Abstract
Flavonoids are polyphenols that are important organic chemicals in plants. The health benefits of flavonoids that result in high commercial values make them attractive targets for large-scale production through bioengineering. Strategies such as engineering a flavonoid biosynthetic pathway in microbial hosts provide an alternative way to produce these beneficial compounds. Escherichia coli, Saccharomyces cerevisiae and Streptomyces sp. are among the expression systems used to produce recombinant products, as well as for the production of flavonoid compounds through various bioengineering approaches including clustered regularly interspaced short palindromic repeats (CRISPR)-based genome engineering and genetically encoded biosensors to detect flavonoid biosynthesis. In this study, we review the recent advances in engineering model microbial hosts as being the factory to produce targeted flavonoid compounds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The diversity of chemical compounds found in many organisms is tremendous, and the extent of their benefits towards human well-being are now extensively investigated [1, 2]. Plants produce various polyphenols that have the potential to serve as valuable fine chemicals. One of the major constituents of polyphenols is flavonoids, particularly flavanones that act as a precursor to many flavonoid compounds. There are more than 9000 flavonoid compounds found in plants, comprising one of the largest families of natural products [3]. Tomatoes, limes and green tea leaves are among easily accessible fruits and plants carrying commercially valuable flavonoids such as quercetin, hesperidin and catechins, respectively [1, 4, 5].
Flavonoids are water soluble plant pigments derived from the phenylpropanoid pathway [6]. The production of these secondary metabolites in plants is highly regulated and tissue-specific [7]. Flavonoids are involved in the secondary antioxidant defence system in plant tissues when exposed to various abiotic and biotic stresses [8]. They also act as UV protectants and activators of nodulation in plants [9]. Many flavonoids have medical and commercial values as nutraceuticals [10], and some exhibit pesticide properties [11]. Flavonoids make their way into the human body through the ingestion of plant-based foods [12], and they help prevent chronic diseases such as cardiovascular problems, cancer and neurodegenerative disorders [13]. Recently, seven flavonoid compounds that include luteolin, myricetin, astragalin, rutin, epigallocatechin gallate, epicatechin gallate and gallocatechin gallate were reported to inhibit the NS2B-NS3 protease of the Zika virus [2]. In addition, they are important nutritional compounds found in foods and beverages that offer various health benefits. Some flavonoids such as anthocyanins in prunes can act as antioxidants. Others are beneficial in many physiological and pharmacological industries by virtue of their anticancer, antiviral, anti-obesity and anti-diabetic activities [2, 14,15,16,17]. Flavonoids also act as anti-infection agents and microbial deterrents in plants [18, 19]. Based on the latest report on the market profitability of flavonoids, their total market value can reach up to USD 1.05 billion in 2021 [20]. Due to this high market value, flavonoid compounds are attractive targets for mass production.
Some of the important flavonoid products are extracted from plant species that are difficult to culture, require long growing seasons, and thus increasing production costs. In addition, the total chemical synthesis of various natural products such as flavonoids that are mostly complex compounds can be commercially infeasible [21] and often require extreme reaction conditions [7]. Another challenge faced by the extraction of flavonoid compounds is the slow growth rates of plants, which limits their large-scale production [6]. Therefore, an alternative technique is needed to overcome this issue in procuring flavonoids from plant extracts or chemical synthesis, and the proposed solution is to engineer the flavonoid biosynthetic pathway into a microbial host. The shortcomings described should be addressed with careful planning, since flavonoid compounds are expensive in nature. For example, the market price for flavonoid compounds such as naringenin used in analytical experimentation can amount to USD 248/g. In commercially available grapefruit juices, naringenin content can only reach up to 0–12.6 mg/100 mL [22]. Thus, the production of a vast amount of these expensive products could positively influence the business sectors dedicated towards flavonoid production. This review highlights the structural and functional properties of different classes of flavonoids as well as their biosynthetic pathways in plants. Alternative strategies to produce flavonoids using microbial hosts such as E. coli, S. cerevisiae and Streptomyces sp. through metabolic engineering, newly emerging CRISPR technology and genetically encoded-biosensors capable of detecting these flavonoids (that are currently available only in E. coli and S. cerevisiae) are also discussed.
Classes of flavonoids and their importance
All flavonoids possess a C-15 carbon skeleton with a C6–C3–C6 carbon framework. Flavonoids can be classified into several groups including flavones, flavonols, flavanones, flavanonols, flavan-3-ols, chalcones, anthocyanidins and isoflavones (Table 1). Basic flavonoid compounds are delineated by the position of the linkage between the B-ring and the C-ring (benzopyrano moiety) [12]. Flavones, a class of flavonoids, are converted from flavanones by flavone synthase (FNS) through the introduction of a single double bond between the second and third carbon atom [23]. These compounds are mostly found in various parts of the plants, such as in the stems, leaves, and roots, and are also responsible for interactions with other organisms, including microorganisms, other plants and insects [3].
Flavonols are the most abundant flavonoids in foods [8] and are present in fruits, vegetables, and teas [24]. The dietary intake of flavonols can lead to positive impacts such as the elevated activity of erythrocyte superoxide dismutase (an enzyme with antioxidant activity present in red blood cells), a reduction in lymphocyte DNA damage, a decrease in 8-hydroxy-2′-deoxyguanosine, a marker of oxidative damage [25], and higher antioxidant capacity of plasma with the capability to scavenge free radicals [25, 26].
Flavanones are present in various fruits at high amounts, such as 98% in grapefruit, 96% in limes and 90% in lemons [5]. They have a saturated heterocyclic C ring, which has no conjugation between both rings A and B [8, 27].Thus, they often act as precursors to other classes of flavonoids and are referred to as the focal point in the biosynthesis of flavonoids [3, 4, 28]. Flavanonols such as taxifolin are types of flavonoids that lack the conjugation provided by the 2,3-double bond with the 4-oxo group, making them weak antioxidants [29]. A functional flavanone, naringenin, is one of the intermediate precursors in plant flavonoid pathway. This compound serves as a key branch point in the synthesis of major classes of flavonoids, which include anthocyanins, flavanones and flavonols [30].
