Abstract
NAM, ATAF, and CUC (NAC) transcription factors comprise a large plant-specific gene family and a few members of this family have been characterized for their roles in plant growth, development, and stress tolerance. In this study, systematic sequence analysis revealed 140 putative NAC or NAC-like genes (ONAC) in rice. Phylogenetic analysis suggested that NAC family can be divided into five groups (I–V). Among them, all the published development-related genes fell into group I, and all the published stress-related NAC genes fell into the group III (namely stress-responsive NAC genes, SNAC). Distinct compositions of the putative motifs were revealed on the basis of NAC protein sequences in rice. Most members contained a complete NAC DNA-binding domain and a variable transcriptional regulation domain. Sequence analysis, together with the organization of putative motifs, indicated distinct structures and potential diverse functions of NAC family in rice. Yeast one-hybrid analysis confirmed that 12 NAC proteins representing different motif compositions can bind the NAC core DNA-binding site. Real-time polymerase chain reaction (PCR) analysis revealed 12 genes with different tissue-specific (such as callus, root, stamen, or immature endosperm) expression patterns, suggesting that these genes may play crucial regulatory roles during growth and development of rice. The expression levels of this family were also checked under various abiotic stresses including drought, salinity, and low temperature. A preliminary check based on our microarray data suggested that more than 40 genes of this family were responsive to drought and/or salt stresses. Among them, 20 genes were further investigated for their stress responsiveness in detail by real-time PCR analysis. Most of these stress-responsive genes belonged to the group III (SNAC). Considering the fact that a very limited number of genes of the NAC family have been characterized, our data provide a very useful reference for functional analysis of this family in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Expression of most eukaryotic genes rely on specific transcription factors that bind to or modulate DNA structure in the regulatory region of genes, which in turn affects the activity of RNA polymerases for initiation of transcription. In plants, more than 50 families of different transcription factors have been identified based on sequence analysis in model species such as Arabidopsis and rice (Riechmann et al. 2000; Xiong et al. 2005; Riano-Pachon et al. 2007), and numerous reports suggest transcription factors are involved in almost all aspects of cellular activities with their related roles in gene expression.
NAM, ATAF, and CUC (NAC) transcription factors comprise a large protein family. Proteins of this family contain a highly conserved N-terminal DNA-binding domain and a variable C-terminal domain (Xie et al. 2000; Duval et al. 2002; Ernst et al. 2004; Olsen et al. 2005). NAC was originally derived from the names of three proteins, no apical meristem (NAM), ATAF1-2, and CUC2 (cup-shaped cotyledon), that contain a similar DNA-binding domain (Souer et al. 1996; Aida et al. 1997). NAC proteins appear to be widespread in land plants but no homolog has been identified thus far in other eukaryotes (Riechmann et al. 2000). The early reported NAC transcription factors are implicated in various aspects of plant development. A few examples are NAM from Petunia (Souer et al. 1996) and CUC1-2 (Aida et al. 1997) from Arabidopsis which have roles in controlling the formation of boundary cells of the meristem; NAP (Sablowski and Meyerowitz 1998) from Arabidopsis which acts as a target gene of AP3/PI and functions in the transition between cell division and cell expansion in stamens and petals; and AtNAC1 which mediates auxin signaling to promote lateral root development (Xie et al. 2000). Recently, a few NAC transcription factors were reported to play an essential role in regulating senescence, cell division, and wood formation (Ishida et al. 2000; Takada et al. 2001; Vroemen et al. 2003; Weir et al. 2004; Kubo et al. 2005; Kim et al. 2006; Zhong et al. 2006; Demura and Fukuda 2007; Ko et al. 2007; Mitsuda et al. 2007; Zhong et al. 2007).
