Abstract
Key message
SbNAC genes (131) encoding 183 proteins were identified from the sorghum genome and characterized. The expression patterns of SbSNACs were evaluated at three sampling time points under post-flowering drought stress.
Abstract
NAC proteins are specific transcription factors in plants, playing vital roles in development and response to various environmental stresses. Despite the fact that Sorghum bicolor is well-known for its drought-tolerance, it suffers from grain yield loss due to pre and post-flowering drought stress. In the present study, 131 SbNAC genes encoding 183 proteins were identified from the sorghum genome. The phylogenetic trees were constructed based on the NAC domains of sorghum, and also based on sorghum with Arabidopsis and 8 known NAC domains of other plants, which classified the family into 15 and 19 subfamilies, respectively. Based on the obtained results, 13 SbNAC proteins joined the SNAC subfamily, and these proteins are expected to be involved in response to abiotic stresses. Promoter analysis revealed that all SbNAC genes comprise different stress-associated cis-elements in their promoters. UTRs analysis indicated that 101 SbNAC transcripts had upstream open reading frames, while 39 of the transcripts had internal ribosome entry sites in their 5′UTR. Moreover, 298 miRNA target sites were predicted to exist in the UTRs of SbNAC transcripts. The expression patterns of SbSNACs were evaluated in three genotypes at three sampling time points under post-flowering drought stress. Based on the results, it could be suggested that some gene members are involved in response to drought stress at the post-flowering stage since they act as positive or negative transcriptional regulators. Following further functional analyses, some of these genes might be perceived to be promising candidates for breeding programs to enhance drought tolerance in crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sorghum [Sorghum bicolor (L.) Moench] is the fifth most important grain crop world-wide (FAO 2015), and it is used for food, animal feed, fiber and biofuel (Rooney et al. 2007). Sorghum belongs to the tribe Andropogoneae of the Poaceae family, and is grown in the arid and semi-arid regions of South Africa, Australia, India and America. It is characterized by C4 photosynthesis, high biomass productivity, adaptation to diverse environments and native to tropical and subtropical climates. While it has a small (818Mbp) and diploid genome (2n = 20) along with a rich germplasm diversity, the availability of its whole genome sequence and annual life cycle make sorghum an ideal model for the study of plant response to abiotic stress and plant genomics research among cereals and Andropogoneae species, respectively (Menz et al. 2004; Casa et al. 2005).
Sorghum is more capable of adaptation to drought stress compared to other cereals, but it is affected by water limitation at both pre- and post-flowering developmental stages (Ludlow and Muchow 1990; Tuinstra et al. 1997; Kebede et al. 2001; Sanchez et al. 2002; Mullet et al. 2002; Blum 2004). Post-flowering drought stress can result in leaf pre-maturation, plant senescence and a significant reduction in crop yield (Rosenow et al. 1997). Post-flowering drought tolerance in sorghum is associated with the ‘stay green’ phenotype, which allows plants to maintain their ability of photosynthesis in their leaves during the grain filling stage (Walulu et al. 1994; Xu et al. 2000; Subudhi et al. 2000).
NAC is one of the most important TF families involved in plant senescence and response to drought stress (Tran et al. 2004; GUO et al. 2004; Wu et al. 2016). NAC is derived from three different proteins that were initially observed to contain highly-conserved DNA-binding domains, namely NAM (No Apical Meristem), ATAF1/2 and CUC2 (cup-shaped cotyledon) (Souer et al. 1996; Aida et al. 1997). The NAC transcription factors, as a gene family, are one of the largest transcription factor families, which are plant-specific and exist in a wide range of species (Riechmann et al. 2000). There are 117, 151, 152, 163, 152 and 26 NAC genes in the Arabidopsis, rice, soybean, poplar, tobacco and citrus, respectively (Rushton et al.; Hu et al. 2010; Nuruzzaman et al. 2010; Le et al. 2011). The NAC protein has a conserved domain which is comprised of nearly 160 amino acid residues in the N-terminal region, and it consists of five subdomains (A–E) and, also, a C-terminal variable domain that functions as a transcriptional activator or repressor (Ooka et al. 2003; Olsen et al. 2005; Kim et al. 2007).
NAC proteins have been involved in a wide range of plant developmental processes, such as leaf senescence (GUO et al. 2004), seed and embryo development (Duval et al. 2002; Sperotto et al. 2009), cell division (Kim et al. 2006), shoot apical meristem formation (Kim et al. 2007), fiber development (Ko et al. 2007), floral morphogenesis (Sablowski and Meyerowitz 1998), lateral root development and auxin signaling (Xie et al. 2000; He et al. 2005), grain nutrient remobilization (Uauy et al. 2006; Waters et al. 2009; Ricachenevsky et al. 2013), shoot branching determination (Mao et al. 2007), plant response to biotic and abiotic stresses such as drought, salinity, cold shock, viral infection and wounding (Ren 2000; Collinge and Boller 2001; Fujita et al. 2004; Jensen et al. 2008).
Several NAC genes from Arabidopsis and other crops have been identified which are involved in the plant response to abiotic and biotic stress, namely SNACs (STRESS-RESPONSIVE NAC). Meanwhile, some of the NAC genes are known to control leaf senescence (Guo and Gan 2006). Overexpression of the ATAF subfamily in Arabidopsis such as ANAC019 (AT1G52890), ANAC055 (AT3G15500) and ANAC072 (AT4G27410) genes significantly enhanced drought tolerance in transgenic plants, whereas ATAF1 (AT1G01720) is reported as a gene that negatively regulates the drought signal transduction pathways (Tran et al. 2004; Lu et al. 2007). Transgenic rice (Oryza sativa) have been observed to overexpress OsNAC5 (Os11g08210) (Jeong et al. 2013), OsNAC6 (Os01g66120) (Nakashima et al. 2012; Rachmat et al. 2014), OsNAC10 (Os11g03300) (Jeong et al. 2010), SNAC1 (Os03g60080) (Hu et al. 2006), and therefore they exhibit improved tolerance to drought. Furthermore, transgenic Arabidopsis overexpressing wheat TaNAC2 and TaNAC69 genes (Xue et al. 2011; Tang et al. 2012; Mao et al. 2012), maize ZmSNAC1 (Lu et al. 2012) and sorghum SbSNAC1 (Lu et al. 2013) displayed an enhanced level of drought tolerance at the seedling stage. AtNAP was found to be involved in cell expansion and senescence in Arabidopsis (Guo and Gan 2006). Also, NAM-B1 in wheat is known to accelerate senescence, while OsNAC5 in rice can control the remobilization of nutrients during grain maturation (Uauy et al. 2006; Ricachenevsky et al. 2013).
In the present study, 131 non-redundant NAC members from sorghum were identified and classified into 15 subfamilies according to the conserved domain sequences of NAC. Moreover, phylogenetic relationships of the NAC proteins in sorghum and Arabidopsis were comprehensively analyzed for functional predictions, while several putative SbNAC genes were identified and hypothesized to play roles in the plants response to stress and in the control of leaf senescence. The expression pattern of SNAC members in sorghum were investigated in flag leaves during the post-flowering stage under drought stress in three genotype of sorghum contrasting in salt tolerance using quantitative real-time RT-PCR (qPCR). Furthermore, assessment of chromosomal locations, gene structures, domain and additional motifs, promoter and UTRs were performed. The results of this study can be beneficial for further functional characterization of the candidate stress responsive NAC genes in sorghum.