A group of flavonoids comprised of monomers is called flavan-3-ols. This class of flavonoids comprises basic units of oligomers and polymers known as proanthocyanidins or condensed tannins [24]. Flavan-3-ols and closely related flavan-3,4-diol analogues can be synthesized by the reduction transformation of dihydroflavonols [31]. Catechin, a flavan-3-ol, present in green tea, was observed to help in the reduction of low-density lipoprotein (LDL), since this lipoprotein is a leading factor for higher risk of heart attacks [1].
Chalcones are a group of flavonoids that are characterized by the presence of two phenolic groups that are connected by an open three carbon bridge [3]. Plants, mostly do not accumulate chalcones as they are quickly isomerized into other compounds, as seen by the isomerization of naringenin chalcone by chalcone isomerase (CHI) into naringenin [4]. The antibacterial activities against methicillin-resistant Staphylococcus aureus [32], anticancer effects against oral carcinogenesis [33] and antioxidant properties [34] of chalcones, proved the potential of this flavonoid group that can be further be exploited for health and economic benefits.
Anthocyanidins and their glycosylated counterparts, anthocyanins, are derived from flavonols and have the basic structure of flavylium ion that is deprived of a ketone oxygen at the 4th-position [35]. Pollinating insects are usually attracted to plants with vibrant colors and here anthocyanidins may play a role [29]. In addition, the use of anthocyanins extracted from plants as natural food colorants are widely practiced [35]. Meanwhile, health benefits of anthocyanidins and anthocyanins are due to their antioxidant, anti-obesity, anti-inflammatory and anti-carcinogenic properties [36]. Among members of this group are cyanidin, delphinidin, malvidin and pelargonidin [17] and are mostly found in fruits such as berries [35], vegetables, olive oil, cocoa and cereals [37].
Side branches of the flavonoid pathway led to the formation of various flavonoids, such as isoflavones, which are also intermediates in anthocyanin formation [4]. Isoflavone synthase (IFS) is responsible in the synthesis of isoflavone and legumes particularly, have unique enzymatic function that is involved in the 2,3 migration of B-ring present in liquiritigenin and naringenin, yielding daidzein and genistein respectively [38]. They are mostly found in leguminous plants [17], with soy being the primary source of isoflavones. Based on several studies conducted, isoflavones have potentials in reducing the risk of breast [39] and prostate cancers [40].
Flavonoid biosynthesis pathway
Secondary metabolites in plants, such as flavonoids, are compounds that are not vital for the immediate survival of plants but provide evolutionary advantages for plant survival and reproduction [21]. Flavonoids are synthesized through the phenylpropanoid pathway. Two of the primary precursors involved in the flavonoid biosynthesis pathway are phenylalanine and tyrosine, which are produced through the shikimate and arogenate pathways [12]. Tyrosine ammonia-lyase (TAL) and phenylalanine ammonia-lyase (PAL) are responsible for converting tyrosine and phenylalanine, respectively, into flavonoid intermediates. However, the pathway of phenylalanine conversion is longer than the pathway that used tyrosine as an initial precursor. Phenylalanine is deaminated to produce cinnamic acid by PAL, and the subsequent cinnamic acid production is oxidised by cinnamate 4-hydroxylase (C4H) to p-coumaric acid. In comparison, TAL bypasses the C4H intermediate and directly proceeds to the formation of p-coumaroyl-CoA. The actions of respective enzymes on phenylalanine and tyrosine resulted in the formation of p-coumaroyl-CoA through the action of 4-coumarate:CoA-ligase (4CL). The addition of 3 units of malonyl-CoA during the conversion of p-coumaroyl-CoA by chalcone synthase (CHS) leads to the production of naringenin chalcone, and the conversion of naringenin chalcone to naringenin can occur through the enzymatic action of chalcone isomerase (CHI) or non-enzymatically under alkaline conditions [41, 42]. The flavanone compound produced is used as a universal flavonoid precursor. Figure 1 includes the basic phenylpopanoid pathway for flavonoid synthesis.
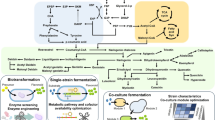
Production of flavonoid in microbial hosts using bioengineering strategies. The strategies for increasing flavonoid production involve overexpression of key flavonoid biosynthesis genes, gene knockdown via CRISPR interference for eliminating of competing enzymes and biosensor-based high-throughput selection of highly productive strains. Recent efforts also focus on using non-natural substrates such as xylose and methanol for flavonoid production. Main enzymes involved in flavonoid biosynthesis are highlighted in blue box. The competing pathway encoded by the listed genes (highlighted in white box) is targeted via CRISPR-dCas9-mediated gene repression. Fluorescent protein-based biosensor approach for increasing flavonoid production via genome engineering is represented by the FdeR transcriptional activator and green fluorescent protein (GFP). Thick arrow indicates increased activity or productivity. Dotted lines indicate multiple enzymatic steps, solid lines represent direct catalytic reaction and dotted line denotes gene repression. Grey color indicates non-natural substrates, yellow color represents native microbial host compounds and green color denotes plant-originated compounds. dCas9 (dead CRISPR-associated protein 9), gRNA (guide RNA), PEP (phosphenolpyruvate), ACC (acetyl-CoA carboxylase), PAL (phenylalanine ammonia-lyase), C4H (trans-cinnamate 4-monooxygenase), TAL (tyrosine ammonia-lyase), 4CL (4-coumarate-CoA ligase), CHS (chalcone synthase), CHI (chalcone isomerase), sucA (2-oxoglutarate dehydrogenase), sucB (dihydrolipoamide acetyltransferase), sucCD (succinyl-CoA synthetase), fabB (3-oxoacyl-acyl carrier protein synthase I), fabf (3-oxoacyl-acyl carrier protein synthase II), fumC (fumarate hydratase), adhE (acetaldehyde dehydrogenase), scpC (propionyl-CoA:succinyl-CoA transferase), eno (phosphopyruvate hydratase). (Color figure online)
Engineering flavonoid production in microbes
Extraction of flavonoid from its native producing plant becomes impractical when large amounts are required due to inherently slow growth of plants, complicated extraction methods and often low natural abundance of high-value flavonoids. Using well-studied and genetically tractable organisms like Escherichia coli and Saccharomyces cerevisiae, we are now able to design and implement heterologous pathways leading to flavonoid compounds in a more economical and sustainable way [6, 43,44,45,46,47,48]. These microbial hosts are unable to produce flavonoids due to the lack of phenylpropanoid pathway in their systems [49, 50]. Early attempts of metabolic engineering for flavonoid overproduction involved: (1) enhancing metabolic flux through the phenylpropanoid pathway by overexpressing heterologous rate limiting enzymes such as PAL, CHS, 4CL and TAL [41, 43], (2) optimizing precursor supply through changes in central carbon metabolism [45, 49] and (3) precursor feeding to the engineered hosts [46, 48].
