamiana using a combination of molecular and genetic approaches. Whether there are other canola NAC genes mediating cell death or ROS signaling awaits to be investigated further. Furthermore, whether and how BnaNAC19 and -82 modulate plant immunity against fungal or bacterial pathogens must also be experimentally determined.
Conclusion
NAC TFs are very important in plant development, abiotic stress and defense responses. So far, function of only a few NAC genes in Arabidopsis, rice, wheat, tomato have been reported (Nakashima et al. 2012; Puranik et al. 2012). In the present study, we described the NAC gene family in the important oilseed crop, canola, through mining of EST databases. We successfully identified 60 NAC genes from canola, and cloned the cDNA sequences of 41 NAC genes (Table 1). An analysis of the phylogenetic relationship of NAC proteins from representative plant species indicated an evident increasing numbers of NAC genes from lower to higher plants. This also suggests that different types of duplications of NAC genes may occur during the evolutionary history. A recent study suggests that NAC TFs play an important role in the adaptation of plants to land (Xu et al. 2014). Further, we studied the transcriptional activity of 33 BnaNAC TFs in yeast and found that 11 of them may function as transcriptional repressors in cells while the others act as transcriptional activators (Fig. 2). A previous report suggested that a hydrophobic LVFY motif in the subdomain D of NAC DNA-binding domain may partially contribute to the repression function of some NAC TFs (Hao et al. 2010). Examination of 41 BnaNAC protein sequences indicated that in the subdomain D, of 27 out of 41 BnaNAC proteins, there was a conserved LVFY motif (Online Resource S1). However, four BnaNAC proteins (BnaNAC10, -84, -97 and -103) did not harbor the LVFY motif and they were transcriptional repressors. On the other hand, more BnaNAC proteins showed transcriptional activation activity although they bore a LVFY motif in subdomain D (Fig. 2; Online Resource 3). Taken together, this suggests that the exact nature or sequences of repressor domain of NAC TFs still needs to be pinpointed. Moreover, we found that there were differences in transcriptional activity of orthologous NAC genes between canola and Arabidopsis as described previously, which highlights the limitations of applying conclusions from model species Arabidopsis to canola.
The responses of 17 BnaNAC genes under abiotic stress and hormone treatments were analyzed through qRT-PCR and, we found that 13 BnaNAC genes could be significantly regulated by at least one treatment, suggesting NAC TFs may participate in the cross-talk of different signaling pathways under stress conditions. More importantly, we identified two novel NACs, BnaNAC19 and -82, which mediated programmed cell death in leaves of N. benthamiana, which is dependent on ROS accumulation (Fig. 5). The role of these two genes in inducing cell death was also confirmed in protoplasts of canola (Fig. 6). A bioinformatic analysis and literature search indicated that these were two novel cell death-mediating members of NAC TF family. How these two BnaNACs elicited cell death are still outstanding questions for further research. Overall, this study of NAC genes in canola lays a solid foundation for further exploration of their roles in abiotic stress and ABA signaling. It also provides important information for genetic manipulation of the abundance and/or activity of related NACs to improve stress tolerance of canola.
References
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18:756–767
Chen YN, Slabaugh E, Brandizzi F (2008) Membrane-tethered transcription factors in Arabidopsis thaliana: novel regulators in stress response and development. Curr Opin Plant Biol 11:695–701
Coll NS, Epple P, Dangl JL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18:1247–1256
Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, Dolferus R (2005) The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J 43:745–757
Derelle E, Ferraz C, Rombauts S, Rouze P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynie S, Cooke R, Saeys Y, Wuyts J, Jabbari K, Bowler C, Panaud O, Piegu B, Ball SG, Ral JP, Bouget FY, Piganeau G, De Baets B, Picard A, Delseny M, Demaille J, Van de Peer Y, Moreau H (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103:11647–11652
Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280:547–563
Faria JA, Reis PA, Reis MT, Rosado GL, Pinheiro GL, Mendes GC, Fontes EP (2011) The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol 11:129
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876
Greve K, La Cour T, Jensen MK, Poulsen FM, Skriver K (2003) Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem J 371:97–108
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612
Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell 17:1376–1386
Hao YJ, Song QX, Chen HW, Zou HF, Wei W, Kang XS, Ma B, Zhang WK, Zhang JS, Chen SY (2010) Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232:1033–1043
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397
Hrazdina G, Zobel AM, Hoch HC (1987) Biochemical, immunological, and immunocytochemical evidence for the association of chalcone synthase with endoplasmic reticulum membranes. Proc Natl Acad Sci USA 84:8966–8970
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Jensen MK, Rung JH, Gregersen PL, Gjetting T, Fuglsang AT, Hansen M, Joehnk N, Lyngkjaer MF, Collinge DB (2007) The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol Biol 65:137–150
Jensen MK, Hagedorn PH, de Torres-Zabala M, Grant MR, Rung JH, Collinge DB, Lyngkjaer MF (2008) Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J 56:867–880
Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K (2010) The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J 426:183–196
Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197
Jeong JS, Kim YS, Redillas MC, Jang G, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK (2012) OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J 11:101–114
Kaneda T, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, Isogai A, Che FS (2009) The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J 28:926–936
