Abstract
Caragana korshinskii Kom., which is widely distributed in the northwest China and Mongolia, is an important forage bush belonging to the legume family with high economic and ecological value. Strong tolerance ability to various stresses makes C. korshinskii Kom. a valuable species for plant stress research. In this study, suitable reference genes for quantitative real-time reverse transcription PCR (qRT-PCR) were screened from 11 candidate reference genes, including ACT, GAPDH, EF1α, UBQ, TUA, CAP, TUB, TUB3, SKIP1, SKIP5-1 and SKIP5-2. A total of 129 samples under drought, heat, cold, salt, ABA and high pH treatment were profiled, and software such as geNORM, NormFinder and BestKeeper were used for reference gene evaluation and selection. Different suitable reference genes were selected under different stresses. Across all 129 samples, GAPDH, EF1α and SKIP5-1 were found to be the most stable reference genes, and EF1α+SKIP5-1 is the most stable reference gene combination. Conversely, TUA, TUB and SKIP1 were not suitable for using as reference genes owing to their great expression variation under some stress conditions. The relative expression levels of CkWRKY1 were detected using the stable and unstable reference genes and their applicability was confirmed. These results provide some stable reference genes and reference gene combinations for qRT-PCR under different stresses in C. korshinskii Kom. for future research work, and indicate that CkWRKY1 plays essential roles in response to stresses in C. korshinskii.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Caragana korshinskii Kom. is a cultivated leguminous shrub belonging to the legume. It widely distributed in the arid and semi-arid regions of northwest China and Mongolia [39, 40]. It is an important forage grass, industrial feedstock and reforestation species with multiple economic and ecological value [12, 15]. As a very strong stress resistant species, it was reported to tolerate drought, cold, saline and other abiotic stresses [36, 37, 40]. In recent years, several stress-related genes were studied in C. korshinskii Kom. [32–34]. In order to better understand the stress tolerance mechanisms of this species, intensive study of the expression patterns of many stress-related genes were urgently required.
Gene expression analyses can help us to thoroughly investigate the complex biological processes, increasing our understanding of signaling and metabolic pathways that underlie environmental responses and development [11]. In the past, Northern blotting and semi-quantitative reverse transcription-PCR were widely used in gene expression studies, but reverse transcription quantitative real-time PCR (qRT-PCR) with high sensitivity and specificity is being widely accepted more and more recently [5, 29]. The stable expression of reference genes used in qRT-PCR significantly impacts the accuracy of data analysis, but the stable ideal reference gene expressed in all tissues or organs, at different developmental stages, under various physiological states and stress conditions is not available. Therefore, screening for suitable reference gene is critical for normalization of the gene expression data [7, 14, 22]. Many studies on reference gene selection in plants under different condition have been reported, such as Arabidopsis [24], tobacco [26], cotton [2], soybean [35], banana [4], poplar [38], rice [16], wheat [28] and potato [3], etc. To our knowledge, there is no any report about systematic evaluation of reference genes in C. korshinskii Kom. for qRT-PCR so far. Caragana intermedia, another species belonging to the Caragana genus, was reported in 2013, but the candidate reference genes they selected were completely different from those used in our study [43].
A total of 11 candidate reference genes were chosen in our research, including 1 each encoding either the actin (ACT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), elongation factor 1-alpha (EF1α), ubiquitin (UBQ), alpha-tubulin (TUA), cyclase associated protein gene (CAP), or 2 genes encoding the beta-tubulins (TUB and TUB3) and 3 genes encoding the F-box protein (SKIP1, SKIP5-1, and SKIP5-2). The expression stability of these reference genes under different stresses were analysed by softwares geNORM, NormFinder and BestKeeper. After comprehensive analysis, the most stable reference gene for normalization of gene expression under different treatments, such as drought, heat, cold, salt, ABA and high pH were identified. Finally, the expression analysis of CkWRKY1, a gene encoding a member of WRKY transcription factor family, was detected to confirm the reliability of the selected reference genes.
Materials and methods
Plant material and different treatments
The seeds of C. korshinskii Kom. were collected from Chifeng County, Inner Mongolia, China. The seeds were sewn in pots containing a soil mixture (rich soil: vermiculite, 1:1, v/v) and grown at conditions with 22 °C, 16-h light/8-h dark. One- month-old seedlings were used in the following experiments.
