Abstract
Reverse transcription quantitative real-time PCR (RT-qPCR) is a sensitive technique for quantifying gene expression, but its success depends on the stability of the reference gene(s) used for data normalization. Only a few studies on validation of reference genes have been conducted in fruit trees and none in banana yet. In the present work, 20 candidate reference genes were selected, and their expression stability in 144 banana samples were evaluated and analyzed using two algorithms, geNorm and NormFinder. The samples consisted of eight sample sets collected under different experimental conditions, including various tissues, developmental stages, postharvest ripening, stresses (chilling, high temperature, and pathogen), and hormone treatments. Our results showed that different suitable reference gene(s) or combination of reference genes for normalization should be selected depending on the experimental conditions. The RPS2 and UBQ2 genes were validated as the most suitable reference genes across all tested samples. More importantly, our data further showed that the widely used reference genes, ACT and GAPDH, were not the most suitable reference genes in many banana sample sets. In addition, the expression of MaEBF1, a gene of interest that plays an important role in regulating fruit ripening, under different experimental conditions was used to further confirm the validated reference genes. Taken together, our results provide guidelines for reference gene(s) selection under different experimental conditions and a foundation for more accurate and widespread use of RT-qPCR in banana.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gene expression analysis has become increasingly important in furthering our understanding of the signaling and metabolic pathways which underlie developmental and cellular processes. Northern blotting, semi-quantitative reverse transcription-PCR, and reverse transcription quantitative real-time PCR (RT-qPCR, Bustin et al. 2009) have been frequently used, and RT-qPCR is the best method available for determining changes in gene expression, due to its higher sensitivity, specificity, and broad quantification range of up to seven orders of magnitude (reviewed in Bustin 2002; Wong and Medrano 2005; Artico et al. 2010). Therefore, RT-qPCR has become the preferred method for the validation of high-throughput or microarray results and the quantitation of gene expression (Chuaqui et al. 2002; Czechowski et al. 2005; Die et al. 2010).
Although RT-qPCR is widely used to quantify biologically relevant changes in mRNA levels, the accuracy of RT-qPCR is influenced by a number of variables, including the variability in RNA samples, extraction protocols (particularly due to the co-purification of inhibitors), and efficiencies of the RT and PCR (Nolan et al. 2006; Die et al. 2010; Schmidt and Delaney 2010). Consequently, a normalization step is an essential pre-requisite. A number of strategies have been proposed to normalize RT-qPCR data but normalization remains one of the most important challenges concerning this technique (Huggett et al. 2005). The most commonly applied approach for normalization for RT-qPCR is the use of one or more reference gene(s), which should be expressed at a constant level across various conditions, such as developmental stages or tissue types, and its expression is assumed to be unaffected by experimental parameters (Radonic et al. 2004; Huggett et al. 2005). However, several reports have demonstrated that there are no universally applicable reference genes with an invariant expression, and if the chosen reference gene exhibits a large expression fluctuation, the normalization will lead to inappropriate biological data interpretation (Czechowski et al. 2005; Gutierrez et al. 2008; Bustin et al. 2009; Artico et al. 2010). Thus, there is an urgent need to systematically evaluate the stability of potential reference genes for every particular experimental system prior to their use in RT-qPCR normalization. Meanwhile, several algorithms, such as geNorm (Vandesompele et al. 2002), NormFinder (Andersen et al. 2004), BestKeeper (Pfaffl et al. 2004), and qBasePlus (Hellemans et al. 2007) have been well developed to validate the most stable reference gene(s) from a panel of potential genes or candidate genes under a given set of experimental conditions.
Recognizing the importance of reference gene(s) in normalization of RT-qPCR data, various reference genes have been evaluated for stable expression under specific conditions in various organisms. Nevertheless, most of these studies mainly deal with human or animal tissues, and little attention has been paid to the validation and/or identification of reference genes in the plant sciences (Gutierrez et al. 2008). At the present time, several stable reference gene(s) have been reported under different conditions for Arabidopsis (Czechowski et al. 2005; Remans et al. 2008), tobacco (Schmidt and Delaney 2010), pea (Die et al. 2010), Petunia (Mallona et al. 2010), chicory (Maroufi et al. 2010), berry (Reid et al. 2006), coffee (Cruz et al. 2009), cotton (Tu et al. 2007; Artico et al. 2010), grass species (Hong et al. 2008; Martin et al. 2008; Dombrowski and Martin 2009; Silveira et al. 2009; Lee et al. 2010), peach (Tong et al. 2009), poplar (Brunner et al. 2004), potato (Nicot et al. 2005), rice (Kim et al. 2003; Jain 2009), soybean (Libault et al. 2008; Hu et al. 2009), sugarcane (Iskandar et al. 2004), tomato (Expósito-Rodríguez et al. 2008), wheat (Paolacci et al. 2009), and longan (Lin and Lai 2010). So far, however, suitability of reference genes for RT-qPCR studies has not yet been validated for banana fruit.