Various strategies have been employed to engineer flavonoid biosynthetic pathways into these systems as mentioned peviously, and recently, increasing carbon flux towards flavonoid production in microbial hosts using the CRISPR interference system [51,52,53], seemed to be the preferred option. The use of genetically encoded biosensor is gaining traction due to their apparent ability in detecting flavonoid compounds from metabolic engineering [54, 55]. A scheme of bioengineering strategies that have been employed for increasing flavonoid production in engineered microbial hosts is depicted in Fig. 1. Various strategies and techniques employed for the engineering of flavonoid biosynthesis pathways in E. coli, S. cerevisiae and Streptomyces sp. systems are summarized in Table 2.
Engineering flavonoid production in Escherichia coli
There have been various studies conducted to produce flavonoids in engineered microbial hosts. Recent discoveries in the microbial biotechnology field have found that expression of either partial or whole metabolic pathways has allowed for the biosynthesis of valuable end products in E. coli [19, 52, 56, 57]. E. coli often the host of choice due to several advantages: (1) it is a widely-recognized and well-studied biological workhorse, (2) requires easily available rich medium, (3) short doubling-time, hence reducing time strain for product extraction. Flavonoid production in engineered E. coli BL21(DE3) using genes involved in the phenylpropanoid pathways in a bacterial host was first investigated 16 years ago by Hwang et al. [43]. The early strategies to produce flavonoid compounds such as pinocembrin and naringenin in E. coli involved the overexpression of three enzymes; PAL, 4CL and CHS [43, 44] in E. coli. The three genes were strategically cloned by placing T7 promoter and ribosome-binding sequences upstream of each cloned genes and this led to the production of pinocembrin and naringenin. However, the titres were observed to be low (0.75 and 0.45 mg/L of pinocembrin and naringenin, respectively) [43].
Flavonoid biosynthesis in microbial hosts require the heterologous expression of various phenypropanoid enzymes such as PAL, C4H, 4CL, CHS and CHI. However, C4H, a P450 cytochrome monooxygenase responsible for hydroxylating trans-cinnamic acid into coumaric acid is nonfunctional in E. coli due to its instability and lack of specific cytochrome P450 reductase [41, 44]. Hence, simultaneous expression of heterologous 4CL and CHS with TAL (which substituted for PAL and C4H) in independent plasmids was utilized, and naringenin at titer of 20.8 mg/L was successfully achieved from this shortened naringenin biosynthetic pathway, 250 times higher than the yield observed in Hwang et al. [43] when tyrosine is not supplemented in the medium [44].
In the efforts to further increase the flavonoid biosynthesis in E. coli, malonyl-CoA (precursor) pool was improved through co-expression of two sub-unit genes of acetyl CoA carboxylase, dtsR1 and accBC in addition to overexpression of PAL, cinnamate/coumarate:CoA ligase (ScCCL), CHS and CHI. The engineered E. coli produced about 3–4 times higher naringenin and pinocembrin titers when compared to the strain that only express these flavonoid biosynthetic enzymes without acetyl-CoA carboxylase [45]. The production of chalcone intermediates was also attributed to the affinity of PAL towards phenylalanine and tyrosine, and also due to the activity ScCCL as these enzymes helped to overcome the need of C4H expression in E. coli. The same strategy was then combined with newly constructed plasmid containing FNS 1, F3H and FLS which encode for flavone synthase, flavanone 3β-hydroxylase and flavonol synthase, respectively [58]. This led to the successful production of apigenin, chrysin, kaempferol and galangin. Meanwhile, various catechin production at low amounts (from 0.01 to 0.36 mg/L) were observed when a cluster of genes encoding; flavanone 3-hydroxylase (F3H), dihydroflavonol reductase (DFR), and leucoanthocyanidin reductase (LCR) from Camilla sinensis were cloned into E. coli and cultivated in the presence of eriodictyol [59].
Based on the above-mentioned discoveries, findings worth highlighting are the production of natural form of 2S-flavanones and increased flavonoid production through incorporation of CHI enzyme in the recombinant host and increment of the malonyl-CoA pool through overexpression of acetyl-CoA carboxylase, respectively [45]. Natural form of (2S)-flavanones are crucial for biosynthesis of various flavonoids, hence addition of CHI into the artificial gene cluster led to production of (2S)-naringenin and (2S)-pinocembrin [45]. Strategies implemented by Miyahisa et al. [45] showed the highest targeted flavonoid production in recombinant E. coli at 57–58 mg/L of flavanone compounds produced.
Engineering flavonoid production in Saccharomyces cerevisiae
Saccharomyces cerevisiae is an attractive microbial host for production of flavonoid owing to the facts that this organism is able to perform posttranslational modifications of eukaryotic proteins that will result in a better expression of plant proteins [6] and, unlike E. coli, yeast can readily express type II P450 hydroxylases, many of which are involved in flavonoid biosynthesis [60]. During the early studies on the ability of recombinant yeast to produce flavonoid compounds, expression of PAL and C4H (the cytochrome P450 enzyme), together with a cytochrome P450 reductase (CPR) were found to be sufficient in channeling carbon flux into the phenylpropanoid pathway [61]. This gave a huge advantage to yeast system over recombinant E. coli strategy. In addition, production of flavonoids was not affected by any undesired modifications to the compounds due to the absence of this pathway in yeasts [49]. However, there is a challenge faced when engineering secondary metabolite pathway using yeast host, as they lack the ability to support multigene (polycistronic) transcriptional units that permit the coordinated expression of many heterologous genes in compact operons when compared to other bacterial hosts [62].