Materials and methods
Identification of NAC TFs in sorghum
To identify NAC gene family members in sorghum (Sorghum bicolor (L.) Moench), four different approaches were applied. First, Plant Transcription Factor Database (TFDB 3.0) (http://planttfdb.cbi.pku.edu.cn) (Jin et al. 2017) and GrassTFDB (http://grassius.org/grasstfdb.php) (Yilmaz et al. 2009) were used to search for members of the NAC gene family. Next, hidden Markov model (HMM) profiles of NAM domain (PF02365) was obtained from Pfam database (http://pfam.xfam.org/) (Finn et al. 2016) and used in order to search putative NAC domains against downloaded protein database of sorghum from ensemble (http://plants.ensembl.org/info/website/ftp/index.html) via HMMER under the Linux software. Then, S. bicolor genome (v3.1) at Phytozome v12 (http://www.phytozome.net/) (Goodstein et al. 2012) and MOROKOSHI (http://sorghum.riken.jp/morokoshi/Home.html) (Makita et al. 2015) were searched by using the NAC Keyword. Finally, NAC protein sequence of Arabidopsis thaliana and Oryza sativa were downloaded from TFDB, and then their conserved NAC domain were used as a query for BlastP analysis in Phytozome database to identify homologous proteins. Redundant and incomplete sequences were removed manually and all protein sequences were checked for presence of the NAC domain through Pfam and SMART databases (http://smart.embl-heidelberg.de/) (Letunic et al. 2015).
Sequence alignment and phylogenetic analysis
The protein sequence of NAC domains were aligned using the ClustalX2 program (Larkin et al. 2007). The unrooted phylogenetic trees were constructed using the Neighbor-Joining (NJ) method with the following parameters: p-distance model, pairwise deletion and 1000 bootstrap replicates by MEGA7 software (http://www.megasoftware.net/) (Kumar et al. 2006).
Chromosomal location and gene structures of SbNAC gene family
The cDNA sequences of SbNACs were used as the queries for the instances of BLASTn search against the Sorghum bicolor genome (V3.1) at Phytozome database. Distribution of SbNAC genes were visualized using the MapChart software (version 2.3) (Voorrips 1994). Genomic and coding sequences of SbNAC members were obtained from Phytozome and were loaded onto the Gene structure display server v2.0 (http://gsds.cbi.pku.edu.cn/) to obtain the intron splicing sites (Hu et al. 2015).
Detection of conserved motifs
The conserved motifs in the SbNAC protein sequences were found using Multiple Expectation Maximization for Motif Elicitation (MEME) program version 4.11.4 (http://meme-suite.org/tools/meme) with the following parameters: maximum number of motifs, minimum and maximum width values were set at 20, 6 and 50, respectively (Bailey et al. 2009). All determined motifs by MEME were searched in InterPro database (http://www.ebi.ac.uk/interpro/) (Finn et al. 2017). Also, the transmembrane motifs were distinguished by TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
Promoter analysis
5′-UTR regions and 1000 bp upstream of the cDNA from each target gene, which were considered as the promoter sequences, were downloaded from Phytozome. Then, Plant cis-acting regulatory element (Plant CARE) database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002) was used in order to discover all cis-acting regulatory DNA elements (cis-elements).
Untranslated regions (UTR) analysis and microRNA target sites prediction
UTR (5′-UTRs and 3′-UTRs) sequences were obtained from Phytozome v12 database (http://www.phytozome.net/). Then, upstream open reading frames (uORFs) and internal ribosome entry sites (IRES) in 5′-UTRs were found by UTRScan tool at UTRdb (Grillo et al. 2010). Furthermore, sRNA targets were predicted using Plant Small RNA Target Analysis Server (psRNATarget) (Dai and Zhao 2011).
Plant growth and drought treatments
The study was carried out in the Seed and Plant Improvement Institute field, Karaj, Iran, as a split-plot on randomized complete block design with three replications during spring 2015.
The main plots consisted of two levels of irrigation (I1: irrigation after 60 mm evaporation from the surface evaporation pan as normal irrigation and I2: water withholding after flowering as severe water stress). The subplots included three genotypes, i.e. Kimia (a drought-tolerant native cultivar), Sepideh (a drought-susceptible native cultivar) and KGS23 (a drought-tolerant promising line) (Kharrazi and Rad 2002; MOSTAFA et al. 2011). The samples were collected quickly from the flag leaves (at least 7 random flag leaves were mixed as a biological replication) of both the well-watered and drought-stressed plants at 14, 21 and 28 days after flowering. Then, plant samples were frozen in liquid nitrogen rapidly and were stored at − 80 °C until RNA isolation.
Specific primers designing, RNA extraction and gene expression analysis
Thirteen specific primer pairs were designed to study the expression patterns of candidate SbNAC genes in sorghum using Oligo software version 7.0 (Rychlik 2007). Moreover, a specific primer pair was specified for Serine/threonine-Protein Phosphatase 2A (PP2A) gene (XM_002453490) as the endogenous control according to the previous studies (Sudhakar Reddy et al. 2016) (Supplementary Table 1).
Total RNA was extracted using the Trizol reagent (Invitrogen, USA) according to the manufacturer’s instruction for each of the samples (2 treatments, 3 genotypes and 3 time points). Qualification and quantification of total RNA was assessed using agarose gel electrophoresis and nanodrop spectrophotometer, respectively. After eliminating the genomic DNA contamination with DNAse I digestion (Geneall, Korea), the first-strand cDNA was synthesized by 1 µg of DNA-free total RNA from each sample as the template using iScript cDNA Synthesis kit (Bio-rad, USA), based on the manufacturer’s protocol. Quantitative real-time PCR was done in three independent biological replicates with the MiniOpticon system (Bio-rad, Cat No. CFB-3120) using the SYBR Green supermix kit (Quanta BioScience, USA) according to the instruction manual. The amplification profiles were carried out under the following conditions: 95 °C for 5 min followed by 40 cycles at 95 °C for 30 s, 57–61 °C (depending on each primer’s annealing temperature) for 30 s, 72 °C for 30 s followed by a melting curve analysis. Relative quantification was calculated by the 2−ΔΔCT method (Schmittgen and Livak 2008). The calibrator used for sorghum genotypes under drought stress were the sorghum genotypes under normal conditions. To analyze the relative expression, a temporal split-plot design was used and the mean values of treatments were calculated by the least significant difference tests. Also, the Pearson’s correlation analysis was used to evaluate the relationship between gene expression of candidate SbNAC genes at 28 days after flowering and yield of these genotypes (Kimia, Sepideh and KGS23) under drought conditions using SAS (P = 0.05) (Afifi et al. 2003).
Results and discussion
Identification of NAC transcription factor gene family in sorghum
The SbNAC members in Sorghum bicolor were identified by keyword (178, 123, 195 and 120 NAC transcripts were discovered at MOROKOSHI, GRASS, TFDB and Ensemble, respectively.), HMM profile (126 transcripts), and the BLAST searches against S. bicolor (v3.1) at Phytozome v12. It is noteworthy that 12 genes were found as “NAC” genes when searching the keyword on the Phytozome database, which were members of another NACs (stands for Nascent polypeptide-associated complex) and not the NAC we were looking for (i.e. NAM, ATAF1/2 and CUC2), or they had other domains such as the Kinesin motor and the NB-ARC domain. After removing the sequence redundancies and the none-NAC transcription factors, 183 protein sequences—encoded by 131 NAC gene loci—were identified in Sorghum bicolor. The NAC genes in S. bicolor are designated as SbNAC followed by the number 001-131 according to the chromosomal location and different transcripts encoded by the same locus showed with an extra decimal part (e.g., SbNAC008.1–SbNAC008.2). Then, 183 protein sequences containing the NAC domain were confirmed by SMART and Pfam, except two protein sequences (SbNAC027.2, SbNAC116.3). Sequences without any NAC domain were not used for further analysis. The Pfam database indicated one NAC domain, while the SMART database indicated two NAC domains for SbNAC086. The detailed information of SbNAC TFs in S. bicolor including locus ID, chromosome distribution, protein length, and position of NAC domain listed in Supplementary Table 2.
Phylogenetic tree reconstruction
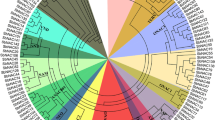
The unrooted phylogenetic tree was constructed based on NAC domains in sorghum, which classified the NAC family into 15 subfamilies (A–O) based on the tree topology (Fig. 1). Group F is the largest subfamily with 25 SbNACs, while group N is the smallest subgroup with only one member in sorghum. To further study the evolutionary relationships, prediction of protein function and abiotic stress-responsive SbNAC genes, another unrooted phylogenetic tree was constructed from alignments of all NAC domains in sorghum and Arabidopsis, along with 8 known NAC domain sequences from other plants such as rice, wheat, maize and tobacco (Supplementary Fig. 1; Supplementary Table 3). Based on the phylogenetic tree of Arabidopsis and sorghum, all NACs can be divided into 19 clades (A–S); thirteen subgroups belong to both trees containing the same NAC members. The results confirmed the existence of a diversified group of SbNAC family members in sorghum with various functions, such as what is reported for other plants (Fang et al. 2008; Nuruzzaman et al. 2010; Le et al. 2011). Interestingly, among the groups identified by phylogenetic analysis, one group only included Arabidopsis NACs, suggesting that NACs from monocots and dicots are evolutionarily distinct.