Banana is the second ranking fruit crop in the world, with an annual world production of 70 million metric tons (Mbéguié-A-Mbéguié et al. 2009). Banana fruits are the staple food of over 400 million people in the developing countries, not only as a popular dessert fruit, but also as a source of vital carbohydrate. However, postharvest problems, such as rapid ripening, particularly susceptible to biotic and abiotic stresses, are generally observed and account for a high percentage of product loss (Chen et al. 2008; Ma et al. 2009; Mbéguié-A-Mbéguié et al. 2009; Tang et al. 2010). The postharvest biology of banana fruit has been the object of many studies at physicochemical, biochemical, and molecular levels (Inaba et al. 2007; Mbéguié-A-Mbéguié et al. 2008, 2009; Roy et al. 2010; Elitzur et al. 2010; Wang et al. 2010). The understanding of expression patterns of some key genes will help understand the mechanisms involved in these processes in banana fruit and improve fruit quality and storage potential. Furthermore, studies of the molecular events associated with the ripening and stress responses of banana fruit to various exogenous regulators may also help elucidate what leads to postharvest deterioration and loss. At present, almost all RT-qPCR studies in banana fruit use the ACT or GAPDH gene as reference (Inaba et al. 2007; Mbéguié-A-Mbéguié et al. 2008, 2009; Elitzur et al. 2010; Wang et al. 2010), even though the stability of these two reference genes has not been verified yet. Thus, RT-qPCR analyses in banana fruit are still limited by the use of inappropriate references genes.
In the present work, we reported the validation of 20 reference genes, including Actin (ACT1, 2), adenine phosphoribosyltransferase (APT), clathrin adaptor complexes medium (CAC), cyclophilin (CYP), DnaJ-like protein (DNAJ), elongation factor 1-alpha (EF1α), eukaryotic initiation factor 5A (EIF5A-2), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), GTP-binding nuclear protein (RAN), ribosomal protein L (RPL4), ribosomal protein S (RPS2, 4), s-adenosyl methionine decarboxylase (SAMDC1, 2), TIP4I-like family protein (TIP4I), alpha-tubulin (TUA), ubiquitin conjugating enzyme (UBCE2), and ubiquitin (UBQ1, 2) to identify the most suitable internal reference gene(s) for normalization of RT-qPCR data obtained in a large set of biological samples representing the different experimental conditions including various tissues, developmental stages, postharvest ripening stages, stresses (chilling, high temperature and pathogen), and hormone treatments of banana fruit. In addition, to illustrate the usefulness of the new reference genes, expression analysis of one gene of interest related to ethylene signal transduction, EIN3-binding F-box protein (MaEBF1) during postharvest banana fruit ripening, was presented. Our data provide a superior set of validated reference genes that are suitable for RT-qPCR analysis in banana fruit under different experimental conditions.
Materials and methods
Plant materials and treatments
Freshly harvested roots, leaves, flowers, and pre-climacteric banana (Musa acuminata AAA Group, cv. Carvendish) fruits were obtained from a commercial plantation near Guangzhou, south-eastern China. Roots, leaves, flowers, peel, and pulp of pre-climacteric banana fruit at the 75–80% plump stage consisted of samples of different tissues. For samples of different development stages, banana fruit were sampled at 3, 6, 8, and 12 weeks after anthesis, and peel and pulp of banana fruit were separated and collected.
For samples of postharvest ripening, stresses, and hormone treatments, pre-climacteric banana fruit at the 75–80% plump stage were harvested. Hands were separated into individual (fruit) fingers. Fruit were selected for freedom from visual defects and for uniformity of weight, shape, and maturity. The selected fruit were first surface sterilized by dipping in a 1% hypochloride solution for 1 min and then immersed in 0.05% Sporgon (with 46% Prochloraz–Mn; Aventis, Valencia, Spain) for 3 min to prevent fungal disease. They were then allowed to air-dry at 25°C for 2 h and treated as follows.
For postharvest ripening samples, the selected banana fruit was treated with 100 μl l−1 ethylene for 18 h and then ripened at 25°C. Postharvest ripening samples were taken at 0, 1, 3, 5, and 7 days after ethylene treatment. Peel and pulp of banana fruit were separated and collected.
For abiotic stress samples, the selected banana fruit were placed into unsealed plastic bags and transferred to 8 and 38°C for chilling and high-temperature stresses, respectively. Samples were taken at 0, 1, 3, 5, and 7 days after treatment and peel of banana fruit collected.
For biotic stress samples, the selected banana fruits were inoculated with 20 μl (105 spores ml−1) of Colletotrichum musae spores in suspension as described by Ma et al. (2009). Fruit were placed into unsealed plastic bags and stored at 25°C. Samples were taken at 0, 1, 5, 9, and 13 days after treatment, and peel of banana fruit collected.
For hormone treatment samples, the selected banana fruit were treated for 30 min in 5 l of distilled water containing 5 mM salicylic acid (SA) and 0.1 mM methyl jasmonate (MeJA), respectively, under a reduced pressure of about 0.1 MPa as described by Chen et al. (2008). Fruit were placed into unsealed plastic bags and stored at 25°C. Samples were taken at 0, 1, 3, 5, and 7 days after treatment, and peel of banana fruit collected.
Data regarding all eight sample sets mentioned above are summarized in Table 1. Samples were frozen in liquid nitrogen immediately after harvesting and finally stored at −80°C until future use.
RNA isolation, quality control, and cDNA synthesis
Frozen tissues were ground in liquid nitrogen using a mortar and a pestle. Total RNA was extracted using the hot borate method of Wan and Wilkins (1994). Potentially contaminating DNA was eliminated by treatment with DNAse I digestion using the RNAse-free kit (Promega, Madison, WI, USA). The RNA concentration and purity were determined using a Biospec-nano spectrophotometer (Shimadzu, Kyoto, Japan). Only RNA samples with 260/280 ratio between 1.9 and 2.1 and 260/230 ratio higher than 2.0 were used for subsequent analyses. The integrity of the RNA samples was also assessed on 2.0% agarose/formaldehyde gel electrophoresis. Two microgram of total RNA was reverse-transcribed using the M-MLV cDNA synthesis kit (Promega) and oligo-dT primers according to the manufacturer’s instructions.