NAM, ATAF, and CUC proteins were also found to participate in plant responses to pathogens, viral infections, and environmental stimuli (Xie et al. 1999; Ren et al. 2000; Collinge and Boller 2001; Kim et al. 2007). In Arabidopsis, three NAC genes, ANAC019, ANAC055, and ANAC072, were induced by drought, salinity, and/or low temperature (Tran et al. 2004), and the transgenic Arabidopsis plants overexpressing these genes showed improved stress tolerance compared to the wild type (Tran et al. 2004). Furthermore, proteins of these genes can bind to a cis-element containing CATGTG motif (Tran et al. 2004). Several NAC genes appeared to be hormone-inducible (Hoth et al. 2002; Xie et al. 2002; Greve et al. 2003). An Arabidopsis NAC gene AtNAC2 that can be induced by high salinity, abscisic acid, aminocyclopropane carboxylic acid, and naphthalene acetic acid has been predicted to be a downstream gene in the ethylene and auxin signal pathways (He et al. 2005). Overexpression of AtNAC2 resulted in altered lateral root development and enhanced salt tolerance (He et al. 2005). Recently, a stress-responsive NAC gene, SNAC1, was also characterized in rice (Hu et al. 2006). Overexpression of this gene in rice resulted in significantly increased stomata closure and drought resistance in drought-stressed field conditions while the photosynthesis rate and yield of transgenic plants was not affected under normal growth conditions (Hu et al. 2006). Another stress-responsive NAC gene, OsNAC6, which is a member of ATAF subfamily (Kikuchi et al. 2000; Ooka et al. 2003), has been reported to be induced by abiotic stresses and jasmonic acid treatment (Ohnishi et al. 2005), and overexpression of this gene in rice resulted in increased stress resistance (Nakashima et al. 2007). The tomato gene StNAC, which belongs to the ATAF subfamily, was induced by pathogen attack and wounding (Collinge and Boller 2001). In Brassica napus, nine members of the NAC family (BnNAC) were identified for their differential expression after flea beetle feeding and cold temperature treatment (Hegedus et al. 2003). A NAC gene, NAM-B1, conferring nutrient remobilization from leaves to developing grains was reported in ancestral wild wheat (Uauy et al. 2006), which further exemplified the functional diversity of NAC gene family.
Although quite a few NAC transcription factors have been characterized for their diverse functions, the functions of the majority of members in this family remain unknown. In the first classification of the NAC family, only eight cDNA genes (OsNAC1–OsNAC8) in rice were included (Kikuchi et al. 2000). The second classification of this family was based on 75 cDNAs representing 55 non-redundant genes in rice and 105 putative genes in Arabidopsis (Ooka et al. 2003). With the completion of genome sequencing in rice, more than 100 members of the NAC family have been proposed for the rice genome (Xiong et al. 2005), but the basic information of this family in rice remains to be discovered. In this study, the number of members in the NAC family of rice was reanalyzed and 140 putative NAC or NAC-like genes were identified. In addition, 12 tissue-specific and 20 stress-responsive genes were identified in this family.
Experimental procedures
Mining and bioinformatic analysis of NAC family
Three methods were used to identify all putative NAC genes in rice. First, the keyword “NAC”, “no apical meristem”, or “NAM” was used as a query to search against the annotation database of rice genome (http://www.tigr.org/, release 5). Second, the conserved NAM DNA-binding domains of known NAC proteins were used to search (BLASTP program) against the predicted protein database of rice genome (http://www.tigr.org/, release 5) with a threshold E value less than 1E-5. Third, the HMM profile of the NAM domain in the Pfam database (http://pfam.sanger.ac.uk/) was used to search the annotated rice protein database. All hits with expected values less than 1.0 were collected. The nonredundant sequences resulted from these three methods were then compared with the NAC family in the rice transcription factor database (RiceTFDB, http://ricetfdb.bio.uni-potsdam.de/v2.1/) (Riano-Pachon et al. 2007) and the previously reported annotation of this family (Ooka et al. 2003). All non-redundant putative NAC protein sequences were manually checked for the NAM domain.
Multiple alignments of the full length protein sequences were performed with CLUSTALX (Thompson et al. 1997) and the conserved NAC domains were extracted for constructing phylogenetic tree with the program MrBayes version 3.0 (Ronquist and Huelsenbeck 2003). The HMM profile of the NAM domain was reconstructed with the HMMbuild program in the HMMER package version 2.1 (Eddy 1998). The program multiple EM for motif elicitation (MEME); (Bailey and Elkan 1994) was used to predict the potential motifs. All motifs discovered by MEME with expected values lower than 1E-30 were searched in the InterPro database with InterProScan (Mulder et al. 2005).
To investigate putative cis-acting regulatory DNA elements (cis-elements) in the promoter regions of the tissue-specific or stress-responsive genes, promoter sequences (500 bp regions upstream the 5′ end of the full-length cDNA or predicted CDS) of these genes were extracted from rice genomic sequences and searched against the promoter database PLACE 26.0 (http://www.dna.affrc.go.jp/PLACE/index.html) (Higo et al. 1999). The promoter analysis and calculation of significance were performed as described previously (Nemhauser et al. 2004).