Group A (NAM) consisted of 14 SbNACs and 13 AtNACs. Studies of this gene group from Arabidopsis (CUC1/NAC054 and CUC2/NAC098) indicated that they may play a role in the development of apical meristems in the shoot (Aida et al. 1997).
Group B (NAC1) has 8 and 3 NAC members of sorghum and Arabidopsis, respectively. The reported NAC gene, NAC1 (ANAC21/ANAC22) from this subfamily can transmit auxin signals to promote lateral root development which act downstream of TIR1 (Xie et al. 2000). Therefore, this group in sorghum may be involved in auxin transportation.
Group C (SND1/VND6) includes 13 members of each SbNACs and AtNACs which could be divided into two subgroups (SND1 and VND6). AtNSTs (ANAC066/NST2 and ANAC043/NST1) and AtSNDs (NAC012) play a key role in secondary wall thickening in fibers that integrated into subgroup C-I (SND1). Other NAC transcription factors from this group are found to be related to vascular development in Arabidopsis and can be classified into subgroup C-II (VND6) (Kubo et al. 2005).
Group D includes only two members in sorghum, which their functions have not been known as yet.
Group E (TIP: Turnip crinkle virus interacting protein) contains the highest member of AtNAC with 14 members and 7 SbNACs. A transcriptional activator, ANAC091 (TIP: TCV-interacting protein) interact with the coat protein of turnip crinkle virus (TCV) to activate the transcription of genes that are involved in the basal resistance in the plant (Kubo et al. 2005). ANAC062 (NTL6) is the positive regulator of pathogen resistance against P. syringae, and it directly binds to a conserved sequence in the promoters of cold-responsive pathogenesis-related genes in the independent action of the salicylic acid pathway (Seo et al. 2010). ANAC089 is a membrane-tethered transcription factor which is localized to the ER and the truncated version (lacking the membrane-bound domain) negatively controls floral initiation, and it suppresses fructose sensitivity when present in the nucleus (Li et al. 2010, 2011).
Group F is the largest cluster of NAC with 25 in sorghum and 2 members in Arabidopsis. Group F does not include any known members of NAC in Arabidopsis.
Group G consists of 8 and 15 members of NAC in Arabidopsis and sorghum, respectively. All members of Group G in the sorghum tree existed by a commonality of the tree in sorghum and Arabidopsis, except for SbNAC108 which joined to the group R. ANAC073/SND2 and ANAC010/SND3 are grouped into this cluster. Therefore, it seems that these genes are involved in secondary wall thickening (Zhong et al. 2008).
Group H consists of 2 members of the SbNACs in the unrooted sorghum tree, but this group is in the unrooted tree of sorghum and Arabidopsis, consisting of three SbNACs. SbNAC070 from K group in the unrooted sorghum tree joined to the group H in the phylogenic tree of sorghum and Arabidopsis.
Group I contains 4 and 3 members of the NAC in sorghum and Arabidopsis, respectively. However, no function has been identified for these members as yet.
Group J (TERN) includes 2 and 5 members in Arabidopsis and sorghum, respectively. TERN (AB021178) is a member of tobacco which is also located in this group, and therefore this group is known as the TERN group (Benson et al. 1997).
Group K has 7 members in the sorghum unrooted tree, while the unrooted phylogenic tree between sorghum and Arabidopsis contains three SbNACs and an AtNAC for which no function has been identified.
Group L includes 11 and 8 member of NAC in sorghum and Arabidopsis, respectively. ANAC042, ANAC036 and LOV1 (ANAC034/35) are involved in the camalexin biosynthesis induction, leaf cell growth and flowering time regulation, respectively (Kato et al. 2010; Saga et al. 2012; Xu et al. 2012). ANAC009 (FEZ) controls the root cap cell division which is negatively regulated by ANAC033 (SOMBRERO) from group C (Willemsen et al. 2008).
Group M (SNAC: Stress- responsive NAC) contains 13 SbNAC, 12 AtNAC and 6 published NAC domains from other plants, and it accounts for 10% of the total NACs in sorghum. The objective of this study was to identify the putative SbNAC genes that are involved in response to various abiotic stresses especially drought. Earlier reports of phylogenetic analyses of the NAC gene family in rice showed that most of the stress-responsive NACs classified into one group which was called the stress-responsive NAC group (SNAC) (Fang et al. 2008). In this study, group M includes all six well-known stress-related NACs based on the phylogenetic analysis in sorghum and Arabidopsis, and it was named the SNAC group in sorghum. Group SNAC with all the published stress-related NAC TFs was further divided into 3 subgroups: NAP, SNAC1 and ATAF. The NAP subfamily is involved in senescence, whereas SNAC1 and ATAF subfamilies have functions in relation to abiotic stress, especially drought tolerance (Tran et al. 2004; Guo and Gan 2006; Hu et al. 2006).
Group N includes a member of sorghum and three membrane-bound members of Arabidopsis.
Overexpression of ANAC016 caused higher levels of drought susceptibility and accelerate senescence, whereas overexpression of ANAC013 led to a reduction in the plants susceptibility to oxidative stress (Hofmann 2013; Sakuraba et al. 2015).
Group O (NAC2) consists of 8 and 4 NAC members in Arabidopsis and sorghum, respectively. The name of this group is attributed to NAC2 based on previous cluster analysis of the NAC family genes in Arabidopsis and rice (Ooka et al. 2003). The dimerization form of ANAC050 and ANAC052 are involved in the transcriptional repression and the control of flowering time by interacting with the histone demethylase JMJ14 (Ning et al. 2015; Zhang et al. 2015). NLT4 transcription factor (ANAC053) regulates the productions of reactive oxygen species (ROS). This regulation occurs by a drought stress pathway mediated by ABA which induces leaf senescence under drought conditions (Lee et al. 2012). Two members of this family, ANAC082 and ANAC078, play roles in the ribosomal stress response and flavonoid biosynthesis, respectively (Morishita et al. 2009; Ohbayashi et al. 2017). Each member in this group seems to have a different function.
Group P consists of 9 AtNAC. Only two membrane-bound transcription factors, ANAC005 and ANAC069/NLT13, were characterized in this family. The overexpression of ANAC069 caused an increasing susceptibility to salt stress, whereas the knockdown of ANAC069 in Arabidopsis plants caused a decreasing susceptibility to osmotic stress. Indeed, ANAC069 negatively regulates salt and osmotic stress tolerance by ROS accumulation and prohibition of proline biosynthesis (He et al. 2017). ANAC005 lacking 20 amino acids in the C-terminal is localized in the nucleus, and regulates vascular development in Arabidopsis (Zhao et al. 2016). All members in group P only belong to the Arabidopsis, indicating that this subfamily may have been lost in sorghum. A similar phenomenon was reported in the phylogenetic analysis of rice and Arabidopsis NAC proteins (Ooka et al. 2003).
Group Q includes SbNAC107 from the group K in the sorghum tree; two members belong to the Arabidopsis and SENU5 belongs to the tomato. Expression of SENU5 gene was up-regulated during the senescence in tomato (John et al. 1997).
Group R contains only SbNAC108 from the group G of the sorghum tree, which joined this group. Group R also contains 3 members in the Arabidopsis.
Group S has two members in Arabidopsis and sorghum. Only one AtNAC (XND1/ANAC104) function has been studied in this group which is involved in lignocellulose synthesis (Zhao et al. 2008). AtNSTs, AtSNDs, and AtXND are master transcriptional switches for the developmental process of the cell wall in vascular plants (Zhong et al. 2006, 2008). Therefore, all members of group C, G and S could be involved in regulating the development of the cell wall.