Selection of banana candidate reference genes
Twenty candidate reference genes based on previous reports were selected for investigation to identify the most stably expressed reference gene(s) to be used in RT-qPCR studies. Except for EIF5A-2 (GeneBank number FJ800963.1) and GAPDH (GeneBank number AY821550) obtained from National Center for Biotechnology Information (NCBI, Bethesda, MD, USA), other eighteen reference gene(s), including ACT1, ACT2, APT, CAC, CYP, DNAJ, EF1α, RAN, RPL4, RPS2, RPS4, SAMDC1, SAMDC2, TIP4I, TUA, UBCE2, UBQ1, and UBQ2 were selected from our transcriptome database obtained by high-throughput Solexa/Illumina sequencing platform (Beijing Genomics Institute, Shenzhen, China).
Design and validation of RT-qPCR primers
Primer pairs were designed based on selected sequences to amplify a large portion of the 3′ untranslated region (3′ UTR) of the 20 candidate reference genes using Primer 3 (version 0.4.0) software under default parameters. Designing primers in 3′-UTR could ensure the specific amplification. All primer pairs were custom-ordered from a commercial supplier (Sangon, Guangzhou, China). Before RT-qPCR, each primer pair was tested via standard RT-PCR to check for size specificity of the amplicon by 2.0% agarose gel electrophoresis and ethidium bromide staining. In addition, target amplicons were sequenced to confirm specificity of the PCR products. The primer specificities were further confirmed with the melting-curve after amplification during the following RT-qPCR analysis. No further analysis was performed on any primer that showed non-single product or product in no template control (NTC). A standard curve which was repeated in four dependent plates using a dilution series of the mixed cDNAs from all tested samples as the template (spanning five orders of magnitude) was made to calculate the gene-specific PCR amplification efficiency (E) and correlation coefficient (R 2) for each gene. The primer sequences and amplicon characteristics including Tm, length, amplification efficiency with standard deviation (SD), and correlation coefficient of the 20 candidate reference genes are listed in Table 2.
RT-qPCR conditions
RT-qPCR was carried out in 96-well plates with a ABI 7500 Real-Time PCR System and 7500 System Software (Applied Biosystems, Alameda, CA, USA) using SYBR Green-based PCR assay. Each reaction mix containing 6 μl diluted cDNAs, 10 μl of SYBR Green PCR Master Mix (Invitrogen, Carlsbad, CA, USA), 0.25 μM of each primer and 0.1 μl ROX to a final volume of 20 μl was subjected to the following conditions: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s in 96-well optical reaction plates (Bio-Rad, Foster City, CA, USA). The melting curves were analyzed at 60–95°C after 40 cycles. Each RT-qPCR analysis was performed in triplicate.
Determination of reference gene expression stability
Two statistical algorithms geNorm (Version 3.5) (http://medgen.ugent.be/~jvdesomp/genorm/) and NormFinder (http://www.mdl.dk/publicationsnormfinder.htm) were used to evaluate the stability of the 20 candidate reference genes under different experimental conditions. Expression levels of the tested reference genes were determined by CT values, the number of amplification threshold cycles needed to reach a specific threshold level of detection. The reference gene with the highest relative quantity (the minimum CT value) was set to 1; the other average CT values of each duplicate reaction of sample for other reference genes were converted to relative quantities data accordingly. These log-transformed data were then exported into geNorm and NormFinder, respectively. The geNorm algorithm first calculates an expression stability value (M) for each gene and then the pairwise variation (V) of this gene with the others. All the tested genes are ranked according to their stability in the tested sample sets and the number of reference genes necessary for an optimal normalization is indicated. The NormFinder algorithm identifies the optimal normalization gene among a set of candidate genes. It ranks the set of candidate normalization genes according to the stability of their expression patterns in a given sample set under a given experimental design. The lowest stability value represents the gene which has the most stable expression within the gene set examined.
Normalization of MaEBF1
EBF (EIN3-binding F-box) proteins have been shown to negatively regulate the ethylene signaling pathway via mediating the degradation of EIN3/EIL proteins (Yang et al. 2009; An et al. 2010). MaEBF1 was cloned from banana fruit by our lab (GeneBank number HQ853255) and was used as a target gene to demonstrate the usefulness of the validated candidate reference genes in RT-qPCR. Gene expression levels of MaEBF1 were quantified during postharvest ripening using the one or two most stable reference gene(s) and the most unstable gene with the lowest or highest M values as determined by geNorm in the same RT-qPCR conditions mentioned above. Primer pairs (forward: 5′-CCTCAATAAGCGATTACGGTG-3′ and reverse: 5′-TCAGACTCCCAAGAGATTCC-3′) of MaEBF1 were also verified as reference genes were done.