Biochemical assay in yeast
Yeast one-hybrid assay was performed using the Matchmaker one-hybrid system (Clontech, Palo Alto, CA, USA). Two pHIS2-cis reporter vectors, containing the NAC binding sequences from promoters of OsERD1 (Tran et al. 2004) and a putative ornithine aminotransferase gene (LOC_Os03g44150, OsOAT), respectively, were constructed. Activation vectors were constructed by fusing the open reading frames of selected NAC genes with different motif compositions to the GAL4 activation domain in the vector pGADT7-Rec2. Reporter and activation vectors were co-transformed into yeast strain Y187 for verification of the DNA–protein interactions. Primers used for yeast one-hybrid constructs were listed in the Supplemental material online.
Plant growth and stress treatment
To identify tissue-specifically expressed genes, seeds of Minghui 63 were grown under normal conditions. Thirteen tissues representing major tissues or organs of rice in an entire life cycle were collected for real-time PCR analysis.
To verify the expression profiles of stress-responsive rice NAC genes identified from DNA chip expression profiling, seedlings of Minghui 63 were grown on sandy-soil (one-third paddy soil mixed with two-thirds river sand). At the four-leaf stage, the seedlings were treated with salt and cold stresses. For high salinity treatment, sodium chloride (NaCl) was added for a final concentration of about 200 mM. For cold stress, the seedlings were transferred to a growth chamber at 4°C with 12 h light/12 h dark for 5 days and then back to normal growth conditions for recovery. Drought stress was induced by stopping watering at about 2 weeks before flowering and leaves were sampled according to the degree of leaf-rolling.
Real-time PCR analysis
Gene-specific primers (Electronic supplementary material Tables 1 and 2) were designed for NAC genes showing tissue-specific or stress-responsive expression in DNA chip analysis. For real-time PCR analysis, first-strand cDNAs were synthesized from DNaseI-treated total RNA using SuperscripIII reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacture instructions. Real-time PCR was performed in an optical 96-well plate with an ABI PRISM 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). Each reaction contained 12.5 μL 2× SYBR Premix® Ex Taq TM (TAKARA), 0.5 μL 50× ROX reference dye II, 1.0 μL cDNA samples, and 200 nM of gene-specific primer in a final volume of 25 μL. The thermal cycle used is as follows: 95°C for 10 s; 45 cycles of 95°C for 5 s, 60°C for 34 s. Rice Actin1 gene (Accession number X16280) was used as internal control with primers 5′-TGGCATCTCTCAGCACATTCC-3′ and 5′-TGCACAATGGATGGGTCAGA-3′. The relative expression levels were determined as described previously (Livak and Schmittgen 2001).
Results and discussion
Annotation update of NAC family in rice
By keyword (“NAC”, “NAM”) search in the rice annotation database (TIGR, release 5, http://www.tigr.org), a total of 118 gene loci with annotations containing one of the three words were browsed. BLASTP search of the predicted rice protein database with the conserved DNA-binding domain of known NAC proteins resulted in 156 gene loci that included those from keyword search. The predicted protein sequences that resulted from keyword and BLASTP searches were further checked with profile Hidden Molkov Model (pHMM) of the NAM domain to remove false sequences (Eddy 1998). Taken together, a total of 138 gene loci were predicted to encode putative NAC or NAC-like proteins (Table 1). Compared to the annotation of NAC family in RiceTFDB (http://ricetfdb.bio.uni-potsdam.de/v2.1/) in which 123 loci were claimed (Riano-Pachon et al. 2007), the 138 loci identified in this study include all the loci in the RiceTFDB database. Among the 75 cDNA clones of the rice NAC family (ONAC001-075 representing 56 loci) annotated by Ooka et al. (2003), all except two clones are included in the 138 loci. The cDNA sequences of the two exceptional clones, AK068153 and AK105493, cannot be mapped to the genomic sequence of rice, although the predicted protein sequences of the two cDNAs have high similarity (>60% in the NAM DNA-binding domain) to other NAC sequences. To keep the nomenclature of this family consistent, the names of reported genes and those of the previous nomenclature system (i.e., ONAC001-075) are kept in this updated nomenclature of NAC family in rice (named ONAC001-140, Table 1). For the reported genes of this family, the published names are referenced by priority and followed by a systematic name. The systematic names of the redundant cDNA clones in the previous annotation were replaced with newly identified members. All the newly identified members were named according to their sequential locations on the chromosomes.