For cold and heat treatments, seedlings were placed at 4 or 45 °C respectively for 0.5, 1, 3, 6, 12, 24 and 48 h. For drought, salinity, high pH and ABA treatments, the seedlings were removed from soil slightly, then washed clean with tap water. In drought treatment, the seedlings were placed on filter paper in the initial conditions for 0.5, 1, 3, 6, 12, 24 and 48 h. For salinity, high pH and hormonal treatments, the seedlings were dipped in solution of 200 mM NaCl, pH 10 solution (adjusted by NaOH) and 100 μM ABA for 0.5, 1, 3, 6, 12, 24 and 48 h respectively. The seedlings without any treated were used as control. The shoots of all samples was collected and frozen immediately in liquid nitrogen for RNA extraction. Each of the samples contained three seedlings respectively and every treatment was repeated three times independently unless otherwise stated.
Totally 129 samples were used in our study, including 126 samples treated by drought, heat, cold, salinity, high pH and ABA respectively and 3 samples as controls.
RNA isolation and cDNA synthesis
Total RNA was extracted from all samples with Trizol Regent (Invitrogen, USA), and then treated with RNase-free DNase (Ambion). The purity and integrity of RNA extracted were detected by A260/280 ratios and electrophoresis on 1 % agarose gel. One microgram of total RNA was reverse transcribed into cDNA with the M-MLV reverse transcriptase kit using oligo (dT) 18 primers (TaKaRa) according to the manufacturer’s instructions. The cDNA acquired by reverse transcription was diluted to 16 folds for qRT-PCR.
Primer design and qRT-PCR
The sequences of the candidate reference genes were obtained from the GeneBank, which were submitted by our lab previously. According to these sequences, Primer Premier 5 was used to design the primers for qRT-PCR. The primers followed the rules which describe by Udvardi et al. [29].
Furthermore, the efficiency was calculated based on the slope of the line (E = 10slope), considering an ideal value range (1.8 ≥ E ≤ 2), this experiment was repeated three times [20].
The cDNA was amplified using SYBR Premix ExTaq (TaKaRa), with a Roche LightCycler 480 Real Time PCR system. The thermal cycling program was 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s and 72 °C for 15 s. The melting curves were analyzed at 60–95 °C after 40 cycles. Each RT-qPCR analysis was performed in triplicate.
Stability analysis of reference genes
Stability analysis of reference genes were performed by geNORM [31], NormFinder [1] and BestKeeper [21], which are the most widely used reference gene screening software for qRT-PCR. The CT values were calculated by LightCycler 480 software and the data were imported into geNorm v3.5, NormFinder and BestKeeper analysis tools, which were used as described in their manuals. Considering the three analytic results obtained by geNorm, NormFinder and BestKeeper together, the most stable reference genes under different stresses for qRT-PCR were selected from C. korshinkii Kom..
Expression analysis of CkWRKY1 under different stresses
A WRKY transcription factor encoding gene, CkWRKY1 (GeneBank number JX987095) was previously cloned from C. korshinkii Kom. in our lab. In order to confirm the usefulness of the validated candidate reference genes in qRT-PCR, the transcript level of CkWRKY1 was quantified under different treatments using the most stable reference gene and the most unstable gene we selected. The 2ΔΔCT method [18, 27] was used to evaluate the relative expression level of CkWRKY1 in all samples. The following primers were used in qRT-PCR analysis:
-
F-CkWRKY1-rt: TACAATACAACATACCAAATAGGGGG
R-CkWRKY1-rt: CCTATAGCATGGAAGACTGAGCTGAG
Results
PCR specificity and efficiency analysis of the candidate reference genes
The specificity of the amplification was confirmed by agarose gel electrophoresis (Supplementary Fig. 1, Online Resource) and melting curve analysis (Supplementary Fig. 2, Online Resource).