Chromosomal distribution and gene structures of the SbNAC members in sorghum
The SbNAC genes are distributed on all 10 chromosomes in sorghum (Fig. 2). Chromosome 3 had the highest number of SbNACs, having 20 of them (15%), followed by 19 SbNACs (14%) on chromosome 1, while chromosome 8 and 4 contain 8 (7%) and 9 (6%) members of SbNACs, respectively. Different group members of the NAC family have been dispersed on different sorghum chromosomes. Major duplication and diversification within the NAC family seems to have occurred in their ancestors. The highest number of the gene in group M (SNAC) is located on chromosomes 3 (4 members) and 2 (3 members), respectively.
It is clear that there are possible mechanisms for the evolution of multigene families, and one prominent mechanism is gene structural diversity (Hu et al. 2010; Shang et al. 2013). To determine intron numbers and the exon–intron structure of the SbNAC genes, they were analyzed according to their cluster in the phylogenic analysis (Fig. 1). In general, most members of the SbNACs in the same group exhibit similar gene structures in terms of their intron number and intron phase. This shows their close evolutionary relationship in each subfamily. Also, gene structure analysis indicated that the number of introns were highly diverse, i.e. ranging from 0 to 14 numbers among SbNAC genes (Supplementary Fig. 2), which is similar to rice (ranging from 0 to 16) and Brachypodium distachyon (from 0 to 14) (Nuruzzaman et al. 2010; You et al. 2015). Furthermore, we discovered that the majority of NAC gene transcripts (85 of 183) in sorghum contain two introns. This observation was also reported for rice, cotton and banana (Nuruzzaman et al. 2010; Shang et al. 2013; Cenci et al. 2014). Additionally, most of the group F (16 members) contained no intron in their open reading frame (ORF), while group G had the maximum number of intron diversities among its members, i.e. from 2 to 14 introns. Also, the longest (SbNAC111) and shortest (SbNAC125) genomic sequence of the SbNACs was 13.87 kb and 510 bp from group G and F in sorghum, respectively.
Identification of additional conserved motifs and trans-membrane domain in SbNACs
To better understand the structural diversity of NAC proteins in sorghum, additional conserved motifs (20 putative motifs) were predicted using the MEME program. Generally, closely related members belong to the same clade in the phylogenetic tree, and they shared the same or very similar motif distributions (Fang et al. 2008). As expected, most of the SbNACs in the same groups contain similar motif compositions (Supplementary Fig. 3), whereby functional similarities are demonstrated. However, the functions of most motifs remain to be clarified when searching against the InterPro database (Supplementary Table 4). Previous studies illustrated that a typical NAC protein consists of five well-conserved subdomains (A-E) in the NAC domain which is located in the N-terminal (Kikuchi et al. 2000; Duval et al. 2002; Ooka et al. 2003). The protein sequences which do not include all of these five subdomains are named NAC-like proteins in rice (Fang et al. 2008). In this study, motif 2 plus 8, 4, 1, 6 plus 3 and 5 correspond to the subdomains A, B, C, D and E, respectively. These were found in most of the NAC family members of sorghum. Furthermore, one to four motifs existed in one member of either groups A, B, C, L, N, along with two members of the group K and all members of the groups F, H, I and J. Therefore, these members may be considered as NAC-like proteins in sorghum.
Similar to the previous studies in rice, potato and Brachypodium distachyon, most of the conserved motifs here were observed to be placed on the N-terminal region of SbNAC proteins and these motifs may play essential roles for the NAC protein functions (Fang et al. 2008; Singh et al. 2013; You et al. 2015). In addition, ten protein transcripts were encoded by seven loci and were predicted as membrane-bound transcription factors (MTFs) of the NAC family in sorghum, and this was performed by using TMHMMServer (Supplementary Fig. 4). All predicted trans-membrane SbNACs were located at the C-terminal, whereas only the TM motif of SbNAC038 was situated at the N-terminal, as reported in Brachypodium distachyon (You et al. 2015). According to the Arabidopsis and sorghum phylogenetic tree (Supplementary Fig. 1), trans-membrane NAC members in sorghum belonged to the group E (SbNAC043, SbNAC096 and SbNAC121), G (SbNAC038), N (SbNAC028) and O (SbNAC095 and SbNAC100), respectively.
Promoter analysis
The promoter region is located mainly in 1000 base pairs (bp) upstream of the transcriptional start site (ATG), and it consists of cis-elements which determine the temporal and spatial expression of the gene (Dean and Schmidt 1995; Ibraheem et al. 2010). Therefore, to find putative stress-responsive cis-elements in the promoter sequence of the SbNAC genes, the PlantCARE database was used in order to scan against 1 kb upstream of the cDNA in addition to the 5′ UTRs of NAC genes (available for 102 members). According to the results, 8 types of stress-related cis-elements were found, including MYB binding sites involved in drought-inducibility (MBS), dehydration-responsive elements (DRE), low temperature-responsive elements (LTR), heat shock elements (HSE), defense and stress-responsive elements (TC-rich repeats), cis-acting regulatory element essential for the anaerobic induction (ARE) and enhancer-like elements involved in anoxic specific inducibility (GC-motif and CGT-motif). Furthermore, some elements possibly involved in hormonal responses, such as ABA, gibberellin, auxin, ethylene (Eth), methyl jasmonate (MeJA) and salicylic acid (SA), were detected in this study (Supplementary Table 5). Previous works suggested that NAC genes with more than 5 cis-elements related to the plants response to stress or hormones in the promoter region might be involved in hormonal and stress response pathways (You et al. 2015). In this study, 131 and 118 SbNAC genes contained at least two and more than five cis-elements, respectively, in their promoter regions, which can contribute to the plants response to stress or hormones. In addition, 419 MeJA-responsive elements (CGTCA-motif and TGACG-motif), 226 ABA responsive elements (ABREs and motif IIb), 143 MBSs, 105 HSEs, 115 enhancer-like elements involved in anoxic specific inducibility (GC-motif and CGT-motif), 111 AREs, 99 gibberellin-responsive elements (TATC-box), 77 SA- responsive elements (TCA-element) and 58 defense and stress responsive elements (TC-rich repeats) were found in promoter regions of SbNAC genes, suggesting that NAC genes in sorghum play vital roles in the plants response to various factors such as MeJA, ABA, drought, heat shock, GA, SA, defense responses, anoxic processes and anaerobic processes. Furthermore, there were 10 and 25 of the SbNAC genes which contained over 5 copies of ABA and MeJA responsive elements in their promoter regions, respectively, suggesting that these SbNAC genes might be involved in the plants response to stress through signal pathway that can be either dependent or independent of ABA. Interestingly, the results revealed that more than five of both the ABA and MeJA responsive elements have been detected in the promoter of SbNAC035, SbNAC066 and SbNAC104 genes which belong to the group M (SNAC).
UTR analysis and microRNA target prediction
The untranslated regions (5′-UTRs and 3′-UTRs) are involved in the post-transcriptional regulation of gene expression, including modulation of the mRNAs nucleo-cytoplasmic transport, control of translation efficiency, subcellular localization of mRNAs and their stability (Jansen 2001; Bashirullah et al. 2001). Therefore, UTR sequences of the SbNAC genes were searched against the UTRdb database. No 3′-UTR and 5′-UTR had been reported for 29 and 30 out of 183 transcripts of SbNAC members, respectively. In general, 5′-UTRs are much shorter than 3′-UTRs and the GC content of 5′-UTRs is greater than that of 3′-UTRs (Pesole et al. 2001). Likewise, the GC content of 5′-UTR (54.74%) was more than that of 3′-UTR (40.17%), and the average length of 5′-UTR (310.01 nt) was less than 3′-UTR (531.33 nt) in the SbNAC family (Supplementary Tables 6, 7). The translational efficiency can be affected by the structural features of the 5′-UTR, including the length and start-site consensus sequences, the presence of a secondary structure, upstream AUGs, upstream open reading frames (uORFs) and internal ribosome entry sites (IRES) (Mignone et al. (2002); Gray and Wickens 1998). IRES is an element within the 5′-UTR that mediates the initiation of translation via cap-independent translation, while uORF within the 5′-UTR reduces the protein synthesis through ribosomal scanning block or through its delay (Martínez-Salas et al. 2012; Ozretić et al. 2012). We could predict 101 uORFs from all SbNACs, while SbNAC111 showed the highest number (7) of uORFs. Furthermore, 39 members of the NAC transcripts include the IRES sequence (Supplementary Table 5).