Generally, the 138 loci of NAC family are evenly distributed among the 12 chromosomes of rice. However, there are eight gene clusters each with two to four genes tandem or closely located in the genome including a cluster containing four genes on chromosome 11 (LOC_Os11g31330, LOC_Os11g31340, LOC_Os11g31360, and LOC_Os11g31380). The sequences of the clustered members are highly similar to each other, suggesting a contribution of gene duplication in the expansion of this family.
Phylogenetic analysis of NAC family
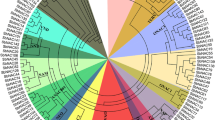
To reveal the evolutionary relationship of NAC genes, all the putative NAC protein sequences of rice were collected for phylogenetic analysis. In addition, seven published NAC proteins from other plants were included as reference sequences. NAC protein sequences. The result suggested that the rice NAC family can be classified into five groups (I–V) and each subfamily has been largely diversified (Fig. 1). Group I has 54 rice NAC members, and can be further classified into five subgroups: I-1 (OsNAC7), I-2 (NAC1), I-3 (NAM/CUC), I-4 (GRAB2), and I-5 (NAC2). All the reported NAC transcription factors related to development were classified into subgroups I-2, I-3, and I-4. Group II also contains a large number of members (54 rice NAC sequences) including four sequences (LOC_Os11g31330, LOC_Os11g31340, LOC_Os11g31360, and LOC_Os11g31380) that are tandem located in chromosome 11 as a gene cluster and can be classified into a few subgroups. However, the tree structure of this group is more complex than that of group I, and group II contains none of the published NAC sequences. Group III, containing 14 ONAC genes, is named stress-related NAC (SNAC) because all the published stress-related genes of NAC family fall in this group. These published genes include SNAC1 (Hu et al. 2006) and OsNAC6 (Nakashima et al. 2007) from rice; ANAC019, ANAC055, and ANAC072 from Arabidopsis (Tran et al. 2004); and StNAC from tomato (Collinge and Boller 2001). TERN (GenBank accession number: AB021178) identified from tobacco (Benson et al. 1997) is also located in this group. Group IV contains 14 NAC members from rice. Group V has two NAC members of rice as well as the reported NAC gene SENU5 identified from tobacco (John et al. 1997). The high sequence diversification of the NAC family suggest that the function of this family has also been diversified, which can be supported by the reported NAC members involved in diverse aspects of plant growth, development, and stress responses as reviewed in the Introduction.
Phylogenetic analysis of NAC protein sequences. Unrooted phylogenetic tree was derived from the program MrBayes version 3.0 (Ronquist and Huelsenbeck 2003) based on the conserved NAC domain of rice and the published NAC genes. The numbers at the branching sites indicated the posterior probability values for nodal support. The names of the reported NAC genes continued to be used in order to keep a consistency with the previous references, while the unpublished NAC genes in rice were designated according to their locus ID from The Institute for Genomic Research (TIGR http://www.tigr.org/)
To further reveal the diversification of NAC family in rice, putative motifs were predicted by the program MEME, and 36 putative motifs were predicted in the NAC family of rice (Table 2). Based on the composition of motifs, the NAC family of rice can be classified into 15 types (A–O, Fig. 2). Most NAC proteins (including 97 rice NAC members in types A–E) contain a complete NAC DNA-binding domain and a diversified C-terminal activation domain. In the previous annotation of this family (Ooka et al. 2003), five motifs were identified in the DNA-binding domain of a typical NAC protein. Our results showed that each of five motifs in the previous report has at least two variants for different subgroups of NAC members (Fig. 2). We noticed that the sequences of type J (three genes), K (6), L (8), M (3), and N (5) do not have all five motifs in the DNA-binding domain, while the sequences of type F (10), G (6), H (6), and I (3) have three to four motifs with locations matching that of the five motifs of the typical NAC DNA-binding domain but the motif sequences are completely different. Therefore, these members may be more properly called NAC-like proteins. In the transcriptional regulation region (TRR), at least ten motifs were identified (Fig. 2). Some motifs are present only in one or two sequences, and for simplicity of classification, these sequences were classified into type E (18 genes), which has a complete DNA-binding domain but a “Variable” TRR, or into type O (11 genes), which contains unclassifiable sequences based on motif composition.