The GenBank numbers of the 11 candidate reference genes, the primer sequences and the amplicon sequences l are listed in Table 1 and Supplementary Table 1 (Online Resource). As can be seen from Table 1, amplicons length vary from 119 bp (CAP) to 173 bp (TUB3). The efficiency for the 11 reference genes varied from 1.934 ± 0.017 for UBQ to 1.990 ± 0.054 for CAP.
Expression profile of the reference genes
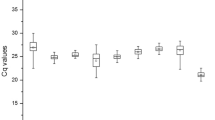
The expression of the reference genes was detected by qRT-PCR in 129 samples. The expression levels of 11 reference genes were analysed across all samples with CT values (Fig. 1). The CT values of the reference genes ranged from 18 to 27 in all studied samples, most of them lying between 19 and 26. Among all the candidate genes, ACT showed the highest expression level (average CT of 19.8), whereas SKIP5-1 showed the lowest expression level (the average CT of 23.8). And EF1α (2.71 cycles), GAPDH (3.12 cycles) and SKIP5-1 (3.09 cycles) displayed the least gene expression variations in all samples, suggesting they might be the suitable reference genes for qRT-PCR. In contrast, TUB (6.57 cycles), CAP (5.53 cycles) and TUA (5.67 cycles) were the most variable ones among the reference genes examined.
Expression stability analysis of the reference genes
Three different algorithms, including geNorm, NormFinder and BestKeeper were used to evaluate the stability of the 11 candidate reference genes.
When analyzed by geNorm, the stability of reference gene was evaluated by the M value [31]. The more stable reference genes have the lower M value, conversely, the more unstable the reference gene, the higher the M value. The results obtained with geNorm algorithm were presented in Fig. 2. Under drought stress, SKIP5-1 and CAP were the most stable genes with the least M value of 0.256, while SKIP1 was the least stable gene with the high M value of 0.748. Under cold, high pH and ABA treatment, TUB and TUB3 were the most stable genes with the M values of 0.155, 0.190 and 0.231 respectively. TUA was the least stable gene with the M values of 0.508, 0.447 and 0.546 respectively. Under heat treatment, the M values of SKIP5-2 and EF1α (0.286) were the lowest, and the M value of TUB (0.990) was the highest. Under salt treatment, the M values of SKIP5-2 and SKIP5-1 (0.161) were the lowest, and the M value of SKIP1 (0.525) was the highest. When all samples were analyzed together, EF1α and SKIP5-1 were the most stable genes with an M value of 0.353. In contrast, TUA was the least stable gene with an M value of 0.826.
The V value was used to determine the optimal number of reference genes required in the analysis by geNorm [31]. A threshold V value of 0.15 was adopted to determine whether the inclusion of an additional reference gene was necessary [17]. For example, if V2/3 was lower than 0.15, this indicated that the combination of the two most stable reference genes was sufficient for target gene normalization under most experimental conditions. On the contrary, if V2/3 was higher than 0.15, the combination of the two most stable reference genes was not enough for target gene normalization and the third reference gene must be added into. In this study, all of V2/3 is less than 0.15 (Fig. 3). According to geNorm, the best combinations among the 11 reference genes were SKIP5-1 and CAP under drought stress (V2/3 = 0.125), TUB and TUB3 under cold (V2/3 = 0.079), high pH (V2/3 = 0.090) and ABA treatment (V2/3 = 0.110), SKIP5-2 and EF1α under heat treatment (V2/3 = 0.098), SKIP5-2 and SKIP5-1 under salt treatment (V2/3 = 0.062). In addition, EF1α and SKIP5-1 was the best combination of reference genes to normalize gene expression data in all samples (V2/3 = 0.115).
The results evaluated by NormFinder were summarized in Table 2. The reference gene has a lower stability values indicating that the gene is more stable in the results obtained by NormFinder [1]. According to NormFinder, the most stable top three genes are CAP, SKIP5-1 and EF1α under drought stress; SKIP5-1, CAP and SKIP5-2 under cold stress; UBQ, SKIP1 and GAPDH under heat stress; EF1α, SKIP5-1 and SKIP5-2 under salt stress; EF1α, CAP and TUB under high pH treatment; EF1α, ACT and SKIP1 under ABA treatment; and EF1α, ACT and GAPDH under all of these stresses. Compared to the results from NormFinder and geNORM, although there are some differences in ranking, but the most stable top five genes almost consistently shared between two results. We also evaluated the data by BestKeeper [21] to confirm the suitability reference genes evaluated by geNORM and NormFinder. The results are summarized in Table 3.