In plants, small RNAs—mainly miRNAs—negatively regulate gene expression at the post-transcriptional level by binding to conserved sequences of target mRNA (Reinhart et al. 2002; Bartel 2004). The miRNAs target genes are involved in diverse physiological processes in sorghum, including growth and development, nutrient homeostasis and various environmental stress responses (Zhang et al. 2011). Using the psRNA Target, 200 and 98 miRNA target sites were found in 3′-UTR and 5′-UTR sequences of the SbNAC gene family transcripts, respectively (Supplementary Tables 8, 9). The miRNAs were predicted to target SbNAC transcription factors, which were revealed to belong to 26 distinct miRNA families.
Expression profiles of the SNAC group members in sorghum under drought stress
There is a growing amount of evidence which shows that NAC genes are involved in various stress responses, senescence and different developmental processes in plants (Guo and Gan 2006; Le et al. 2011; Shiriga et al. 2014; Ha et al. 2014). Also, many stress-related NAC genes are categorized in SNAC groups according to the previous description (Fang et al. 2008). Accordingly, the SNAC group in sorghum was selected for gene expression analysis. ‘Kimia’ cultivar and ‘KGS23’ promising line are both sorghum drought-tolerant genotypes, whereas the ‘Sepideh’ cultivar is susceptible genotype to drought stress (Yield data are available in Supplementary Table 10). The gene expression patterns were evaluated in the flag leaves of these three aforementioned genotypes under drought stress at three time points (i.e. 14, 21 and 28 days after flowering) through quantitative real-time PCR. Analysis of variance revealed that the effects of genotypes and time points were significant on the relative gene expression in all SbSNACs (P ≤ 0.05 or P ≤ 0.01) under severe drought stress, except for SbNAC005 and SbNAC041, respectively. Furthermore, significant interactions were identified between the cultivars and the time points, affecting the relative gene expression of all genes, excluding SbNAC005 and SbNAC035 (P ≤ 0.05 or P ≤ 0.01) (Supplementary Table 11).
As previously described, the SNAC group in sorghum can be further classified into three clusters (NAP, ATAF and SNAC1) based on phylogenetic analysis (Supplementary Fig. 1). The NAP subgroup contains seven SbNACs (SbNAC014, SbNAC034, SbNAC035, SbNAC037, SbNAC041, SbNAC066 and SbNAC104) which were clustered with senescence-associated NACs, including AtNAP (AJ222713) (Guo and Gan 2006), OsNAP (Os03g0327800) (Chen et al. 2014) and NAM-B1 (DQ869673) (Uauy et al. 2006). Based on the results of gene expression analysis, the relative expression of SbNAC066 in flag leaves was not detected at 14 days after flowering in all tree genotypes, whether susceptible or tolerant, while it was up-regulated significantly under water-deficit treatments at 21 and 28 days after flowering. The relative expression of SbNAC104 was significantly up-regulated in the susceptible cultivar, but the pattern of expression did not change in the tolerant cultivars at all three time points. Under drought stress, SbNAC066 and SbNAC104 showed significantly higher expression levels in flag leaves of the susceptible cultivar compared to the drought-tolerant cultivars. However, the gene expression pattern of these genes was not significantly different when comparing the two tolerant genotypes (Fig. 3a). The AtNAP plays a crucial role in leaf senescence in Arabidopsis, and the T-DNA insertion of this gene causes a significant delay in leaf senescence (GUO et al. 2004; Guo and Gan 2006). As SbNAC066 and SbNAC104 joined the AtNAP on the phylogenic tree in sorghum and Arabidopsis, and their expression patterns match under drought stress (Supplementary Fig. 1), it seems that these two genes play roles in drought-induced senescence in the post flowering stage of sorghum.
Relative expression profiles of 13 SbNAC genes (SNAC group) in three genotypes of sorghum under post- flowering drought stress. Transcript level of each gene in every genotype under water control conditions (control) was used as the calibrator of each time points and y-axis shows the fold change. Sorghum PP2A gene was chosen as internal control for normalization. Each experiment was performed three times independently, and error bars represent standard deviation. a Relative expression of AtNAP clade genes (SbNAC066 and SbNAC104) in phylogenic tree, b relative expression of OsNAP clade genes (SbNAC014, SbNAC035 and SbNAC041) in phylogenic tree, c relative expression of NAM-B1 clade genes (SbNAC034 and SbNAC037) in phylogenic tree, d relative expression of ATAF clade genes (SbNAC052, SbNAC073 and SbNAC116) in phylogenic tree, and e relative expression of SNAC1 clade genes (SbNAC005, SbNAC021 and SbNAC050) in phylogenic tree
The relative gene expression of SbNAC014, SbNAC035 and SbNAC041 had an increasing trend in all the genotypes during drought stress; however, their expression levels were different among the cultivars after 28 days of the flowering onset. A significant up-regulation was observed for SbNAC014 in the tolerant cultivars, especially in ‘Kimia’, after long-term drought stress (Fig. 3b). The phylogenetic analysis showed that these three genes belong to the OsNAP cluster (Supplementary Fig. 1). The OsNAP in rice is the homologue of AtNAP, and genes expression of OsNAP were significantly induced by ABA and abiotic stress, e.g. high salinity, drought and low temperature (Chen et al. 2014). Overexpression of OsNAP in rice caused a significant decrease in the rate of water loss. It enhanced drought tolerance via an ABA-mediated pathway, and improved grain yields under drought stress at the flowering stage (Chen et al. 2014). Based on the expression pattern of SbNAC014, SbNAC035 and SbNAC041 genes, it seems that they act as transcriptional activators similar to OsNAP, and play roles in response to post-flowering drought stress in sorghum. It can be hypothesized that when the plants are exposed to drought stress at the post-flowering stage, these genes are up-regulated to a greater extent in the tolerant cultivars, compared to the susceptible one. Thus, the rate of water loss could be reduced in the tolerant cultivars by controlling their stomatal closure which can ultimately delay leaf senescence.
The expression of SbNAC034 was exclusively up-regulated in the tolerant cultivars in response to drought stress, while the expression of SbNAC037 was up-regulated in all three cultivars, especially in the ‘Kimia’ (tolerant cultivar) (Fig. 3c). Based on the phylogenic analysis, SbNAC034 joined NAM-B1, and SbNAC037 which belonged to the NAP subgroup was orthologous of ANAC025 based on Blast x (Supplementary Fig. 1). As previously reported, the NAM-B1 gene expression is up-regulated after the anthesis in wheat flag leaves and acts as a positive regulator of senescence and nutrient remobilization during grain maturation (Uauy et al. 2006; Ricachenevsky et al. 2013). There is a close relationship between wheat NAM-B1 and Arabidopsis NACs (ANAC025, ANAC018 and ANAC056), and all of them belong to the NAP subfamily (Podzimska-Sroka et al. 2015). Taken together, these results suggest that SbNAC034 and SbNAC037 might have a similar function in nutrient remobilization.
Under the drought stress treatment, SbNAC052 and SbNAC073 were significantly up-regulated only in the susceptible cultivar. An additive trend was observed in transcript levels of SbNAC073 and SbNAC116 in ‘Sepideh’ by elongating drought stress during the days after flowering. SbNAC116 were up-regulated in both susceptible and tolerant cultivars at the third time point. However, lower relative expression levels of SbNAC116 were detected in ‘Kimia’ and ‘KGS23’ (Fig. 3d). These three genes (SbNAC052, SbNAC073 and SbNAC116) belong to the ATAF subgroup based on the phylogenetic analysis of sorghum and Arabidopsis NACs. The ATAF subfamily can be divided into two clades, as formerly reported (Ooka et al. 2003). Among Arabidopsis NAC genes, ANAC019, ANAC055 and ANAC072 can be categorized into the same clade, and all of them appear to be positive regulators in drought stress (Tran et al. 2004). In contrast, ANAC002 (ATAF1) was clustered in another clade with ANAC102, ANAC081 (ATAF2) and ANAC032, while ATAF1 is assumed to be a negative regulator in drought signaling pathways via modulating osmotic stress-responsive gene expression (Lu et al. 2007). According to the phylogenetic tree and the gene expression patterns of SbNAC052, SbNAC073 and SbNAC116, it is suggested that those genes might negatively regulate the expression of stress responsive genes under drought stress in sorghum (Supplementary Fig. 1).