Classification of NAC family in rice based on motif composition. Putative motifs shared among a subset of NAC protein sequences were resulted from the analysis with MEME program (see Materials and methods). Numbered boxes represent different putative motifs (annotations are listed in Table 2). The number of NAC genes belonging to each type was indicated in the bracket after the type. The majority of NAC family proteins have a typical NAC DNA-binding domain containing five kinds of motifs. In the type E, diverse motifs were identified in the activation domain and each was present only in one to two members, thus designated as “Variable” for simplicity. The members in the type O share little structural similarity with other NAC proteins. M motif; TRR transcriptional regulation region
In general, most of the closely related members in the phylogenetic tree have the same or very similar motif composition (Table 1). However, the classification based on motif composition does not completely match the phylogenetic classification. This is not very surprising since the phylogenetic tree is based on the alignments of DNA-binding domain sequences, whereas the classification of motif composition is based on the combination of different motifs. Although the functions of most of the motifs remain to be determined (most of them do not have homologous sequences in the InterPro database, http://www.ebi.ac.uk/interpro/, release 6.1), the motif composition of these unknown NAC (or NAC-like) sequences may provide clues for further function analysis of these genes.
Yeast one-hybrid assay of ONAC proteins
Our previous work showed that SNAC1 and SNAC2 can bind the NAC recognition site (NACRS)-like sequence in the promoter of OsERD1 (Hu et al. 2006; Hu et al. 2008). To determine whether other ONAC proteins with different motif compositions can bind the NAC DNA binding sequence, activation constructs (pGADT7-ONAC) containing the opening reading frames of 12 ONAC genes (representing almost all the motif composition types) the fused to the GAL4 activation domain were co-transformed with pHIS-cis construct (containing the NAC DNA binding sequences in the promoters of OsERD1 gene or OsOAT gene) into the yeast strain Y187. The result showed that all the co-transformants of pGADT7-ONAC and pHIS-cis, but not the negative control, could grow very well on the SD/Leu-/Trp-/His-medium with 30 mM 3-AT (Fig. 3), suggesting that these ONAC proteins with different motif compositions could bind the NAC DNA binding sequence in yeast.
Yeast one-hybrid assay of ONAC proteins with different motif compositions. a The pGAD-ONAC and the pHIS-cis reporter constructs used for co-transformation of yeast strain Y187. b The positive transformants were examined by growth performance on the SD/Leu−/Trp− plate and the SD/Leu−/Trp−/His− plate containing 30 mM 3-AT. P positive control (p53HIS2 plus pGAD-Rec2-53); N negative control (p53HIS2 plus pGAD-ONAC); 1–4 four different colonies of the co-transformants
Identification of tissue-specifically expressed NAC genes in rice
Increasing evidence suggest tissue-specifically expressed transcription factors play critical roles in plant growth and development (Mitsuda et al. 2007; Yoo et al. 2007). Nevertheless, tissue-specific NAC transcription factor has not been reported in rice. A preliminary check of our whole-genome expression profile data of rice for different tissues suggested that 17 genes showed tissue-specific expression (i.e. expression signal was detected only in 1–3 tissues of more than 30 tissues being investigated) (Wang et al., unpublished data). Real-time PCR was then performed to confirm the tissue-specific expression patterns of 12 genes in 13 representative tissues from an entire life cycle of rice. The results suggested that all the 12 genes examined showed tissue-specific expression in only 1–2 tissues (Fig. 4). Among these tissue-specific genes, seven genes (LOC_01g29840, LOC_02g12310, LOC_05g34310, LOC_11g31340, LOC_11g31330, LOC_11g31360 and LOC_11g31380) are specifically expressed in immature endosperm, three genes (LOC_02g15340, LOC_06g33940 and LOC_11g03370) are specifically expressed in callus (including one that is callus and root specific), two genes (LOC_07g37920 and LOC_12g22630) are specifically expressed in stamen. These tissue-specific expressed genes may deserve special notice for further investigation of their function since quite a few members of the NAC family in plants have been proven to play important roles in regulating growth and development.