Results
Verification of amplicons, primers specificity, and PCR amplification efficiency
Agarose gel electrophoresis (Fig. 1) revealed that all primer pairs amplified a specific PCR product with the expected size, except for UBQ2, which showed a very slight non-specific amplification. Specific amplifications were also confirmed by the presence of a single peak in the melting curve obtained after 40 cycles of amplification (a representative trace is shown as Supplemental Fig. S1). Furthermore, sequence analysis of cloned amplicons revealed that all sequenced amplified fragments were identical or nearly identical to the sequences used for primer design, with 1–3 bp mismatched (but the sequences of amino acids were fully identical). The PCR amplification efficiency for the 20 reference genes varied from 91.940% for ACT2 to 106.218% for RAN, and correlation coefficients ranged between 0.982 and 0.999 for RAN and RPS2 or GAPDH, respectively (Table 2).
Expression profile of the reference genes
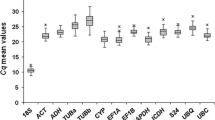
Analysis of the raw expression levels across all samples identified some variation amongst reference genes (Fig. 2). The cycle threshold (CT, Bustin et al. 2009) values of the 20 reference genes studied ranged from 21.46 to 28.41 in all tested samples, while the majority of these values were between 25.31 and 28.41 (Fig. 2). CYP, EIF5A-2, RPS2, and GAPDH showed higher expression levels than other genes in all tested samples, with the CT values ranging from 21.46 to 22.26, while RPS4, RAN, APT, DNAJ, and ACT2 presented lower expression levels, with the CT values ranging from 28.0 to 28.41. In addition, with regard to individual reference gene expression variation, EF1a (7.15 cycles), APT (8.89), UBQ1 (7.4), and UBCE2 (9.30 cycles) showed the higher values, while TIP4I (2.78 cycles), RPS-2 (1.75 cycles), and UBQ2 (1.74 cycles) showed the lower values in all tested samples. These results indicated that none of the selected genes had a constant expression in different banana samples, and it was therefore extremely important to evaluate a suitable reference gene(s) for gene expression normalization under given experimental conditions in banana.
Expression stability of candidate reference genes
Since the 20 candidate reference genes showed wide variations in expression levels in different sample sets, it was necessary to use statistical methods to rank the stabilities of the 20 genes and determine the number of reference genes necessary for accurate gene-expression profiling under the given experimental conditions. The two most widely used algorithms, geNorm and NormFinder, were used in the following analysis:
In geNorm program, the average expression stability (M) value for each gene is calculated based on the average pairwise variation between all genes tested. Stepwise exclusion of the least stable gene allows the genes to be ranked according to their M value (the lower the M value, the higher the gene’s expression stability). The results obtained with geNorm algorithm were presented in Fig. 3 and summarized in Supplemental Table S1. Among the 20 candidate reference genes used for analysis, not all most stable reference genes were identical in the different sample sets (Fig. 3 and Supplemental Table S1). The RAN and RPS2 genes ranked highest in different tissue samples with an M value of 0.220 (Fig. 3a). The CAC and UBQ2 genes proved to be the best candidates for normalization in peel of banana fruit samples at different development stage with an M value of 0.219 (Fig. 3b), while the RAN and RPS2 genes were most stably expressed in pulp of banana fruit samples at different development stage with an M value of 0.132 (Fig. 3c). For peel or pulp of banana fruit samples during postharvest ripening, the most stable genes were CAC and SAMDC1 with an M value of 0.180, and DNAJ and RPS4 with an M value of 0.297, respectively (Fig. 3d, e). The ACT1 and EIF5A-2 genes, or EIF5A2 and GAPDH genes ranked high in banana fruit samples under abiotic or biotic stress conditions, respectively (Fig. 3f, g). For the hormone-treated samples, the most stable genes were UBQ2 and RAN with an M value of 0.135 (Fig. 3h). When all sample sets were analyzed together, the RPS2 and UBQ2 were the two most stably expressed genes with an M value of 0.379 and might be widely used as a single reference gene for multiple samples (Fig. 3i). In contrast, APT was the least stable among the genes examined. These results highlight the fact that it is probably better to choose different reference genes depending on experimental conditions.
Average expression stability values (M) of the candidate reference genes. Average expression stability values (M) of the reference genes were measured during stepwise exclusion of the least stable reference genes. A lower M value indicated more stable expression, as analyzed by the geNorm software in banana sample sets under different experimental conditions, including different tissue (a), developmental stages (peel) (b), developmental stages (pulp) (c), postharvest ripening (peel) (d), postharvest ripening (pulp) (e), abiotic stress (f), biotic stress (g), and hormone treatment (h). The M values calculated for all banana samples examined were also given (i)
The geNorm program was also used to calculate the optimal number of reference genes required for accurate normalization in the different sample sets. The software determines the pairwise variation Vn/n + 1, which measures the effect of adding further reference genes on the normalization factor (that is calculated as the geometric mean of the expression values of the selected reference genes). It is advisable to add additional reference genes to the normalization factor until the added gene has no significant effect. Vandesompele et al. (2002) used 0.15 as a cut-off value, below which the inclusion of an additional reference gene is not required. Pairwise variation analysis (Fig. 4) showed that normalization required the use of three reference genes in all samples since the V2/3 value (0.21) was higher than 0.15. Therefore, it would be ideal to include RPS2, UBQ2, and RAN as reference genes to normalize gene expression data in all samples. While in other sample sets, including different tissues, peel or pulp of banana fruit at developmental stages, peel or pulp of banana fruit during postharvest ripening, abiotic- or biotic- stressed, and hormone-treated samples, only two genes would be sufficient since the V2/3 values in these eight sample sets were inferior to the 0.15 cut-off level. Thus, according to geNorm, the best combinations for these eight sample sets were RAN + RPS2, CAC + UBQ2, RAN + RPS2, CAC + SAMDC1, DNAJ + RPS4, ACT1 + EIF5A-2, EIF5A-2 + GAPDH, RAN + UBQ2, respectively.