Kim SY, Kim SG, Kim YS, Seo PJ, Bae M, Yoon HK, Park CM (2007) Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res 35:203–213
Kim SG, Lee S, Seo PJ, Kim SK, Kim JK, Park CM (2010) Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95:56–65
Kjaersgaard T, Jensen MK, Christiansen MW, Gregersen P, Kragelund BB, Skriver K (2011) Senescence-associated barley NAC (NAM, ATAF1,2, CUC) transcription factor interacts with radical-induced cell death 1 through a disordered regulatory domain. J Biol Chem 286:35418–35429
Lanier W, Moustafa A, Bhattacharya D, Comeron JM (2008) EST analysis of Ostreococcus lucimarinus, the most compact eukaryotic genome, shows an excess of introns in highly expressed genes. PLoS One 3:e2171
Larue CT, Wen J, Walker JC (2009) A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J 58:450–463
Li J, Zhang J, Wang X, Chen J (2010) A membrane-tethered transcription factor ANAC089 negatively regulates floral initiation in Arabidopsis thaliana. Sci China Life Sci 53:1299–1306
Liang W-W, Yang B, Yu B-J, Zhou Z-Z, Li C, Sun Y, Zhang Y, Jia M, Wu F-F, Zhang H-F, Wang B-Y, Deyholos M, Jiang Y-Q (2013) Identification and analysis of MKK and MPK gene families in Canola (Brassica napus L.). BMC Genom 14:392
Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63:289–305
Mallory AC, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14:1035–1046
Mendes GC, Reis PA, Calil IP, Carvalho HH, Aragao FJ, Fontes EP (2013) GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc Natl Acad Sci USA 110:19627–19632
Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17:2993–3006
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:97–103
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465:30–44
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247
Ostergaard L, King GJ (2008) Standardized gene nomenclature for the Brassica genus. Plant Methods 4:10
Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381
Puranik S, Sahu PP, Mandal SN, Venkata Suresh B, Parida SK, Prasad M (2013) Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS One 8:e64594
Rechsteiner M, Rogers SW (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21:267–271
Redillas MC, Jeong JS, Kim YS, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK (2012) The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J 10:792–805
Sablowski RW, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92:93–103
Seo PJ, Kim SG, Park CM (2008) Membrane-bound transcription factors in plants. Trends Plant Sci 13:550–556
Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park CM (2010) Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J 61:661–671
Song SY, Chen Y, Chen J, Dai XY, Zhang WH (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234:331–345
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85:159–170
Sun L, Yang ZT, Song ZT, Wang MJ, Sun L, Lu SJ, Liu JX (2013) The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J 76:274–286
Sun Y, Wang C, Yang B, Wu F, Hao X, Liang W, Niu F, Yan J, Zhang H, Wang B, Deyholos M, Jiang Y-Q (2014) Identification and functional analysis of mitogen-activated protein kinase kinase kinase (MAPKKK) genes in canola (Brassica napus L.). J Exp Bot 65:2171–2188
Takada S, Hibara K, Ishida T, Tasaka M (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128:1127–1135
Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K (2010) The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics 284:173–183
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57:503–516
Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15:1563–1577
Wang X, Basnayake BM, Zhang H, Li G, Li W, Virk N, Mengiste T, Song F (2009) The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol Plant Microbe Interact 22:1227–1238
Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Yang C, Xie Q (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19:1279–1290
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036
Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419:167–170
Xu B, Ohtani M, Yamaguchi M, Toyooka K, Wakazaki M, Sato M, Kubo M, Nakano Y, Sano R, Hiwatashi Y, Murata T, Kurata T, Yoneda A, Kato K, Hasebe M, Demura T (2014) Contribution of NAC transcription factors to plant adaptation to land. Science 343:1505–1508
Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22:1249–1263
Yang B, Srivastava S, Deyholos MK, Kav NNV (2007) Transcriptional profiling of canola (Brassica napus L.) responses to the fungal pathogen Sclerotinia sclerotiorum. Plant Sci 173:156–171
Yang B, Jiang Y, Rahman MH, Deyholos MK, Kav NN (2009) Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol 9:68
Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23:2155–2168
Yang ZT, Wang MJ, Sun L, Lu SJ, Bi DL, Sun L, Song ZT, Zhang SS, Zhou SF, Liu JX (2014) The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet 10:e1004243
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Zhang H, Yang B, Liu WZ, Li H, Wang L, Wang B, Deng M, Liang W, Deyholos MK, Jiang YQ (2014) Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol 14:8
Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18:3158–3170
Zhong H, Guo QQ, Chen L, Ren F, Wang QQ, Zheng Y, Li XB (2012) Two Brassica napus genes encoding NAC transcription factors are involved in response to high-salinity stress. Plant Cell Rep 31:1991–2003
Acknowledgments
This work was supported by a startup funding and in part by the National Natural Science Foundation of China (No. 31471153 to Y-Q. J and 31301648 to B.Y.). We would like to acknowledge assoc. prof. Liu X–Y (NWAFU) for providing the pBS-mCherry vector and, prof. Michael K. Deyholos (University of Alberta, Canada) for providing the canola seeds and critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Boya Wang, Xiaohua Guo, Chen Wang and Jieyu Ma have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, B., Guo, X., Wang, C. et al. Identification and characterization of plant-specific NAC gene family in canola (Brassica napus L.) reveal novel members involved in cell death. Plant Mol Biol 87, 395–411 (2015). https://doi.org/10.1007/s11103-015-0286-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0286-1