Real-time PCR of genes showing tissue-specific expression. x-axes are representative tissues and y-axes are scales of relative expression level. Thirteen representative tissues are as follows: Cal callus at 15 days after subculture; Sd1 young shoots (3 days after germination); Sd2 three-leaf seedlings; Lf1 leaf from plants with two tillers; Rt root from plants with two tillers; Lf2 leaf from plants with young panicle 4–5 cm in length; Sh sheath from plants with young panicle 4–5 cm in length; Pan young panicle 4–5 cm in length; Stm stem from plants at 5 days before flowering; Sta stamen at 1 day before flowering; Esp1 endosperm at 7 days after pollination (DAP); Esp2 endosperm at 14 DAP; Esp3 endosperm at 21 DAP. The bars are standard deviations of technical repeats
Expression profiles of the NAC family under different stresses
To obtain an overview of the expression level changes of the rice NAC family under stress conditions, stress/nonstress signal ratios of the ONAC genes were extracted from our microarray data of ‘Minghui 63’ for drought and salt stresses at tillering stage (Zhou et al. 2007). In the leaves of drought-stressed plants at tillering stage, the stress/nonstress signal ratios of 20 genes were higher than 2.0 [with the range of 2.0–21.40 (Electronic supplementary material Figure 1)]. In the leaves of salt-stressed plants at tillering stage, 21 genes showed induction ratios higher than 2.0 (with the range of 2.1–7.5), and 14 of them also showed induction ratios higher than 2.0 under drought stress (Electronic supplementary material Figure 1). Compared to the number of induced genes, only a few genes showed suppressed expression (three and two genes with stress/nonstress signal ratio less than 0.4 under drought and salinity stress, respectively). At whole-genomic level, about 7% genes in the rice genome were induced by drought or salt stresses (Zhou et al. 2007), whereas in the ONAC family, at least 15% (21/140) genes were induced by one of the two stresses. These results suggest a significant portion of the NAC family genes in rice are responsive to abiotic stresses. Interestingly, we noticed that most of the genes in the group III of the phylogenetic tree (12 of 14 rice genes) are responsive to at least one of the stresses, while other stress-responsive genes are random distributed in other phylogenetic groups.
It should be noted that the number of stress-responsive genes might be underestimated with a twofold expression level change because some genes are also significantly induced or suppressed by the stresses based on statistical analysis, even if their expression level changes are less than twofold. Here we still take twofold as a threshold to estimate the stress-responsive genes because more than 95% of the genes with twofold or higher expression changes can be confirmed by real-time PCR analysis. For example, we checked 20 genes with expression level changes higher than twofold in the microarray data by real-time PCR analysis, and the results showed that almost all of them can be confirmed for their stress-responsive expressions (Fig. 5). Among the 20 genes, 5 genes were induced by drought (LOC_Os05g34830, LOC_Os11g03300, LOC_Os01g48130, LOC_Os06g04090, and LOC_Os02g34970), 19 genes (all except LOC_Os06g04090) were induced by salt, and 16 genes (all except four genes LOC_Os12g29330, LOC_Os06g46270, LOC_Os05g34830, and LOC_Os12g41680) were induced by cold. A few genes showed a slight difference in drought induction patterns between real-time PCR and microarray analysis, which may be resulted mainly from different batches of stressed samples used for real-time PCR and microarray analysis (the degree of drought stress in field conditions is difficult to control to the same degree between replications at different times).
Real-time PCR analysis of stress-responsive NAC genes. Stress-inducible expression patterns of the stress-responsive NAC genes are represented. x-axes are time courses of abiotic stress treatments and y-axes are scales of relative expression level. D drought; S salt; C cold. For drought stress, rice leaves were sampled before stress (D0) and after stress at slight leaf-rolling (D1), moderate leaf-rolling (D2), and severe leaf-rolling (D3) stages. For salt and cold stress, seedlings were sampled at 0 (S0), 1 (S1), 6 (S2), and 12 h (S3) and 0 (C0), 1 (C1), 3 (C2), 10 h (C3) after treatment, respectively. The bars are standard deviations of technical repeats
To date, a number of transcription factor genes from different families such as DREB (Liu et al. 1998; Kasuga et al. 1999; Yamaguchi-Shinozaki and Shinozaki 2001; Haake et al. 2002), MYB (Abe et al. 1997), bZIP (Uno et al. 2000), and zinc finger (Mukhopadhyay et al. 2004) have been reported as having an effect on improving stress tolerance. Increasing evidence suggest that some members of the NAC family also contribute to abiotic stress tolerance in Arabidopsis (Fujita et al. 2004; Tran et al. 2004). So far, only two genes of the NAC family in rice have been functionally characterized for their roles in abiotic stress tolerance. One is a stress-responsive gene, SNAC1 (ONAC002), reported in our previous study (Hu et al. 2006). This gene is induced specifically in guard cells by drought stress, and overexpression of this gene in rice can promote stomata closure and significantly improve drought resistance under the field conditions. Another is OsNAC6, which is also induced by various stresses, and transgenic rice plants overexpressing this gene showed significantly improved tolerance of dehydration stresses (Nakashima et al. 2007). In this study, more than 20 genes of NAC family in rice were identified as responsive to drought, salinity, or cold stress. These stress-responsive genes have large diversity both in stress-induced expression patterns and protein structures (based on sequence/motif composition), suggesting that they may participate in the regulation of a wide spectrum of responses to different abiotic stresses. Since rice is one of the most important crops and the model monocot species, functional characterization of these genes has important significance in genetic improvement of rice for abiotic stress tolerance.