Pairwise variation (V) analysis of the candidate reference genes. The pairwise variation (Vn/Vn + 1) was analyzed between the normalization factors NFn and NFn+1 by the geNorm software to determine the optimal number of reference genes required for RT-qPCR data normalization. Arrow indicates the optimal number of genes for normalization in each sample sets
To further confirm the results obtained by the geNorm program, the data were also evaluated with NormFinder algorithm. Different from geNorm, NormFinder takes into account intra- and intergroup variations for normalization factor (NF) calculations. This algorithm ranks the set of candidate reference genes according to the stability of their expression patterns in a given sample set analyzed in a given experimental design. The results of the Normfinder analysis applied to our data sets were summarized in Supplemental Table S2. It was noteworthy that definition of sample subgroups had a notable effect on NormFinder output. However, the NormFinder output with different sample subgroups and no subgroups exhibited almost the same top five stable genes, but with the slight changes in ranking orders. When the outcome of geNorm and NormFinder were compared, only few but relevant, differences were observed (Supplemental Table S1, S2) and the consensus of the results obtained by geNorm and NormFinder analysis were listed in Table 3. In most sample sets, geNorm or NormFinder analysis exhibited almost the same top five stable genes, but with the slight changes in ranking orders, except for banana pulp samples at different developmental stages or during postharvest ripening that showed great differences. In addition, no matter how the order changed, the most unstable gene would almost remain the same in all sample sets.
Reference gene validation
To demonstrate the usefulness of the validated candidate reference genes in RT-qPCR, the relative expression level of one banana fruit gene, MaEBF1 that belongs to EBF (EIN3-binding F-box) family was investigated in the peel or pulp of banana fruit during postharvest ripening, using one or two of most stable reference genes, and the most unstable gene for normalization, which had been validated above by geNorm or NormFinder (Supplemental Table S1, S2 and Table 3; Fig. 3). The analysis revealed that the expression level of MaEBF1 in fruit peel decreased progressively during postharvest ripening and showed similar change patterns with slight difference when using SAMDC1 alone and the combination of CAC + SAMDC1 as reference gene (s) for normalization (Fig. 5a). MaEBF1 expression level in pulp remained constant during postharvest ripening and also showed similar change patterns when using ACT1 alone and the combination of DNAJ + RPS4 as reference gene (s) for normalization (Fig. 5b). In addition, RT-qPCR results for MaEBF1 expressions were confirmed by Northern-blot analysis (data not shown). However, these change patterns were completely obscured during normalization using the least stable reference gene (APT) in peel (Fig. 5a) or EF-1α in pulp (Fig. 5b). These effects were clearly due to low stability of APT or EF-1α expression across these samples. This illustrated the adverse effect of using an unsuitable reference gene for normalization.
Relative quantification of MaEBF1 expression using validated reference genes for normalization under different experimental conditions. a The validated reference gene(s) used as normalization factors were one (CAC) or two (CAC + SAMDC1) most stable reference genes, and the most unstable one (APT) in postharvest ripening (peel) sample sets. b The validated reference gene(s) used as normalization factors were one (ACT1) or two (DNAJ + RPS4) most stable reference genes, and the most unstable one (EF-1α) in postharvest ripening (pulp) sample sets. The expression level in peel or pulp sampled on day 0, which was set to 1. Each value represented the means of three replicates, and vertical bars indicate the standard deviations (SD)
Discussion
RT-qPCR has become a powerful tool for accurate gene expression profiling because of its high throughput, sensitivity, and accuracy (Lee et al. 2010). However, quantification of gene expression is affected by several factors, such as the quantity of the initial material, the quality of the RNA, the efficiency of cDNA synthesis, primer performance, and the methods to be used for statistical analysis (Maroufi et al. 2010). Normalization of the expression level of a target gene against a stably expressed reference gene can compensate for all these kinds of variations (Pfaffl et al. 2004). The ideal reference gene should have similar expression regardless of experimental conditions, including different tissues, developmental stages, and sample treatments. However, no one gene has a stable expression under every experimental condition (Artico et al. 2010). Therefore, it is advisable to validate the expression stability of candidate reference genes under specific experimental conditions prior to their use in RT-qPCR normalization, rather than using reference genes published elsewhere (Remans et al. 2008; Lee et al. 2010). In addition, validation of reference genes has been simplified with the design of statistical algorithms, such as geNorm and NormFinder, which not only test the expression stability of reference genes, but can also determine the number of reference genes required to provide accurate normalization (Vandesompele et al. 2002; Andersen et al. 2004).
ACT, UBQ, GAPDH, EF, CYP, and TUA are commonly used reference genes for gene expression studies in many plant species (Jian et al. 2008; Tong et al. 2009; Hu et al. 2009; Maroufi et al. 2010). However, recent studies indicate that the traditional reference genes are not always stably expressed when tested in other species or in a wider range of experimental treatments (Mukesh et al. 2006; Jian et al. 2008; Expósito-Rodríguez et al. 2008; Hu et al. 2009; Artico et al. 2010). For example, Nicot et al. (2005) demonstrated that ACT did not appear to be the best gene to use as reference gene during the different treatments. In addition, Gutierrez et al. (2008) also found high variability in the relative expression of common reference genes, including ACT, TUB, UBQ, and EF during various developmental stages in Arabidopsis.