Heterologous expression of PAL, 4CL, and CHS from different sources resulted in 7 mg/L and 0.8 mg/L of naringenin and pinocembrin, respectively [6] (Table 2). This production was almost 10 times more than the yield obtained from flavonoid biosynthesis study in E. coli by Hwang et al. [43]. Higher naringenin and pinocembrin titers (28.3 mg/L and 16.3 mg/L, respectively) were produced when CHS, C4H, Pc4cL-2 and CHI-A were engineered under the control of a species-specific GAL1 promoter and fed with phenylpropanoid precursors such as; cinnamic acid, p-coumaric acid, caffeic acid, and ferulic acid [46].
Engineered yeast strains harboring various combinations of flavonoid biosynthetic genes; PAL and CPR from P. trichocarpa x P. deltoides, C4H, 4CL, IFS, CHS, CHI and F3′H from Glycine max and FLS from Solanum tuberosum were later investigated for the production of various stilbenoids and flavonoids [18]. This study was the first to successfully express all flavonoid biosynthetic genes and to engineer the complete pathway involved for the synthesis of stilbenoid and other flavonoid compounds in S. cerevisiae host. The yield of products such as resveratrol was reported to be lower if compared to previous works when phenylalanine was utilized as the starting biosynthetic precursor. This is due to the substrates fluxes’ that could not achieve the metabolon at constant saturating rates as to warrant maximal activity of enzyme involved in the flavonoid biosynthetic pathway [18]. It was also discovered that when the number of flavonoid biosynthetic genes in engineered microbial host is increased, the performance of heterologous biosynthesis system is reduced. Trantas et al. [18] hypothesized that there is one or more enzymatic steps where the substrate saturation of key enzymes is suboptimal, hence leading to lower yield of end-products than expected.
In another strategy, flavonoid biosynthetic genes from a single organism, A. thaliana were transferred [63], in order to achieve optimal synergistic activity [64]. In addition to that, reduction in phenylethanol formation, which is a byproduct from yeast metabolism (by deletion of decarboxylase-encoding genes), genes’ codon optimization and improvement of precursors supply (phenylalanine and tyrosine) resulted in high-titer of naringenin formation at 400 µM (more than 100 mg/L) [63], the highest titre obtained so far by using recombinant yeast host. A recent report by Eichenberger et al. [65] demonstrated the de novo production of phloretin from S. cerevisiae targeting the formation of dihydrochalcones (DHCs) with various commercial interests such as antioxidants and anti-diabetics. Overexpression of enoyl-CoA-reductase that is involved in the reduction of double bonds from p-coumaroyl-CoA, coupled with expression of PAL, C4H, CPR, 4CL and CHS resulted in the reduction of naringenin level and shifted the focus towards DHCs formation, whereby a yield of 42.7 mg/L phloretin formation was achieved [65].
Co-cultivation strategy of S. cerevisiae and E. coli
Another interesting bioengineering approach involves the use of co-cultivation system as this platform allowed for the expression of complementary and complete biosynthetic pathways carried by individual microbial strains [49]. Co-culturing of engineered S. cerevisiae and E. coli carrying various flavonoid biosythetic genes; PAL, ScCCL, CHS, CHI and IFS, successfully yielded 5.8 ± 0.3 mg/L of genistein in the presence of tyrosine [49]. A co-culture system consisted of a tyrosine-producing E. coli and a naringenin-producing S. cerevisiae from D-xylose was recently developed, where optimization of the medium components, inoculation size and the inoculation ratio of the two microorganisms were focused on to produce a high naringenin titer. The optimized culture produced up to 21.16 ± 0.41 mg/L of naringenin, and this gave an eightfold increment from the monoculture of the engineered yeast strains [57].
Engineering flavonoid production in Streptomyces sp.
Aside from E. coli and S. cerevisiae being used as biological factories, actinomycetes have emerged as a potential host in manufacturing various polyketides [47]. However, Streptomyces venezuelae, a commonly used host for flavonoid production as it is a fast growing actinomycete, ease of genetic manipulation and most importantly possesses abundant supplies of important substrates involved in biosynthesis of polyketides, such as malonyl-CoA [47, 66,67,68]. However, the culture period of Streptomyces venezuelae is considered longer, between 3 and 4 days [66] when compared to the cultivation time of bacterial hosts (approximately 24–48 h), hence making it challenging to utilise these actinomycetes for flavonoid production.
Flavonoid and stilbene productions in Streptomyces sp. were first reported by Park et al. [47] which employed several strategies such as supplementation with initial precursors; 4-coumaric acid or cinnamic acid, placement of individual promoters for each phenylpropanoid biosynthetic gene (ScCCL, CHS, CHI, and STS), as well as codon-optimization of the plant genes [47]. Naringenin, pinocembrin, resveratrol, and pinosylvin at titers of 4.0 mg/L, 6.0 mg/L, 0.4 mg/L and 0.6 mg/L were produced, respectively, from engineered S. venezuelae DHS2001, a pikromycin polyketide synthase devoid strain. This strain could hinder the structural modifications of post-PKS modifying enzymes such as PikC hydroxylase. The engineered strain was later utilized for heterologous expression of codon-optimized FNSI, F3H and FLS to produce various flavones and flavonols such as apigenin, chrysin, kaempferol and galangin [69].