In silico cis-element analysis of OsNAC genes
To identify putative cis-acting regulatory DNA elements enriched in OsNAC genes, promoter sequences upstream the 5′ end of the full-length cDNA of OsNAC genes were extracted and subjected to search against the PLACE database. The statistical analysis showed that 17 cis-elements are enriched in rice NAC family genes (Table 3). Among these cis-elements, four elements (ARFAT, PYRIMIDINEBOXHVEPB1, PYRIMIDINEBOXOSRAMY1A, and WBBOXPCWRKY1) are known to be responsive to plant hormones (auxin, GA and SA). Four elements (BOXIINTPATPB, GT1CORE, IBOXCORE, and LTRE1HVBLT49) are responsive to environmental stimulus such as light and cold. Three elements (RYREPEATLEGUMINBOX, RYREPEATVFLEB4, and RYREPEATBNNAPA) are related to seed-specific expression.
Numerous cis-elements have been reported for their essential roles in determining the tissue-specific or stress-induced expression patterns of the genes. In this study, 12 ONAC genes were identified to be expressed in specific tissues or organs (Fig. 4), and 20 genes were confirmed to be induced by different stresses including drought, salt, and cold treatments (Fig. 5). These results promoted us to inspect the relationship of the expression pattern and existence of putative tissue-specific or stress-responsive elements of these genes. Almost all the genes with tissue-specific expression patterns contain at least one of the cis-elements related to the corresponding expression pattern (ESM Table 1). All of the 20 stress-inducible genes contain at least one of the stress responsive cis-elements such as ABRE (ABA-responsive element), DRE (dehydration-responsive element), and LIRE (low-temperature-responsive element) (Yamaguchi-Shinozaki and Shinozaki 1994; Shinwari et al. 1998; Narusaka et al. 2003) (ESM Table 2). In general, the genes containing predicted stress-responsive cis-elements are actually induced by the corresponding stresses. However, this is not always the case. For example, six ONAC genes (LOC_Os05g34830, LOC_Os02g36880, LOC_Os01g48130, LOC_Os02g34970, LOC_Os07g37920, and LOC_Os12g41680) are not induced by cold based on the result of real-time PCR, but they actually contain at least one of the cold-responsive cis-elements in their promoter regions. Such discrepancy might be due to the sequence diversity in the promoter regions between different rice varieties since indica rice Minghui 63 was used for real-time PCR analysis while the promoter sequences analyzed were from japonica rice Nipponbare. This is true at least for three of the six genes (LOC_Os05g34830, LOC_Os02g36880 and LOC_Os01g48130) that are responsive to cold (>twofold) in japonica rice Zhonghua 11 in our previous microarray analysis (Hu et al. 2008). It is remarkable that the expressions of two ONAC genes (Os11g03370 and Os07g37920) are both tissue-specific and stress-responsive. The gene Os11g03370 is expressed specifically in callus and root and induced by salt and cold, while the gene Os07g37920 is stamen-specific and induced by drought and salt. Interestingly, both tissue-specific and stress-responsive cis-elements are presented in their promoter regions (ESM Tables 1 and 2).
In conclusion, this study has provided not only an updated annotation of the NAC family in rice, but also the identification of many tissue-specifically expressed or stress-responsive genes. These data provide a very useful reference as well as a starting point for revealing the function of NAC family genes in rice, especially for those genes involved in the regulation of growth and development at specific stages and stress tolerance.