This means that even the most stable reference gene(s) reported in a species, the using of them in other species under study or in a new experimental set-up should be validated. Accordingly, for RT-qPCR studies in banana, the stability of reference genes needs to be verified prior to use in RT-qPCR. Almost all current RT-qPCR studies in banana fruit are using ACT or GAPDH as the reference gene (Inaba et al. 2007; Mbéguié-A-Mbéguié et al. 2008, 2009; Wang et al. 2010; Elitzur et al. 2010). The direct transfer of traditional and recently proposed novel candidate reference genes by Czechowski et al. (2005) to non-model plants such as banana is hampered by the limited availability of genomic sequences. We thus selected a series of candidate reference genes for which such sequence information could be obtained from our transcriptome sequencing database for banana fruit. In the present work, we developed a RT-qPCR method for 20 candidate reference genes, including EIF5A-2, GAPDH, RAN, EF1α, APT, RPL4, UBQ1, UBQ2, DNAJ, UBCE2, ACT2, CAC, TIP4I, TUA, SAMDC1, SAMDC2, ACT1, CYP, RPS2 and RPS4, and MaEBF1 as the target gene. The specificity of the RT-qPCR primer pairs was confirmed by agarose gel electrophoresis (Fig. 1), melting curves analysis (Supplemental Fig. S1), and sequencing of the amplicons. The PCR amplification efficiency was estimated, and the reference genes were ranked according to their expression level stability under different experimental conditions using geNorm and NormFinder algorithms. It has been reported that primer- or amplification-specificity must be validated empirically with direct experimental evidence (electrophoresis gel, melting profile, DNA sequencing, amplicon size, and/or restriction enzyme digestion) (Bustin et al. 2009; Derveaux et al. 2010). However, it should be pointed out that most of the reference genes tested in the present study are probably members of large gene families and so obtaining specific primers is difficult. This problem is compounded by the large size and limited sequencing of the banana genome. Other measures to identify specific amplicons could include analyzing the primer annealing regions for SNPs and modeling the secondary structure of the amplicon (using, for example, UNAFold software) (Derveaux et al. 2010). In addition, limiting the number of cycle to 30–35 in RT-qPCR analysis might also improve the specificity of PCR amplification. Regardless of these issues, the results from the stability analyses remain valid for the primer pairs and conditions described.
When all banana samples were tested, RPS2 and UBQ2 were overall the most stable and best candidates for the normalization of general gene expression for banana. But different sets of samples had their own best reference genes (Fig. 3, Supplemental Table S1, S2). For example, RAN and RPS2 genes ranked higher in different tissue samples and banana pulp samples at different developmental stages, whereas CAC and UBQ2 did better than RAN and RPS2 when studying samples harvesting from banana peel at different developmental stages. CAC and SAMDC1, or DNAJ and RPS4 were the best reference genes for banana peel or pulp samples during postharvest ripening, while DNAJ or SAMDC1 appeared to be the least stable reference gene in banana peel samples at different developmental stages or abiotic stress samples. For abiotic or biotic stress samples, the best reference genes were ACT1 and EIF5A2, or EIF5A2 and GAPDH, respectively. Finally, UBQ2 and RAN were considered as the most stable genes for hormone treatment samples. Our analysis indicated that each experimental condition tested demanded a specific set of reference genes. This result emphasized the importance of reference genes validation for each experimental condition, especially when samples belong to very different sets. Similar results were also reported in other studies, including perennial ryegrass (Lee et al. 2010), coffee (Barsalobres-Cavallari et al. 2009), and pea (Die et al. 2010). More importantly, our results showed that ACT and GAPDH, which were commonly used in RT-qPCR analysis in banana at present, were not the most suitable reference genes under most experimental conditions, suggesting that other more suitable reference genes than ACT and GAPDH should be taken into account in RT-qPCR analysis in banana in future studies. However, it should be pointed out that validations of 20 reference genes by the same procedures used in this work not always give support to their frequent use in other plants, as there were many reports suggested that the reference genes were regulated differently in different plant species and might exhibit differential expression patterns (Hu et al. 2009; Artico et al. 2010; Maroufi et al. 2010). For instance, UBQ2 gene showed highly stable expression in banana (present work), peach (Tong et al. 2009), grass (Hong et al. 2008), and Arabidopsis (Czechowski et al. 2005), whereas its putative homologue had been shown unsuitable for normalization in rice (Jain et al. 2006), soybean (Jian et al. 2008), cucumber (Wan et al. 2010), and chicory (Maroufi et al. 2010).
Increasing evidences have shown that a single reference gene cannot be used to accurately normalize RT-qPCR data and that the combination of multiple reference genes is preferred. Although increasing the number of reference genes for normalization will improve the accuracy of the analysis, as mentioned above, this is expensive and time consuming. Therefore, the number of internal controls should be taken into account if the amount of RNA is limited or a large number of samples need to be analyzed (Lin and Lai 2010). It has been suggested that the number of reference genes needed to be employed is dependent on the considerations of a researcher’s purpose (Hu et al. 2009). Use of two stable reference genes is a valid normalization strategy in most experimental conditions, and the present study also suggested that only two genes would be sufficient to get more accurate and reliable normalization compared with the use of a single reference gene in most sample sets, except for consideration all tested samples (Fig. 4). In addition, our results also showed that the choice of the best combination of reference genes depends on the experimental conditions.