Production of flavonoids in Streptomyces sp. was further enhanced by the integration of matB and matC genes encoding for malonyl-CoA synthetase and the putative dicarboxylate carrier protein, respectively (from S. coelicolor) into the chromosomal DNA of S. venezuelae mutant DHS2001 and with exogenous supplementation of malonate precursor [70]. This led to the production of naringenin and pinocembrin at 35.6 mg/L and 44.1 mg/L which showed a 7- and 6-fold increase from the engineered strain lacking matB and matC. Meanwhile, apigenin and chrysin production seen an increment of up to ninefold, to 15.3 mg/L and 30.9 mg/L in the mutant carrying the additional FNS gene specific for flavone synthesis [70]. Marin et al. [48] recently reported on the production of apigenin, luteolin and eriodictyol in engineered actinomycete S. albus, where various aspects were manipulated such as spore conditioning and precursor feeding, in efforts to increase the amount of targeted product formations. However, small yields of 0.384 mg/L of apigenin, 0.002 mg/L of eriodictyol and 0.09 mg/L of luteolin were detected [48].
Genome engineering using CRISPR-Cas9 system for flavonoid production in microbial hosts
CRISPR/Cas9 system is a recently discovered and preferred tool for metabolic engineering due to its apparent ability to speed up cell factory construction and to act as a target specific synthetic transcriptional regulator [53, 71]. The implementation of CRISPR/Cas9 tools is often initiated in E. coli as genome modification that includes gene deletion and insertion often reached efficiency of nearly 100% in this model organism [72]. Recent approaches towards flavonoid bioengineering using E. coli as a platform, are making use of this (CRISPR)-based genome engineering strategies.
Catalytically-inactive or dead CRISPR-associated protein 9 (dCas9) has been utilized for transcriptional repression of targeted genes via user-defined guide RNA-directed binding at the targeted promoter region that resulted in gene knockdown effect due to the transcription interference. This approach termed as CRISPRi, has been exploited in acquiring improved flavonoid production via genome-wide repression of gene in competing pathways [51, 52]. The use of CRISPR-Cas9 system in genome editing was first explored by Jiang et al. [73], where almost 65% of recombinant E. coli recovered, carried introduced mutations. This finding paved way for a newer and a more efficient DNA recombineering technology using E. coli for various biotechnological purposes.
By developing the CRISPathBrick system, the repression of competing genes such as sucABCD, fumC and scpC in E. coli led to a 2.5-fold improvement in the naringenin concentration (18.9 mg/L) compared to a non-targeted control strain [51]. Another CRISPR-dCas9 interference system was developed to increase the supply of malonyl-CoA, an important precursor in the flavonoid production. The multiple targeted repression of the fabB, fabF, fumC, sucC and adhE genes increased the naringenin titer up to 7.4-fold (421.6 mg/L) without affecting the growth of the engineered bacterial strains [52]. Repression of adhE and eno on top of the abovementioned genes increased pinocembrin titer to 525.8 mg/L [56] (a 20-fold increase).
Production of O-methylated anthocyanin (peonidin 3-O-glucoside or P3G) was demonstrated through CRISPR interference in E. coli [74]. Silencing of metJ, a ligand-responsive transcriptional repressor, deregulated methionine and increased accumulation of S-adenosyl-l-methionine (SAM), a precursor important for methylation leading to the targeted P3G product. Eventually, 21-fold increment of product titer of up to 56 mg/L (in scale-up to shake flask condition) was achieved from this CRISPRi strategy, when compared to non-targeting CRISPRi control strain [74]. In another study, overexpression of At2g03770, a sulfotransferase (ST) from A. thaliana and repression of cysH in E. coli BL21(DE3) enhanced the production of naringenin 7-sulfate (derivative of naringenin) at 47.7 mg/L of titer. The repression of cysH inhibited the intracellular 3′-phosphoadenosine-5′-phosphosulfate consumption for sulfur metabolism, hence increasing the pool of sulfate donor in engineered E. coli [75].
Saccharomyces cerevisiae has an excellent homologous recombination capability, hence contributing to its suitability as a recombinat host in genetic engineering [76], particularly by using CRISPR-Cas9 system as a tool [77]. A modular and tunable approach for flavonoid biosynthesis was also demonstrated through the development of a CRISPR-Cas9-based SWITCH system to control the genetic engineering and pathway control states of naringenin production in engineered yeast [53]. This switchable system enables the engineered yeast strain to fine tune naringenin production based on CasX-mediated recombination events that governed the genetic engineering and regulation states. The development of this switchable metabolic state in the engineered strain further illustrates the advancement and feasibility in the production of targeted product via microbial synthetic biology.
Genetically encoded biosensors for rapid flavonoid detection in E. coli and S. cerevisiae
In recent years, there has been significant growth in the development of genetically encoded devices and systems for the detection of secondary metabolites particularly in microbial engineering. The design of these biosensors are based on several principles, (i) through implementation of heterologous regulator into targeted hosts; (ii) engineering of regulator variants for different inducer specificity; (iii) development of a novel synthetic inducible system by reconstruction of novel transcriptional factors through fusion of unrelated protein modules [78]. The basic idea is to allow for screens and selections by these biosensors that are sensitive towards intracellular metabolite concentration and translates this to gene expression of reporter proteins [79].
The development of transcription factor (TF)-based biosensor for the detection of plant secondary metabolites has aided in progress of high-throughput metabolite and strain screening process using fluorescence-activated cell sorting (FACS) platform. A naringenin-responsive transcriptional activator FdeR from Herbaspirillum seropedicae SmR1 was combined with green fluorescent protein (GFP) to serve as a genetically encoded biosensor for detecting and reporting intracellular naringenin level [80]. By integrating targeted genome-wide mutagenesis with the naringenin-reporting biosensor, naringenin production was increased up to 61 mg/L in engineered E. coli [54].
Jang et al. [81] had successfully developed riboswitches constructed using Systematic Evolution of Ligands by Exponential Enrichment (SELEX). These in vivo naringenin-sensitive artificial riboswitches were capable of activating reporter gene expression (tetA-sGFP) in E. coli. An increase of up to 2.91 fold in fluorescence was successfully detected from the supplementation of 200 mg/L of naringenin into the growth media [81]. Meanwhile, Xiu et al. [82] also applied the RNA riboswitch-based biosensor module in E. coli co-cultures that is responsive to naringenin. High correlation was seen between the specific fluorescence of the biosensor strain and titer of heterologous naringenin produced by the second E. coli in this co-culture system. They have successfully showed that the biosensor strain can lessen the metabolic burden encountered by this co-culture system and permitted module-specific optimization. In addition, library generation, screening and selection of naringenin high-producing E. coli strains were simplified as metabolite production is not affected by biosensor strain removal [82].