Based on the phylogenic tree, the SNAC1 subgroup contains three NAC genes in sorghum (SbNAC005, SbNAC021 and SbNAC052) and some known NAC genes including rice SNAC1 (Os03g60080), maize ZmSNAC1 (JQ217429.1), wheat TaNAC02 (AY625683.1) and TaNAC067 (KF646593.1).
Overexpression of ZmSNAC1 (Lu et al. 2012), TaNAC02 (Mao et al. 2012), TaNAC067 (Mao et al. 2014), SbSNAC1 (named SbNAC005 in this report) (Lu et al. 2013) and SbNAC052 (Kadier et al. 2017) (named SbNAC050 in this report) led to enhanced tolerance to drought at the seedling stage in Arabidopsis (Supplementary Fig. 1). Overexpression of SNAC1 caused the improvement of drought tolerance at the vegetative and reproductive stage in transgenic rice under severe drought stress treatments (Hu et al. 2006). The transcript levels of SbNAC005, SbNAC021 and SbNAC052 were not changed notably, i.e. changes that were not more than twofold, in flag leaves of the tolerant cultivars of this study (Fig. 3e). This observation suggests that the latter genes might not be involved in the response to drought stress and the drought during the post-flowering stage that affects the studied sorghum cultivars. They may be expressed in other tissues or developmental stages, or might be regulated at post-transcriptional levels.
Pearson’s correlation coefficient was used to establish the relationship between gene expression of SNAC group in sorghum’s flag leaves and yield. The results indicated significant negative correlation (at least r = − 0.73) between the yield and expression level of SbNAC104, SbNAC052, SbNAC116, SbNAC073, SbNAC005 and SbNAC066, whereas significant positive correlation (r = 0.71) were found between gene expression of SbNAC041 and yield under the drought stress. However, no significant correlations were observed between the expression level of other SNAC genes and yield (Supplementary Table 12).
Conclusion
A genome-wide analysis of the NAC TF gene family in sorghum enabled the identification of 131 members which can be classified into 19 subfamilies based on the unrooted phylogenic tree analysis of SbNACs and AtNACs. Group M (SNAC) was found to be the stress-responsive group, and it is assumed to contain 13 NAC members that may act as candidate TFs in response to abiotic stresses in sorghum. Furthermore, the expression profiles in tolerant and susceptible cultivars by qRT-PCR showed that 5 members (SbNAC014, SbNAC034 SbNAC035, SbNAC037 and SbNAC041) and 3 members (SbNAC052, SbNAC073 and SbNAC116) from the NAP and ATAF subgroups of the NAC genes may act as positive and negative regulators, respectively, in response to post-flowering drought stress in sorghum. In addition, the expression level of some SbNAC genes was significantly correlated with the sorghum cultivar’s yield under drought conditions in the field, indicating their likely roles in post-flowering drought tolerance and yield maintenance. Therefore, further researches are highly recommended to confirm promising candidates for genetic engineering or breeding programs to enhance drought tolerance in crops.
Author contribution statement
SS conducted the experiments and drafted the manuscript. ZSS conceived the project, supervised and coordinated the research. MS supervised the plant culture and physiological assessments. ZSS, RZK and MS revised the manuscript. All authors have read and approved the manuscript.
References
Afifi A, May S, Clark V (2003) Computer-aided multivariate analysis. CRC Press, London
Aida M, Ishida T, Fukaki H et al (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857. https://doi.org/10.1105/tpc.9.6.841
Bailey TL, Boden M, Buske FA et al (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. https://doi.org/10.1016/S0092-8674(04)00045-5
Bashirullah A, Cooperstock RL, Lipshitz HD (2001) Spatial and temporal control of RNA stability. Proc Natl Acad Sci USA 98:7025–7028. https://doi.org/10.1073/pnas.111145698
Benson DA, Boguski MS, Lipman DJ, Ostell J (1997) GenBank. Nucleic Acids Res 25:1–6. https://doi.org/10.1093/nar/25.1.1
Blum A (2004) Sorghum physiology. In: Nguyen HT, Blum A (eds) Physiology and biotechnology integration for plant breeding. CRC Press, Boca Raton, pp 141–223
Casa AM, Mitchell SE, Hamblin MT et al (2005) Diversity and selection in sorghum: Simultaneous analyses using simple sequence repeats. Theor Appl Genet 111:23–30. https://doi.org/10.1007/s00122-005-1952-5
Cenci A, Guignon V, Roux N, Rouard M (2014) Genomic analysis of NAC transcription factors in banana (Musa acuminata) and definition of NAC orthologous groups for monocots and dicots. Plant Mol Biol 85:63–80. https://doi.org/10.1007/s11103-013-0169-2
Chen X, Wang Y, Lv B et al (2014) The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol 55:604–619. https://doi.org/10.1093/pcp/pct204
Collinge M, Boller T (2001) Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol 46:521–529. https://doi.org/10.1023/A:1010639225091
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:W155–W159. https://doi.org/10.1093/nar/gkr319
Dean C, Schmidt R (1995) Plant genomes: a current molecular description. Annu Rev Plant Physiol Plant Mol Biol 46:395–418. https://doi.org/10.1146/annurev.pp.46.060195.002143
Duval M, Hsieh T-F, Kim S-Y, Thomas TL (2002) Molecular characterization of AtNAM: a member of theArabidopsis NAC domain superfamily. Plant Mol Biol 50:237–248
Fang Y, You J, Xie K et al (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genom 280:547–563. https://doi.org/10.1007/s00438-008-0386-6
Finn RD, Coggill P, Eberhardt RY et al (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. https://doi.org/10.1093/nar/gkv1344
Finn RD, Attwood TK, Babbitt PC et al (2017) InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199. https://doi.org/10.1093/nar/gkw1107
Fujita M, Fujita Y, Maruyama K et al (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876. https://doi.org/10.1111/j.1365-313X.2004.02171.x
Goodstein DM, Shu S, Howson R et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:1178–1186. https://doi.org/10.1093/nar/gkr944
Gray NK, Wickens M (1998) Control of translation initiation in animals. Annu Rev Cell Dev Biol 14:399–458. https://doi.org/10.1146/annurev.cellbio.14.1.399
Grillo G, Turi A, Licciulli F et al (2010) UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 38:D75–D80. https://doi.org/10.1093/nar/gkp902
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612. https://doi.org/10.1111/j.1365-313X.2006.02723.x
Guo Y, Cai Z, Gan S (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27:521–549. https://doi.org/10.1111/j.1365-3040.2003.01158.x
Ha Van C, Nasr Esfahani M, Watanabe Y et al (2014) Genome-wide identification and expression analysis of the CaNAC family members in chickpea during development, dehydration and ABA treatments. PLoS One 9:e114107. https://doi.org/10.1371/journal.pone.0114107
He X-J, Mu R-L, Cao W-H et al (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916. https://doi.org/10.1111/j.1365-313X.2005.02575.x
He L, Shi X, Wang Y et al (2017) Arabidopsis ANAC069 binds to C[A/G]CG[T/G] sequences to negatively regulate salt and osmotic stress tolerance. Plant Mol Biol 93:369–387. https://doi.org/10.1007/s11103-016-0567-3
Hofmann N (2013) Endoplasmic reticulum–localized transcription factors and mitochondrial retrograde regulation. Plant Cell 25:10–1105
Hu H, Dai M, Yao J et al (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992. https://doi.org/10.1073/pnas.0604882103
Hu R, Qi G, Kong Y et al (2010) Comprehensive analysis of NAC domain transcription factor gene family in populus trichocarpa. BMC Plant Biol 10:145. https://doi.org/10.1186/1471-2229-10-145
Hu B, Jin J, Guo A-Y et al (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Ibraheem O, Botha CEJ, Bradley G (2010) In silico analysis of cis-acting regulatory elements in 5′ regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput Biol Chem 34:268–283. https://doi.org/10.1016/j.compbiolchem.2010.09.003