Some studies that have utilized both geNorm and NormFinder have reported minor changes in gene stability ranking (Le Bail et al. 2008; Cruz et al. 2009; Lee et al. 2010), while others have observed relatively substantial changes, i.e., up to 15 places between the two methods (Paolacci et al. 2009; Lin and Lai 2010). In the present work, only few but relevant, differences were observed between the two methods (Supplemental Table S1, S2). In addition, no matter how the order changed, the most unstable gene would almost remain the same in all sample sets, which had been also observed in other studies (Expósito-Rodríguez et al. 2008; Lin and Lai 2010; Artico et al. 2010; Wan et al. 2010). These discrepancies between the results were expected since the geNorm and NormFinder are based on distinct statistical algorithms. GeNorm selects two genes with a low intra-group variation and approximately the same non-vanishing inter-group variation. In comparison, NormFinder selects the two best genes with minimal combined inter- and intra-group expression variation (Andersen et al. 2004), which could have a notable effect on the subsequent gene stability ranking (Expósito-Rodríguez et al. 2008). Therefore, both methods could provide a stability value for each gene and select the best reference gene for normalization, and the fact that the ranking of candidate reference genes by NormFinder is not always identical to that defined by geNorm is not surprising.
To illustrate the suitability of the reference genes revealed in the present study, using different reference genes for normalization, the relative MaEBF1 mRNA expression levels in the peel or pulp of banana fruit during postharvest ripening using different reference genes for normalization were compared. The results showed that normalization using the least stable reference gene led to misinterpretation of the MaEBF1 expression level (Fig. 5a, b), more importantly, MaEBF1 expression levels analyzed by Northern blot also showed similar change patterns with RT-qPCR analysis using the most stable reference gene for normalization (data not shown). These results suggest that the selection of stable reference genes represents a crucial issue for the correct normalization of RT-qPCR data.
In summary, to the best of our knowledge, this is the first attempt to validate a set of candidate reference genes in banana for the normalization of gene expression analysis using RT-qPCR. Our results suggested that different suitable reference genes or combination of reference genes for normalization should be selected according to the different experimental conditions. The expression analysis of MaEBF1 emphasized the importance of validating reference genes to achieve accurate RT-qPCR results. These results provide a foundation for the more accurate and widespread use of RT-qPCR in the analysis of gene expression in banana.
Abbreviations
- ACT :
-
Actin
- APT :
-
Adenine phosphoribosyltransferase
- CAC :
-
Clathrin adaptor complexes medium
- CYP :
-
Cyclophilin
- DNAJ :
-
DnaJ-like protein
- EBF :
-
EIN3-binding F-box protein
- EF1α :
-
Elongation factor 1-alpha
- EIF5A :
-
Eukaryotic initiation factor 5A
- GAPDH :
-
Glyceraldehyde-3-phosphate dehydrogenase
- RAN :
-
GTP-binding nuclear protein
- RPL :
-
Ribosomal protein L
- RPS :
-
Ribosomal protein S
- RT-qPCR:
-
Reverse transcription quantitative real-time PCR
- SAMDC :
-
s-Adenosyl methionine decarboxylase
- TIP4I :
-
TIP4I-like family protein
- TUA :
-
Alpha-tubulin
- UBCE :
-
Ubiquitin conjugating enzyme
- UBQ :
-
Ubiquitin
References
An FY, Zhao Q, Ji YS, Li WY, Jiang ZQ, Yu XC, Zhang C, Han Y, He WR, Liu YD, Zhang SQ, Ecker JR, Guo HW (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-Box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22:2384–2401
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Artico S, Nardeli SM, Brilhante O, Grossi-de-Sa MF, Alves-Ferreira M (2010) Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol 10:49
Barsalobres-Cavallari CF, Severino FE, Maluf MP, Maia IG (2009) Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Mol Biol 10:1
Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4:14
Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR RT-PCR: trends and problems. Mol Endocrinol 29:23–29
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Chen JY, He LH, Jiang YM, Wang Y, Joyce DC, Ji ZL, Lu WJ (2008) Role of phenylalanine ammonia-lyase in heat pretreatment-induced chilling tolerance in banana fruit. Physiol Plant 132:318–328
Chuaqui RF, Bonner RF, Best CJM, Gillespie JW, Flaig MJ, Hewitt SM, Phillips JL, Krizman DB, Tangrea MA, Ahram M, Linehan WM, Knezevic V, Emmert-Buck MR (2002) Post-analysis follow-up and validation of microarray experiments. Nat Genet 32:509–514
Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J, Barros L, Romano E, Grossi-de-Sá MF, Vaslin M, Alves-Ferreira M (2009) Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol Breeding 23:607–616
Czechowski TSM, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Derveaux S, Vandesompele J, Hellemans J (2010) How to do successful gene expression analysis using real-time PCR. Methods 50:227–230
Die JV, Román B, Nadal S, González-Verdejo CI (2010) Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 232:145–153
Dombrowski J, Martin R (2009) Evaluation of reference genes for quantitative RT-PCR in Lolium temulentum under abiotic stress. Plant Sci 176:390–396
Elitzur T, Vrebalov J, Giovannoni JJ, Goldschmidt EE, Friedman H (2010) The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. J Exp Bot 61:1523–1535
Expósito-Rodríguez M, Borges A, Borges-Pérez A, Pérez J (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19