Abbreviations
- NAC:
-
NAM, ATAF, and CUC transcription factor
- PCR:
-
Polymerase chain reaction
- MEME:
-
Multiple EM for motif elicitation
- Y1H:
-
Yeast one-hybrid
- ERD1:
-
Early responsive to drought 1
- ABA:
-
Abscisic acid
- GA:
-
Gibberellin
- SA:
-
Salicylic acid
References
Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9:1859–1868
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36
Benson DA, Boguski MS, Lipman DJ, Ostell J (1997) GenBank. Nucleic Acids Res 25:1–6
Collinge M, Boller T (2001) Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol 46:521–529
Demura T, Fukuda H (2007) Transcriptional regulation in wood formation. Trends Plant Sci 12:64–70
Duval M, Hsieh TF, Kim SY, Thomas TL (2002) Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol Biol 50:237–248
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
Ernst HA, Olsen AN, Larsen S, Lo Leggio L (2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5:297–303
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876
Greve K, La Cour T, Jensen MK, Poulsen FM, Skriver K (2003) Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem J 371:97–108
Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130:639–648
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115:4891–4900
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Ishida T, Aida M, Takada S, Tasaka M (2000) Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol 41:60–67
John I, Hackett R, Cooper W, Drake R, Farrell A, Grierson D (1997) Cloning and characterization of tomato leaf senescence-related cDNAs. Plant Mol Biol 33:641–651
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291
Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Nagato Y, Matsusoka M, Hirano HY (2000) Molecular analysis of the NAC gene family in rice. Mol Gen Genet 262:1047–1051
Kim SG, Kim SY, Park CM (2007) A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226:647–654
Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM (2006) A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18:3132–3144
Ko JH, Yang SH, Park AH, Lerouxel O, Han KH (2007) ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J 50:1035–1048
Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19:1855–1860
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280
Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101:6309–6314
Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bradley P, Bork P, Bucher P, Cerutti L, Copley R, Courcelle E, Das U, Durbin R, Fleischmann W, Gough J, Haft D, Harte N, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lonsdale D, Lopez R, Letunic I, Madera M, Maslen J, McDowall J, Mitchell A, Nikolskaya AN, Orchard S, Pagni M, Ponting CP, Quevillon E, Selengut J, Sigrist CJ, Silventoinen V, Studholme DJ, Vaughan R, Wu CH (2005) InterPro, progress and status in 2005. Nucleic Acids Res 33:D201–D205
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34:137–148
Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2:E258
Ohnishi T, Sugahara S, Yamada T, Kikuchi K, Yoshiba Y, Hirano HY, Tsutsumi N (2005) OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet Syst 80:135–139
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247
Ren T, Qu F, Morris TJ (2000) HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12:1917–1926
Riano-Pachon DM, Ruzicic S, Dreyer I, Mueller-Roeber B (2007) PlnTFDB: an integrative plant transcription factor database. BMC Bioinform 8:42
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sablowski RW, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92:93–103
Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K (1998) An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun 250:161–170
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85:159–170
Takada S, Hibara K, Ishida T, Tasaka M (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128:1127–1135
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314:1298–1301
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97:11632–11637
Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15:1563–1577
Weir I, Lu J, Cook H, Causier B, Schwarz-Sommer Z, Davies B (2004) CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development 131:915–922
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036
Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419:167–170
Xie Q, Sanz-Burgos AP, Guo H, Garcia JA, Gutierrez C (1999) GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol Biol 39:647–656
Xiong Y, Liu T, Tian C, Sun S, Li J, Chen M (2005) Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol 59:191–203
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6:251–264
Yamaguchi-Shinozaki K, Shinozaki K (2001) Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Novartis Found Symp 236:176–186 discussion 186–179
Yoo SY, Kim Y, Kim SY, Lee JS, Ahn JH (2007) Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLoS ONE 2:e642
Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18:3158–3170
Zhong R, Richardson EA, Ye ZH (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19:2776–2792
Zhou J, Wang X, Jiao Y, Qin Y, Liu X, He K, Chen C, Ma L, Wang J, Xiong L, Zhang Q, Fan L, Deng XW (2007) Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol 63:591–608
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China, the National Program on the Development of Basic Research, the National Program on High Technology Development, and the EU FP6 INCO-MPC2 project CEDROME (INCOCT-2005-015468).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Shirasu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fang, Y., You, J., Xie, K. et al. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280, 547–563 (2008). https://doi.org/10.1007/s00438-008-0386-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-008-0386-6