Jansen R-P (2001) mRNA localization: message on the move. Nat Rev Mol Cell Biol 2:247–256. https://doi.org/10.1038/35067016
Jensen MK, Hagedorn PH, de Torres-Zabala M et al (2008) Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J 56:867–880. https://doi.org/10.1111/j.1365-313X.2008.03646.x
Jeong JS, Kim YS, Baek KH et al (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197. https://doi.org/10.1104/pp.110.154773
Jeong JS, Kim YS, Redillas MCFR et al (2013) OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J 11:101–114. https://doi.org/10.1111/pbi.12011
Jin J, Tian F, Yang DC et al (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45:D1040–D1045. https://doi.org/10.1093/nar/gkw982
John I, Hackett R, Cooper W et al (1997) Cloning and characterization of tomato leaf senescence-related cDNAs. Plant Mol Biol 33:641–651. https://doi.org/10.1023/A:1005746831643
Kadier Y, Zu Y, Dai Q et al (2017) Genome-wide identification, classification and expression analysis of NAC family of genes in sorghum [Sorghum bicolor (L.) Moench]. Plant Growth Regul 83:301–312. https://doi.org/10.1007/s10725-017-0295-y
Kato H, Motomura T, Komeda Y et al (2010) Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J Plant Physiol 167:571–577. https://doi.org/10.1016/j.jplph.2009.11.004
Kebede H, Subudhi PK, Rosenow DT, Nguyen HT (2001) Quantitative trait loci influencing drought tolerance in grain sorghum (Sorghum bicolor L. Moench). TAG Theor Appl Genet 103:266–276. https://doi.org/10.1007/s001220100541
Kharrazi M, Rad M (2002) Evaluation of sorghum genotypes under drought stress conditions using some stress tolerance indices. Afr J Biotechnol 10(61):13086–13089
Kikuchi K, Ueguchi-Tanaka M, Yoshida KT et al (2000) Molecular analysis of the NAC gene family in rice. MGG Mol Gen Genet 262:1047–1051. https://doi.org/10.1007/PL00008647
Kim Y-S, Kim S-G, Park J-E et al (2006) A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18:3132–3144. https://doi.org/10.1105/tpc.106.043018
Kim HS, Park BO, Yoo JH et al (2007) Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J Biol Chem 282:36292–36302. https://doi.org/10.1074/jbc.M705217200
Ko J-H, Yang SH, Park AH et al (2007) ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J 50:1035–1048. https://doi.org/10.1111/j.1365-313X.2007.03109.x
Kubo M, Udagawa M, Nishikubo N et al (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19:1855–1860. https://doi.org/10.1101/gad.1331305
Kumar S, Nei M, Dudley J, Tamura K (2006) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. October 88:559–566. https://doi.org/10.1093/bib/bbn017.MEGA
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Le DT, Nishiyama R, Watanabe Y et al (2011) Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res 18:263–276. https://doi.org/10.1093/dnares/dsr015
Lee S, Seo PJ, Lee HJ, Park CM (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70:831–844. https://doi.org/10.1111/j.1365-313X.2012.04932.x
Lescot M, Déhais P, Thijs G et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. https://doi.org/10.1093/nar/30.1.325
Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43:D257–D260. https://doi.org/10.1093/nar/gku949
Li JQ, Zhang J, Wang XC, Chen J (2010) A membrane-tethered transcription factor ANAC089 negatively regulates floral initiation in Arabidopsis thaliana. Sci China Life Sci 53:1299–1306. https://doi.org/10.1007/s11427-010-4085-2
Li P, Wind JJ, Shi X et al (2011) Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane-bound domain. Proc Natl Acad Sci 108:3436–3441. https://doi.org/10.1073/pnas.1018665108
Lu PL, Chen NZ, An R et al (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63:289–305. https://doi.org/10.1007/s11103-006-9089-8
Lu M, Ying S, Zhang D-F et al (2012) A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep 31:1701–1711. https://doi.org/10.1007/s00299-012-1284-2
Lu M, Zhang D-F, Shi Y-S et al (2013) Expression of SbSNAC1, a NAC transcription factor from sorghum, confers drought tolerance to transgenic Arabidopsis. Plant Cell Tissue Organ Cult 115:443–455. https://doi.org/10.1007/s11240-013-0375-2
Ludlow MM, Muchow RC (1990) A critical evaluation of traits for improving crop yields in water-limited environments. Adv Agron 43:107–153
Makita Y, Shimada S, Kawashima M et al (2015) MOROKOSHI: transcriptome database in sorghum bicolor. Plant Cell Physiol 56:e6. https://doi.org/10.1093/pcp/pcu187
Mao C, Ding W, Wu Y et al (2007) Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytol 176:288–298. https://doi.org/10.1111/j.1469-8137.2007.02177.x
Mao X, Zhang H, Qian X et al (2012) TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J Exp Bot 63:2933–2946. https://doi.org/10.1093/jxb/err462
Mao X, Chen S, Li A et al (2014) Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS One. https://doi.org/10.1371/journal.pone.0084359
Martínez-Salas E, Piñeiro D, Fernández N (2012) Alternative mechanisms to initiate translation in eukaryotic mRNAs. Comp Funct Genomics 2012:391546
Menz MA, Klein RR, Unruh NC et al (2004) Genetic diversity of public inbreds of sorghum determined by mapped AFLP and SSR markers. Crop Sci 44:1236. https://doi.org/10.2135/cropsci2004.1236
Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biol 3(3):reviews0004-1
Morishita T, Kojima Y, Maruta T et al (2009) Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol 50:2210–2222. https://doi.org/10.1093/pcp/pcp159
Mostafa M, Shahbazi M, Khazaei A (2011) Effect of post-flowering water stress on yield and physiological characters of grain sorghum genotypes. Iran J Plant Physiol 2(1):341–344
Mullet JE, Klein RR, Klein PE (2002) Sorghum bicolor—an important species for comparative grass genomics and a source of beneficial genes for agriculture. Curr Opin Plant Biol 5:118–121. https://doi.org/10.1016/S1369-5266(02)00232-7
Nakashima K, Takasaki H, Mizoi J et al (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta Gene Regul Mech 1819:97–103. https://doi.org/10.1016/j.bbagrm.2011.10.005
Ning YQ, Ma ZY, Huang HW et al (2015) Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res 43:1469–1484. https://doi.org/10.1093/nar/gku1382
Nuruzzaman M, Manimekalai R, Sharoni AM et al (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465:30–44. https://doi.org/10.1016/J.GENE.2010.06.008
Ohbayashi I, Lin C, Shinohara N, Matsumura Y (2017) Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in Arabidopsis. Plant Cell. https://doi.org/10.1105/tpc.17.00255
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87. https://doi.org/10.1016/J.TPLANTS.2004.12.010
Ooka H, Satoh K, Doi K et al (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247
Ozretić P, Bisio A, Inga A, Levanat S (2012) The growing relevance of cap-independent translation initiation in cancer-related genes. Period Biol 114(4):471–478
Pesole G, Mignone F, Gissi C et al (2001) Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276:73–81. https://doi.org/10.1016/S0378-1119(01)00674-6
Podzimska-Sroka D, O’Shea C, Gregersen P, Skriver K (2015) NAC transcription factors in senescence: from molecular structure to function in crops. Plants 4:412–448. https://doi.org/10.3390/plants4030412
Rachmat A, Nugroho S, Sukma D, Aswidinnoor H (2014) Overexpression of OsNAC6 transcription factor from Indonesia rice cultivar enhances drought and salt tolerance. Emir J Food Agric 26:519–527. https://doi.org/10.9755/ejfa.v26i6.17672
Reinhart BJ, Weinstein EG, Rhoades MW et al (2002) MicroRNAs in plants. Genes Dev 16:1616–1626. https://doi.org/10.1101/gad.1004402
Ren T (2000) HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12:1917–1926. https://doi.org/10.1105/tpc.12.10.1917