By comprehensive analysis of these three results obtained by geNORM, NormFinder and BestKeeper, the most stable gene and least stable gene were selected. The most stable five genes using these three or two methods are presented in Table 4.
Expression analysis of CkWRKY1 under different treatments
To demonstrate the usefulness of the stable reference genes selected by geNORM, NormFinder and BestKeeper under our conditions, the relative expression level of CkWRKY1 was investigated under different stresses (Fig. 4). The most stable reference gene and gene combination (SKIP5-1 and SKIP5-1+CAP under drought stress, GAPDH and EF1α+SKIP5-2 under heat stress, CAP and TUB+TUB3 under cold stress, EF1α and SKIP5-1+SKIP5-2 under salt stress, EF1α and TUB+TUB3 under high pH, EF1α and TUB+TUB3 under ABA treatment) and the least stable reference gene (SKIP1 under drought stress, TUB under heat stress, CAP under salt stress, TUA under cold, high pH and ABA treatments) were used in this study. The analysis revealed that the expression level of CkWRKY1 increased after drought, heat, cold, high pH and ABA treatment, inversely, the transcript level of CkWRKY1 decreased after heat treatment. When using SKIP5-1 and the combination of SKIP5-1+CAP as reference gene(s) under drought stress, the expression of CkWRKY1 showed similar fluctuation patterns with slight difference. On the contrary, when using SKIP1 as reference gene, the expression of CkWRKY1 showed different change patterns compared to SKIP5-1 and SKIP5-1+CAP as reference gene(s). A similar performance were seen under heat, cold, salt, high pH and ABA treatments (Fig. 4). These results indicate that the stable reference genes selected by our study are reliable for qRT-PCR.
Discussion and conclusion
Although a powerful technology to study gene expression at the transcriptional level, there are several factors that can affect quantification of gene expression by qRT-PCR, such as the quality of the RNA, the efficiency of cDNA synthesis, PCR efficiencies, the methods used for statistical analysis and the reference gene used in normalization [14, 41]. Using different reference gene in qRT-PCR is important to normalization of gene expression analysis. Many research results showed that there was no ideal reference gene stably expressed in all tissues and organs, developmental stages, physiological states and under stress conditions [13]. The stability of the reference gene is relative, different reference gene expression in different conditions is usually not constant, in addition, the reference gene in different species has no absolute versatility [6, 16, 19, 23, 24]. Therefore, the screening of the reference gene for different plants under different conditions is necessary.
The present study selected a set of credible reference genes in C. korshinkii Kom. for the normalization of gene expression analysis using RT-qPCR for the first time. We compared 11 potential candidate reference genes for C. korshinkii Kom. across 129 biological samples under 6 experimental conditions. The PCR amplification efficiency (Table 1) was estimated by a standard curve and it is high enough for qRT-PCR. The data were analyzed by geNorm to select the stable reference gene and confirmed by NormFinder and BestKepper. Although there are some differences between the three results attained by different methods, but the general trend is consistent (Fig. 2; Table 2 and Table 3). Considering the results analyzed by three methods, the most stable reference genes and the least stable genes were summarized in Table 4. Across all the 129 samples under 6 treatments, GAPDH, EF1α and SKIP5-1 are the most stable genes and ranked in top five in three analytical methods, EF1α+SKIP5-1 is the most stable gene combination, SKIP1 and TUB3 are the least stable genes. Similarly, SKIP5-1 and CAP under drought stress, GAPDH1, SKIP1 and SKIP5-1 under heat stress, CAP, ACT, SKIP5-1 and SKIP5-2 under cold stress, EF1α, SKIP5-1 and SKIP5-2 under salt stress, EF1α and ACT under ABA treatment, EF1α under high pH stress are the most stable reference genes. It should be noted that there is no single gene that can be ranked in the top five analyzed by three methods. SKIP5-1+CAP under drought stress, SKIP5-2+EF1α under heat stress, SKIP5-1+SKIP5-2 under salt stress, TUB+TUB3 under cold, ABA and high pH stresses are the most stable gene combinations. SKIP1 and TUB3 under drought stress, TUB under heat stress, CAP and SKIP1 under salt stress, TUA under cold, ABA and high pH stresses are the least stable reference genes.