On the other hand, novel biosensor approach in S. cerevisiae employing prokaryotic LysR-type transcriptional regulators (LTTRs) was demonstrated to be instrumental in increasing naringenin biocatalysis via high throughput strain screening [55]. By expressing naringenin-responsive FdeR and inclusion of the corresponding LTTR operator sequence in the yeast regulatory components, the coupling of this specially-designed yeast biosensor with FACS screening strategies has increased naringenin production up to 12.25 mg/L. Hence, advances in synthetic biology have generated more powerful and higher throughput techniques that enabled the rapid selection of flavonoid-overproducing engineered microbial strains.
Conclusions
In the near future, the production of flavonoids will not solely depend on plant extracts but can also be manufactured through biotechnological approaches via heterologous pathway integration into targeted hosts, as well as genome editing. This metabolic engineering strategies in combination with growth optimization steps will become an effective way to meet the increasing market demands of various flavonoids for nutraceutical industry. The growth of flavonoid production could contribute substantially to the business sector at the global scale.
References
Kim A, Chiu A, Barone MK, Avino D, Wang F, Coleman CI, Phung OJ (2011) Green tea catechins decrease total and low-density lipoprotein cholesterol: a systematic review and meta-analysis. J Am Diet Assoc 111(11):1720–1729
Lim HJ, Nguyen TTH, Kim NM, Park JS, Jang TS, Kim D (2016) Inhibitory effect of flavonoids against NS2B-NS3 protease of ZIKA virus and their structure activity relationship. Biotech Lett 39(3):415–421
Martens S, Mithöfer A (2005) Flavones and flavone synthases. Phytochemistry 66(20):2399–2407
Schijlen EGWM, Ric De Vos CH, Van Tunen AJ, Bovy AG (2004) Modification of flavonoid biosynthesis in crop plants. Phytochemistry 65(19):2631–2648
Peterson JJ, Beecher GR, Bhagwat SA, Dwyer JT, Gebhardt SE, Haytowitz DB, Holden JM (2006) Flavanones in grapefruit, lemons, and limes: a compilation and review of the data from the analytical literature. J Food Compos Anal 19:S74–S80
Jiang H, Wood KV, Morgan JA (2005) Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Appl Environ Microbiol 71(6):2962–2969
Wang Y, Chen S, Yu O (2011) Metabolic engineering of flavonoids in plants and microorganisms. Appl Microbiol Biotechnol 91:949–956
Kumar S, Pandey AK (2013) Chemistry and biological activities of flavonoids: an overview. Sci World J 2013:16
Simkhada D, Kurumbang NP, Lee HC, Sohng JK (2010) Exploration of glycosylated flavonoids from metabolically engineered E coli. Biotechnol Bioprocess Eng 15(5):754–760
Tapas A, Sakarkar D, Kakde R (2008) Flavonoids as nutraceuticals: a review. Trop J Pharm Res 7(3):1089–1099
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62(8):2465–2483
Pandey RP, Parajuli P, Koffas MAG, Sohng JK (2016) Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology. Biotechnol Adv 34(5):634–662
Williams RJ, Spencer JPE, Rice-Evans C (2004) Flavonoids: antioxidants or signalling molecules? Free Radical Biol Med 36(7):838–849
Ferry RD, Smith A, Malkhandi J, Fyfe DW, deTakats PG, Anderson D et al (1996) Phase I clinical trial of the flavonoid quercetin: pharmacokinetics and evidence for in vivo tyrosine kinase inhibition. Clin Cancer Res 2:659–668
Harmon AW, Patel YM (2003) Naringenin inhibits phosphoinositide 3-kinase activity and glucose uptake in 3T3-L1 adipocytes. Biochem Biophys Res Commun 305(2):229–234
Bucolo C, Leggio GM, Drago F, Salomone S (2012) Eriodictyol prevents early retinal and plasma abnormalities in streptozotocin-induced diabetic rats. Biochem Pharmacol 84(1):88–92
Hossain MK, Dayem AA, Han J, Yin Y, Kim K, Saha SK et al (2016) Molecular mechanisms of the anti-obesity and anti-diabetic properties of flavonoids. Int J Mol Sci 17(4):569
Trantas E, Panopoulos N, Ververidis F (2009) Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng 11(6):355–366
Trantas E, Koffas MAG, Xu P, Ververidis F (2015) When plants produce not enough or at all: metabolic engineering of flavonoids in microbial hosts. Front Plant Sci 6:1–16
Global-Flavonoids-Market-will-reach-USD-104763-million-in-2021-Zion-Market-Research-457514 @ http://www.econotimes.com (n.d.) http://www.econotimes.com/Global-Flavonoids-Market-will-reach-USD-104763-million-in-2021-Zion-Market-Research-457514. Accessed 10 May 2017
Chemler JA, Koffas MAG (2008) Metabolic engineering for plant natural product biosynthesis in microbes. Curr Opin Biotechnol 19(6):597–605. https://doi.org/10.1016/j.copbio.2008.10.011
Zhang J (2007) Flavonoids in grapefruit and commercial grapefruit juices: concentration, distribution, and potential health benefits. Proc Fla State Hortic Soc 120(863):288–294
Bilgimol CJ, Priya S, Satheeshkumar PK (2012) Detection of flavones in plants using a PCR based approach. J Nat Prod Plant Res 2(2):298–301
Thilakarathna SH, Rupasinghe HPV (2013) Flavonoid bioavailability and attempts for bioavailability enhancement. Nutrients 5(9):3367–3387
Heneman K, Zidenberg-Cherr S (2008) Some facts about flavonol. https://nutrition.ucdavis.edu/sites/g/files/dgvnsk426/files/content/infosheets/fact-pro-flavonol.pdf. Accessed 7 Mar 2017