Ricachenevsky FK, Menguer PK, Sperotto RA (2013) kNACking on heaven’s door: how important are NAC transcription factors for leaf senescence and Fe/Zn remobilization to seeds? Front Plant Sci. https://doi.org/10.3389/fpls.2013.00226
Riechmann JL, Heard J, Martin G et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110. https://doi.org/10.1126/science.290.5499.2105
Rooney WL, Blumenthal J, Bean B, Mullet JE (2007) Designing sorghum as a dedicated bioenergy feedstock. Biofuels Bioprod Biorefining 1:147–157. https://doi.org/10.1002/bbb.15
Rosenow D, Ejeta G, Clark L, Gilbert M et al (1997) Breeding for pre- and post-flowering drought stress resistance in sorghum. In: Proceedings of the international conference on genetic improvement of sorghum and pearl millet. INSORMIL, Lincoln, pp 400–411
Rushton P, Bokowiec M, Han S et al (2008) Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant physiol 147(1):280–295
Rychlik W (2007) OLIGO 7 primer analysis software. In: PCR primer design. Humana Press, New York, pp 35–59
Sablowski RW, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM Is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92:93–103. https://doi.org/10.1016/S0092-8674(00)80902-2
Saga H, Ogawa T, Kai K et al (2012) Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Mol Plant-Microbe Interact 25:684–696. https://doi.org/10.1094/MPMI-09-11-0244
Sakuraba Y, Kim Y-S, Han S-H et al (2015) The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27:1771–1787. https://doi.org/10.1105/tpc.15.00222
Sanchez AC, Subudhi PK, Rosenow DT, Nguyen HT (2002) Mapping QTLs associated with drought resistance in sorghum (Sorghum bicolor L. Moench). Plant Mol Biol 48:713–726. https://doi.org/10.1023/A:1014894130270
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. https://doi.org/10.1038/nprot.2008.73
Seo PJ, Kim MJ, Park JY et al (2010) Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J 61:661–671. https://doi.org/10.1111/j.1365-313X.2009.04091.x
Shang H, Li W, Zou C, Yuan Y (2013) Analyses of the NAC transcription factor gene family in Gossypium raimondii Ulbr.: chromosomal location, structure, phylogeny, and expression patterns. J Integr Plant Biol 55:663–676. https://doi.org/10.1111/jipb.12085
Shiriga K, Sharma R, Kumar K et al (2014) Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene 2:407–417. https://doi.org/10.1016/J.MGENE.2014.05.001
Singh A, Sharma V, Pal A (2013) Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). Dna …
Souer E, van Houwelingen A, Kloos D et al (1996) The No apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85:159–170. https://doi.org/10.1016/S0092-8674(00)81093-4
Sperotto RA, Ricachenevsky FK, Duarte GL et al (2009) Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230:985–1002. https://doi.org/10.1007/s00425-009-1000-9
Subudhi PK, Rosenow DT, Nguyen HT (2000) Quantitative trait loci for the stay green trait in sorghum (Sorghum bicolor L. Moench): consistency across genetic backgrounds and environments. TAG Theor Appl Genet 101:733–741. https://doi.org/10.1007/s001220051538
Sudhakar Reddy P, Srinivas Reddy D, Sivasakthi K et al (2016) Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Front Plant Sci 7:1–14. https://doi.org/10.3389/fpls.2016.00529
Tang Y, Liu M, Gao S et al (2012) Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol Plant 144:210–224. https://doi.org/10.1111/j.1399-3054.2011.01539.x
Tran LP, Nakashima K, Sakuma Y et al (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 Promoter. Plant Cell 16:2481–2498. https://doi.org/10.1105/tpc.104.022699.Shinozaki
Tuinstra MR, Grote EM, Goldsbrough PB, Ejeta G (1997) Genetic analysis of post-flowering drought tolerance and components of grain development in Sorghum bicolor (L.) Moench. Mol Breed 3:439–448. https://doi.org/10.1023/A:1009673126345
Uauy C, Distelfeld A, Fahima T et al (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314:1298–1301. https://doi.org/10.1126/science.1133649
Voorrips RE (2002) Computer note MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Walulu RS, Rosenow DT, Wester DB, Nguyen HT (1994) Inheritance of the stay green trait in sorghum. Crop Sci 34:970. https://doi.org/10.2135/cropsci1994.0011183X003400040026x
Waters BM, Uauy C, Dubcovsky J, Grusak MA (2009) Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J Exp Bot 60:4263–4274. https://doi.org/10.1093/jxb/erp257
Willemsen V, Bauch M, Bennett T et al (2008) The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev Cell 15:913–922. https://doi.org/10.1016/j.devcel.2008.09.019
Wu X-Y, Hu W-J, Luo H et al (2016) Transcriptome profiling of developmental leaf senescence in sorghum (Sorghum bicolor). Plant Mol Biol 92:555–580. https://doi.org/10.1007/s11103-016-0532-1
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036. https://doi.org/10.1101/gad.852200
Xu W, Subudhi PK, Crasta OR et al (2000) Molecular mapping of QTLs conferring stay-green in grain sorghum (Sorghum bicolor L. Moench). Genome 43:461–469. https://doi.org/10.1139/g00-003
Xu B, Sathitsuksanoh N, Tang Y et al (2012) Overexpression of AtLOV1 in switchgrass alters plant architecture, lignin content, and flowering time. PLoS One. https://doi.org/10.1371/journal.pone.0047399
Xue G-P, Way HM, Richardson T et al (2011) Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4:697–712. https://doi.org/10.1093/MP/SSR013
Yilmaz A, Nishiyama MY, Fuentes BG et al (2009) GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol 149:171–180. https://doi.org/10.1104/pp.108.128579
You J, Zhang L, Song B et al (2015) Systematic analysis and identification of stress-responsive genes of the NAC gene family in Brachypodium distachyon. PLoS One 10:e0122027. https://doi.org/10.1371/journal.pone.0122027
Zhang L, Zheng Y, Jagadeeswaran G et al (2011) Identification and temporal expression analysis of conserved and novel microRNAs in Sorghum. Genomics 98:460–468. https://doi.org/10.1016/J.YGENO.2011.08.005
Zhang S, Zhou B, Kang Y et al (2015) C-terminal domains of histone demethylase JMJ14 interact with a pair of NAC transcription factors to mediate specific chromatin association. Cell Discov 1:15003. https://doi.org/10.1038/celldisc.2015.3
Zhao C, Avci U, Grant EH et al (2008) XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J 53:425–436. https://doi.org/10.1111/j.1365-313X.2007.03350.x
Zhao J, Liu J-S, Meng F-N et al (2016) ANAC005 is a membrane-associated transcription factor and regulates vascular development in Arabidopsis. J Integr Plant Biol 58:442–451. https://doi.org/10.1111/jipb.12379
Zhong R, Demura T, Ye Z-H (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell Online 18:3158–3170. https://doi.org/10.1105/tpc.106.047399
Zhong R, Lee C, Zhou J et al (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Online 20:2763–2782. https://doi.org/10.1105/tpc.108.061325
Acknowledgements
This study was supported by Agricultural Biotechnology Research Institute of Iran (Project Number: 3-03-0305-93122), Education and Extension Organization (AREEO). University of Guilan and the Ministry of Science, Research and Technology are also acknowledged for supporting the project. The authors are grateful for the kind cooperation of Dr. Azim Khazaei and Mr. Mojtaba Jowkar (to plant culture in the field), Dr. Behzad Sorkhi (to provide real-time PCR facility) and Mr. Mohammad Jedari (to create the artworks).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Kan Wang.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sanjari, S., Shirzadian-Khorramabad, R., Shobbar, ZS. et al. Systematic analysis of NAC transcription factors’ gene family and identification of post-flowering drought stress responsive members in sorghum. Plant Cell Rep 38, 361–376 (2019). https://doi.org/10.1007/s00299-019-02371-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02371-8