A CkWRKY1 gene was previously cloned in our lab from a suppression subtractive hybridization library (SSH) of C. korshinskii Kom. under drought stress [39]. In order to illustrate the suitability of the reference genes selected in this study, the relative CkWRKY1 transcriptional expression levels under different stresses using different reference genes for normalization were compared. The research results show that the least stable reference gene will lead to inaccurate analysis results compared with using the most stable gene or gene combinations used. The WRKY proteins are an important superfamily of transcription factors in plants [9]. Many studies revealed that members in this transcription factor family were involved in signaling pathways in plant resistance to both biotic and abiotic stresses [8, 10, 25, 30]. CkWRKY1 did respond to a variety of stresses in our study, suggesting that it may play an essential role in defense mechanisms of C. korshinskii Kom.
The stable reference genes selected in this study will enable more accurate normalization and quantification of gene expression in C. korshinskii Kom. under different stress conditions for qRT-PCR analysis.
Abbreviations
- qRT-PCR:
-
Reverse transcription quantitative real-time PCR
- ACT:
-
Actin
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- EF1α:
-
Elongation factor 1-alpha
- UBQ:
-
Ubiquitin
- TUA:
-
Alpha-tubulin
- CAP:
-
Adenylyl cyclase-associated protein
- TUB:
-
Beta-tubulin
- TUB3:
-
Tubulin beta-3
- SKIP1:
-
SKP1/ASK-interacting protein 1
- SKIP5-1/SKIP5-2:
-
SKP1/ASK-interacting protein 5
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64(15):5245–5250
Artico S, Nardeli SM, Brilhante O, Grossi-de-Sa MF, Alves-Ferreira M (2010) Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol 10(1):49
Castro-Quezada P, Aarrouf J, Claverie M, Favery B, Mugniéry D, Lefebvre V, Caromel B (2013) Identification of reference genes for normalizing RNA expression in potato roots infected with cyst nematodes. Plant Mol Biol Rep. doi:10.1007/s11105-013-0566-3
Chen L, Zhong HY, Kuang JF, Li JG, Lu WJ, Chen JY (2011) Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234(2):377–390. doi:10.1007/s00425-011-1410-3
Chi X, Hu R, Yang Q, Zhang X, Pan L, Chen N, Chen M, Yang Z, Wang T, He Y, Yu S (2012) Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol Genet Genomics 287(2):167–176. doi:10.1007/s00438-011-0665-5
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139(1):5–17
Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37:112–119
Dong J, Chen C, Chen Z (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51(1):21–37
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5(5):199–206
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10(4):366–371
Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131. doi:10.1186/1471-2229-8-131
Fang X, Li J, Xiong Y, Xu D, Fan X, Li F (2007) Responses of Caragana korshinskii Kom. to shoot removal: mechanisms underlying regrowth. Ecol Res 23(5):863–871. doi:10.1007/s11284-007-0449-x
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6(6):609–618
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6(4):279–284
Huijuan G (2012) Spatial distribution of arbuscular mycorrhiza and glomalin in the rhizosphere of Caragana korshinskii Kom. in the Otindag sandy land, China. Afr J Microbiol Res 6(28):5745–5753. doi:10.5897/ajmr11.1560
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345(2):646–651
Li H, Qin Y, Xiao X, Tang C (2011) Screening of valid reference genes for real-time RT-PCR data normalization in Hevea brasiliensis and expression validation of a sucrose transporter gene HbSUT3. Plant Sci 181(2):132–139. doi:10.1016/j.plantsci.2011.04.014
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 ΔΔCT method. Methods 25:402–408
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56(421):2907–2914
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26(6):509–515
Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313(4):856–862
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6(1):27
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227(6):1343–1349
Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ (2011) WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J 10(1):2–11