Williamson G, Manach C (2005) Bioavailability and bioefficacy of polyphenols in humans. II. Review of 93 intervention studies. Am J Clin Nutr 81:243S–255S
Rice-Evans CA, Miller NJ, Paganga G (1996) Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radical Biol Med 20(7):933–956
Khan MK, Huma ZE, Dangles O (2014) A comprehensive review on flavanones, the major citrus polyphenols. J Food Compos Anal 33(1):85–104
Pietta PG (2000) Flavonoids as antioxidants. J Nat Prod 63(7):1035–1042
Kang J-H, Mcroberts J, Shi F, Moreno JE, Jones AD, Howe GA (2014) The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol 164:1161–1174
Marais JPJ, Deavours B, Dixon RA, Ferreira D (2006) The stereochemistry of flavonoids. In: Grotewold E (ed) The science of flavonoids. Springer, New York, pp 1–46
Alcaráz LE, Blanco SE, Puig ON, Tomás F, Ferretti FH (2000) Antibacterial activity of flavonoids against methicillin-resistant Staphylococcus aureus strains. J Theor Biol 205(2):231–240
Makita H, Tanaka T, Fujitsuka H, Tatematsu N, Satoh K, Hara A, Mori H (1996) Chemoprevention of 4-nitroquinoline 1-oxide-induced rat oral carcinogenesis by the dietary flavonoids chalcone, 2-hydroxychalcone, and quercetin. Can Res 56(21):4904–4909
Patil CB, Mahajan SK, Katti SA (2009) Chalcone: a versatile molecule. J Pharm Sci Res 1(3):11–22
Khoo HE, Azlan A, Tang ST, Lim SM (2017) Anthocyanidins and anthocyanins: colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr Res 61(1):1361779
Galvano F, La Fauci L, Vitaglione P, Fogliano V, Vanella L, Felgines C (2007) Bioavailability, antioxidant and biological properties of the natural free-radical scavengers cyanidin and related glycosides. Annali dell’Istituto Superiore di Sanita 43(4):382–393
Lila MA (2004) Anthocyanins and human health: an in vitro investigative approach. J Biomed Biotechnol 2004(5):306–313
Yu O, Jung W, Shi J, Croes RA, Fader GM, McGonigle B, Odell JT (2000) Production of the isoflavones genistein and daidzein in non-legume dicot and monocot tissues. Plant Physiol 124(2):781–794
Yamamoto S, Sobue T, Kobayashi M, Sasaki S, Tsugane S (2003) Soy, isoflavones, and breast cancer risk in Japan. J Natl Cancer Inst 95(12):906–913
Hedlund TE, Johannes WU, Miller GJ (2003) Soy isoflavonoid equol modulates the growth of benign and malignant prostatic epithelial cells in vitro. Prostate 54(1):68–78
Kaneko M, Hwang EI, Ohnishi Y, Horinouchi S (2003) Heterologous production of flavanones in Escherichia coli: potential for combinatorial biosynthesis of flavonoids in bacteria. J Ind Microbiol Biotechnol 30(8):456–461
Mol JNM, Robbinst MP, Dixon RA, Veltkamp E (1985) Spontaneous and enzymic rearrangement of naringenin chalcone to flavanone. Phytochemistry 24(10):2267–2269
Hwang EI, Kaneko M, Ohnishi Y, Horinouchi S (2003) Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl Environ Microbiol 69(5):2699–2706
Watts KT, Lee PC, Schmidt-Dannert C (2004) Exploring recombinant flavonoid biosynthesis in metabolically engineered Escherichia coli. ChemBioChem 5(4):500–507
Miyahisa I, Kaneko M, Funa N, Kawasaki H, Kojima H, Ohnishi Y, Horinouchi S (2005) Efficient production of (2 S)-flavanones by Escherichia coli containing an artificial biosynthetic gene cluster. Appl Microbiol Biotechnol 68(4):498–504
Yan Y, Kohli A, Koffas MAG (2005) Biosynthesis of natural flavanones in Saccharomyces cerevisiae. Appl Environ Microbiol 71(9):5610–5613
Park SR, Yoon JA, Paik JH, Park JW, Jung WS, Ban YH et al (2009) Engineering of plant-specific phenylpropanoids biosynthesis in Streptomyces venezuelae. J Biotechnol 141(3–4):181–188
Marín L, Gutiérrez-del-Río I, Yagüe P, Manteca Á, Villar CJ, Lombó F (2017) De novo biosynthesis of apigenin, luteolin, and eriodictyol in the actinomycete Streptomyces albus and production improvement by feeding and spore conditioning. Front Microbiol 8:1–12
Katsuyama Y, Miyahisa I, Funa N, Horinouchi S (2007) One-pot synthesis of genistein from tyrosine by coincubation of genetically engineered Escherichia coli and Saccharomyces cerevisiae cells. Appl Microbiol Biotechnol 73(5):1143–1149
Katsuyama Y, Funa N, Miyahisa I, Horinouchi S (2007) Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem Biol 14(6):613–621
Cress BF, Toparlak OD, Guleria S, Lebovich M, Stieglitz JT, Englaender JA et al (2015) CRISPathBrick: Modular combinatorial assembly of Type II-A CRISPR arrays for dCas9-mediated multiplex transcriptional repression in E. coli. ACS Synth Biol 4(9):987–1000
Wu J, Du G, Chen J, Zhou J (2015) Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Scientific Reports. https://doi.org/10.1038/srep13477
Vanegas KG, Lehka BJ, Mortensen UH (2017) SWITCH: a dynamic CRISPR tool for genome engineering and metabolic pathway control for cell factory construction in Saccharomyces cerevisiae. Microb Cell Factories 16(1):25
Raman S, Rogers JK, Taylor ND, Church GM (2014) Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci USA 111(50):17803–17808
Skjoedt ML, Snoek T, Kildegaard KR, Arsovska D, Eichenberger M, Goedecke TJ et al (2016) Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. Nat Chem Biol 12(11):951–958
Wu J, Zhang X, Zhou J, Dong M (2016) Efficient biosynthesis of (2 S)-pinocembrin from D-glucose by integrating engineering central metabolic pathways with a pH-shift control strategy. Biores Technol 218:999–1007