Hong SY, Seo PJ, Yang MS, Xiang F, Park CM (2008) Exploring valid reference genes for gene expression studies in Brachypodium distacyon by real-time PCR. BMC Plant Biol 8:112
Hu R, Fan C, Li H, Zhang Q, Fu Y-F (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10:93
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalization, strategies and considerations. Genes Immun 6:279–284
Inaba A, Liu X, Yokotani N, Yamane M, Lu WJ, Nakano R, Kubo Y (2007) Differential feedback regulation of ethylene biosynthesis in pulp and peel tissues of banana fruit. J Exp Bot 58:1047–1057
Iskandar HM, Simpson RS, Casu RE, Bonnet GD, Maclean DJ, Manners JM (2004) Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene in sugarcane. Plant Mol Biol Rep 22:325–337
Jain M (2009) Genome-wide identification of novel internal control genes for normalization of gene expression during various stages of development in rice. Plant Sci 176:702–706
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Jian B, Liu B, Bi Y, Hou W, Wu C, Han T (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59
Kim B, Nam H, Kim S, Chang YJ (2003) Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol Lett 25:1869–1872
Le Bail A, Dittami S, de Franco PO, Rousvoal S, Cock M, Tonon T, Charrier B (2008) Normalisation genes for expression analyses in the brown alga model Ectocarpus siliculosus. BMC Mol Biol 9:75
Lee JM, Roche JR, Donaghy DJ, Thrush A, Sathish P (2010) Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Mol Biol 11:8
Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, Stacey G (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1:44–54
Lin YL, Lai ZX (2010) Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci 178:359–365
Ma BC, Tang WL, Ma LY, Li LL, Zhang LB, Zhu SJ (2009) The role of chitinase gene expression in the defense of harvested banana against anthracnose disease. J Amer Soc Hort Sci 134:379–386
Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10:4
Maroufi A, Bockstaele EV, Loose MD (2010) Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol Biol 11:15
Martin RC, Hollenbeck VG, Dombrowski JE (2008) Evaluation of reference genes for quantitative RT-PCR in Lolium perenne. Crop Sci 48:1881–1887
Mbéguié-A-Mbéguié D, Hubert O, Fils-Lycaon B, Chillet M, Baurens FC (2008) EIN3-like gene expression during fruit ripening of Cavendish banana (Musa acuminata cv. Grande naine). Physiol Plant 133:435–448
Mbéguié-A-Mbéguié D, Hubert O, Baurens FC, Matsumoto T, Chillet M, Fils-Lycaon B, Sidibé-Bocs S (2009) Expression patterns of cell wall-modifying genes from banana during fruit ripening and in relationship with finger drop. J Exp Bot 60:2021–2034
Mukesh J, Aashima N, Akhilesh KT, Jitendra PK (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914
Nolan T, Hands RE, Bustin SA (2006) Quantification of mRNA using real-time RTPCR. Nat Protocols 1:1559–1582
Paolacci AR, Tanzarella OA, Porceddu EP, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10:11
Pfaffl MWTA, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515
Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 23:856–862
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6:27
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227:1343–1349
Roy CS, Roy S, Singh SK, Sengupta DN (2010) Molecular characterization and differential expression of beta-1, 3-glucanase during ripening in banana fruit in response to ethylene, auxin, ABA, wounding, cold and light-dark cycles. Plant Cell Rep 29:813–828
Schmidt GW, Delaney SK (2010) Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics 283:233–241
Silveira ED, Alves-Ferreira M, Guimarães LA, Rodrigues da Silva F, Carneiro V (2009) Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol 9:84
Tang WL, Zhu SJ, Li LL, Liu DJ, Irving DE (2010) Differential expressions of PR1 and chitinase genes in harvested bananas during ripening, and in response to ethephon, benzothiadizole and methyl jasmonate. Postharvest Biol Technol 57:86–91
Tong Z, Gao Z, Wang F, Zhou J, Zhang Z (2009) Selection of reliable reference genes for gene expression studies in peach using realtime PCR. BMC Mol Biol 10:71
Tu L, Zhang X, Liu D, Jin S, Cao J, Zhu L, Deng F, Tan J, Zhang C (2007) Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chin Sci Bull 52:3110–3117
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalisation of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:1–11
Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223:7–12
Wan HJ, Zhao ZG, Qian CT, Sui YH, Malik AA, Chen JF (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399:257–261
Wang Y, Wu J, Xu BY, Liu JH, Zhang JB, Jia CH, Jin ZQ (2010) Cloning of an ADP-ribosylation factor gene from banana (Musa acuminata) and its expression patterns in postharvest ripening fruit. J Plant Physiol 167:989–995
Wong ML, Medrano JF (2005) Real-time PCR for mRNA quantitation. BioTechniques 39:75–85
Yang YG, Wu Y, Pirrello J, Regad F, Bouzayen M, Deng W, Li ZG (2009) Silencing Sl-EBF1 and Sl-EBF2 expression causes constitutive ethylene response phenotype, accelerated plant senescence, and fruit ripening in tomato. J Exp Bot 61:697–708
Acknowledgments
We thank Dr. Yuhai Cui (Agriculture and Agri-Food Canada, Southern Crop Protection and Food Research Centre, London, Canada) for his helpful discussion and revising. We also thank the three reviewers for their helpful suggestions. This study was financially supported by the National Natural Science Foundation of China (Grant Nos. 30800772; 30972068) and Modern Agro-industry Technology Research System (Grant No. nycytx-33).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, L., Zhong, Hy., Kuang, Jf. et al. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234, 377–390 (2011). https://doi.org/10.1007/s00425-011-1410-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-011-1410-3