Schmidt GW, Delaney SK (2010) Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics 283(3):233–241
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3(6):1101–1108. doi:10.1038/nprot.2008.73
Scholtz JJ, Visser B (2012) Reference gene selection for qPCR gene expression analysis of rust-infected wheat. Physiol Mol Plant Pathol 81:22–25
Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20(7):1736–1737. doi:10.1105/tpc.108.061143
Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7(5):491–498
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):research0034
Wang X, Chen X, Liu Y, Gao H, Wang Z, Sun G (2011) CkDREB gene in Caragana korshinskii is involved in the regulation of stress response to multiple abiotic stresses as an AP2/EREBP transcription factor. Mol Biol Rep 38(4):2801–2811. doi:10.1007/s11033-010-0425-3
Wang X, Dong J, Liu Y, Gao H (2010) A novel dehydration-responsive element-binding protein from Caragana korshinskii is involved in the response to multiple abiotic stresses and enhances stress tolerance in transgenic tobacco. Plant Mol Biol Rep 28(4):664–675. doi:10.1007/s11105-010-0196-y
Wang X, Wang Z, Dong J, Wang M, Gao H (2009) Cloning of a 9-cis-epoxycarotenoid dioxygenase gene and the responses of Caragana korshinskii to a variety of abiotic stresses. Genes Genet Syst 84(6):397–405
Wang Y, Yu K, Poysa V, Shi C, Zhou Y (2012) Selection of reference genes for normalization of qRT-PCR analysis of differentially expressed genes in soybean exposed to cadmium. Mol Biol Rep 39(2):1585–1594
Wang Z, Gao H, Wu Y, Han J (2007) Genetic diversity and population structure of Caragana korshinskii revealed by AFLP. Crop Sci 47(4):1737–1743
Wu Y, Hu X, Wang Y (2009) Growth, water relations, and stomatal development of Caragana korshinskii Kom. and Zygophyllum xanthoxylum (Bunge) Maxim. seedlings in response to water deficits. N Z J Agric Res 52(2):185–193
Xu M, Zhang B, Su X, Zhang S, Huang M (2011) Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal Biochem 408(2):337–339
Yang Q, Zhang T, Wang Y, Li G, Yin JJ, Han XM, Qi LW, Li GJ, Wang RG (2013) Construction of a suppression subtractive hybridization library of Caragana korshinskii Under Drought Stress and Cloning of CkWRKY 1 Gene. Scientia Silvae Sinicae 49(7):62–68
Zhang H, Tang M, Chen H, Tian Z, Xue Y, Feng Y (2009) Communities of arbuscular mycorrhizal fungi and bacteria in the rhizosphere of Caragana korshinkii and Hippophae rhamnoides in Zhifanggou watershed. Plant Soil 326(1–2):415–424. doi:10.1007/s11104-009-0022-1
Zheng Y, Xie Z, Gao Y, Jiang L, Shimizu H, Tobe K (2004) Germination responses of Caragana korshinskii Kom. to light, temperature and water stress. Ecol Res 19(5):553–558
Zhong HY, Chen JW, Li CQ, Chen L, Wu JY, Chen JY, Lu WJ, Li JG (2011) Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep 30(4):641–653. doi:10.1007/s00299-010-0992-8
Zhu J, Zhang L, Li W, Han S, Yang W, Qi L (2013) Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PloS One 8(1):e53196
Acknowledgments
We deeply appreciate Dr. Mark Goettel, Editor-in-Chief of Biocontrol Science & Technology, for polishing the manuscript carefully. This work was financially supported by National Natural Science Foundation of China (No. 31360169), Chinese National Programs for High Technology Research and Development (No. 2011AA100203), the Program for New Century Excellent Talents of University (No. ECNT-11-1020) from Ministry of Education (China), and the Innovative Research Group Fund (No. NDPYTD2010-3) from Inner Mongolia Agricultural University (China).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Qi Yang, Jiajia Yin and Gao Li contributed equally to this work.
Key message: The stable reference genes for gene expression evaluation of Caragana korshinskii under drought, heat, cold, salt, ABA and high pH treatments were selected respectively from 11 candidate genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, Q., Yin, J., Li, G. et al. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol Biol Rep 41, 2325–2334 (2014). https://doi.org/10.1007/s11033-014-3086-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3086-9