Zhang W, Liu H, Li X, Liu D, Dong XT, Li FF et al (2017) Production of naringenin from D-xylose with co-culture of E. coli and S. cerevisiae. Eng Life Sci 17(9):1021–1029
Miyahisa I, Funa N, Ohnishi Y, Martens S, Moriguchi T, Horinouchi S (2006) Combinatorial biosynthesis of flavones and flavonols in Escherichia coli. Appl Microbiol Biotechnol 71(1):53–58
Umar KM, Abdulkarim SM, Radu S, Abdul Hamid A, Saari N (2012) Engineering the production of major catechins by Escherichia coli carrying metabolite genes of Camellia sinensis. Sci World J 2012:1–7
Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450 s in optimized redox environments. Methods Enzymol 272:51–64
Ro D-K, Douglas CJ (2004) Reconstitution of the entry point of plant phenylpropanoid metabolism in yeast (Saccharomyces cerevisiae): implications for control of metabolic flux into the phenylpropanoid pathway. J Biol Chem 279(4):2600–2607
Siddiqui MS, Thodey K, Trenchard I, Smolke CD (2012) Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res 12(2012):144–170
Koopman F, Beekwilder J, Crimi B, van Houwelingen A, Hall RD, Bosch D et al (2012) De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb Cell Factories 11:155
Winkel-Shirley B (1999) Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant 107(1):142–149
Eichenberger M, Lehka BJ, Folly C, Fischer D, Martens S, Simón E, Naesby M (2017) Metabolic engineering of Saccharomyces cerevisiae for de novo production of dihydrochalcones with known antioxidant, antidiabetic, and sweet tasting properties. Metab Eng 39(2017):80–89
Jung WS, Lee SK, Hong JSJ, Park SR, Jeong SJ, Han AR et al (2006) Heterologous expression of tylosin polyketide synthase and production of a hybrid bioactive macrolide in Streptomyces venezuelae. Appl Microbiol Biotechnol 72(4):763–769
Park JW, Jung WS, Park SR, Park BC, Yoon YJ (2008) Analysis of intracellular short organic acid-coenzyme A esters from actinomycetes using liquid chromatography-electrospray ionization-mass spectrometry. J Mass Spectrom 42:1136–1147
Yoon YJ, Beck BJ, Kim BS, Kang HY, Reynolds KA, Sherman DH (2002) Generation of multiple bioactive macrolides by hybrid modular polyketide synthases in Streptomyces venezuelae. Chem Biol 9(2):203–214
Park SR, Paik JH, Ahn MS, Park JW, Yoon YJ (2010) Biosynthesis of plant-specific flavones and flavonols in Streptomyces venezuelae. J Microbiol Biotechnol 20(9):1295–1299
Park SR, Ahn MS, Han AR, Park JW, Yoon YJ (2011) Enhanced flavonoid production in Streptomyces venezuelae via metabolic engineering. J Microbiol Biotechnol 21(11):1143–1146
Bayat H, Omidi M, Rajabibazl M, Sabri S, Rahimpour A (2017) The CRISPR growth spurt: from bench to clinic on versatile small RNAs. J Microbiol Biotechnol 27(2):207–218
Cho S, Shin J, Cho BK (2018) Applications of CRISPR/Cas system to bacterial metabolic engineering. Int J Mol Sci 19(4):1089
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31(3):233–239
Cress BF, Leitz QD, Kim DC, Amore TD, Suzuki JY, Linhardt RJ, Koffas MAG (2017) CRISPRi-mediated metabolic engineering of E coli for O-methylated anthocyanin production. Microb Cell Factories 16(10):1–14
Chu LL, Dhakal D, Shin HJ, Jung HJ, Yamaguchi T, Sohng JK (2018) Metabolic engineering of Escherichia coli for enhanced production of naringenin 7-sulfate and its biological activities. Front Microbiol 9:1–13
Stovicek V, Holkenbrink C, Borodina I (2017) CRISPR/Cas system for yeast genome engineering: advances and applications. FEMS Yeast Res 17(5):1–16
Dicarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41(7):4336–4343
de Frias UA, Pereira GKB, Guazzaroni M-E, Silva-Rocha R (2018) Boosting secondary metabolite production and discovery through the engineering of novel microbial biosensors. Biomed Res Int 2018:1–11
Rogers JK, Taylor ND, Church GM (2016) Biosensor-based engineering of biosynthetic pathways. Curr Opin Biotechnol 42:84–91
Siedler S, Stahlhut SG, Malla S, Maury JÔ, Neves AR (2014) Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab Eng 21:2–8
Jang S, Jang S, Xiu Y, Kang TJ, Lee SH, Koffas MAG, Jung GY (2017) Development of artificial riboswitches for monitoring of naringenin in vivo. ACS Synth Biol 6(11):2077–2085
Xiu Y, Jang S, Jones JA, Zill NA, Linhardt RJ, Yuan Q et al (2017) Naringenin-responsive riboswitch-based fluorescent biosensor module for Escherichia coli co-cultures. Biotechnol Bioeng 114(10):2235–2244
Acknowledgements
The work is supported through the Special Allocation for Agencies Under Academy of Sciences Malaysia-Ministry of Science, Technology and Innovation Grant (PKA0514F004, 6300824) and Putra Graduate Initiative (GP/IPS/2016/9482200), Universiti Putra Malaysia. Fatin Lyana Azman Shah is supported by a scholarship from Jabatan Perkhidmatan Awam (JPA) Malaysia.
Author information
Authors and Affiliations
Contributions
FLAS, SS, ABR designed and wrote the paper. SNB, HHG, NMN, TCL, and SNO contributed ideas and helped in the evaluation and correction of the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shah, F.L.A., Ramzi, A.B., Baharum, S.N. et al. Recent advancement of engineering microbial hosts for the biotechnological production of flavonoids. Mol Biol Rep 46, 6647–6659 (2019). https://doi.org/10.1007/s11033-019-05066-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-05066-1