Abstract
A consensus genetic map for chicory (2n = 2x = 18) was obtained after the integration of molecular marker data of two industrial chicory progenies (K28K59, Rubis118) and one witloof chicory progeny (BR). As a limited number of co-dominant markers was available at the beginning of this work, three different microsatellite-enriched libraries were produced from genomic DNA, resulting in 420, 719 and 1,251 sequences, respectively. The level of informative Simple Sequence Repeat (SSR) sequences from the three libraries ranged from 28 to 40%, thus defining a set of 730 SSR markers available for polymorphism screening. A subset of 81 Sequence-Tagged Sites (STS) developed from EST, cDNA, genes, and non-coding sequences was screened through Single Strand Conformational Polymorphism (SSCP) analysis, leading to 46 polymorphic loci integrated in the genetic maps. Markers were grouped and ordered on 9 homologous Linkage Groups (LG) for each of the three maps: 274 markers in K28K59, 282 markers in Rubis118, 178 markers in BR. Co-linear regions between maps were identified through 193 ‘bridge’ markers that allowed the integration of the 9 homologous LG in a consensus map containing 472 markers and covering 878 cM. Comparison across maps revealed the presence of 4 conserved regions with significant distorted markers, also defined as Segregation Distortion Regions (SDR), affected by gametic or zygotic selection factors. Marker distribution was not always uniform; 6 LG possessed homologous clustered regions in all maps. The consensus map could be the starting point for the identification and the cloning of major genes and QTL in fundamental and applied genetic areas in chicory.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chicory (Cichorium intybus L.) is a diploid plant species (2n = 18), belonging to the Asteraceae family, subfamily Cichoriodeae, tribe Lactuceae or Cichorieae (Bremer 1994; Panero and Funk 2002). This species is naturally allogamous, due to a sporophytic self-incompatibility system (Eenink 1981; Varotto et al. 1995). In addition, outcrossing is favoured by a floral morphology (i.e., protandry) unfavourable to selfing in the absence of pollinators (Pécaut 1962; Desprez et al. 1994) and by pollen competition (Desprez and Bannerot 1980; Eenink 1982). Long appreciated as a medical plant (Wittop Koning and Leroux 1972), Cichorium intybus L. is now cultivated for numerous different applications, and can be divided into varieties or cultigroups according to their use:
-
‘industrial’ or ‘root’ chicory, mainly cultivated in northwestern Europe (northern France, Belgium, The Netherlands) in India, South Africa and Chile. Its taproot is used for the production of a coffee substitute or for the extraction of inulin and fructans which are known as prebiotics with promising features in the food industry (Kaur and Gupta 2002) or in preventive treatment of human diseases (Pool-Zobel 2005). Breeder’s practices have evolved from mass selection to the use of a small set of selected families to obtain high-yielding synthetic varieties, and to hybrid production by the use of a cytoplasmic male sterility system (Rambaud et al. 1993; Desprez et al. 1994).
-
‘Brussels’ or ‘witloof’ chicory, commonly cultivated in the same European area as industrial chicory, and of which the roots are used for the production of etiolated buds or chicons by forcing. Witloof chicory is considered a derivative from the Magdeburg type of root chicory. In contrast to other chicory cultigroups, breeding efforts have led to the release of inbred lines with reduced inbreeding depression for the production of highly homogeneous F1 hybrids adapted to hydroponic forcing (Bannerot and de Coninck 1970; Eenink 1980).
-
‘leaf’ chicory, used as fresh or cooked vegetables, like the green-leaved varieties of the ‘Pain de Sucre’ and ‘Catalogne’ cultigroups and the ‘blond’, ‘red’ or ‘variegated’ varieties of the ‘Radicchio’ cultigroup from northern Italy (Lucchin et al. 2008).
-
‘forage’ chicory, initially derived from wild chicory commonly found along roadsides and waste areas, and used since the mid-1970s to increase herbage availability in perennial pastures for grazing livestock. Owing to the selection of varieties that meet nutritional needs and stand performance (Li and Kemp 2005), the application of forage chicory is increasing worldwide (New Zealand, Australia, America, and Europe).
During the last decade, the development of dominant molecular marker systems, essentially RAPD and AFLP, have largely contributed to classify germplasms and discriminate the genetic diversity within different species and cultigroups of Cichorium (Bellamy et al. 1996; Koch and Jung 1997; Kiers et al. 2000; van Stallen et al. 2000; Barcaccia et al. 2003; van Cutsem et al. 2003).
The same kind of markers was used to construct two genetic linkage maps for chicory. The first one was based on an interspecific F1 outbred population between a ‘variegated’ chicory and an inbred line of C. endivia L., a self-compatible species with chromosome structure (2n = 18) closely related to chicory (Rick 1953). Following a ‘pseudo-testcross’ scheme, firstly described by Grattapaglia and Sederoff (1994), de Simone et al. (1997) used dominant PCR-based markers (RAPD, AFLP, SAMPL) specific for C. intybus L. to construct a map that covered 1,204 cM with 351 markers dispersed over 13 LG. The second map was based on an intraspecific F2 population derived from a cross between two inbred lines of witloof chicory varieties. This map covered 609 cM with 129 RAPD markers on 9 LG (van Stallen et al. 2003), later completed by AFLP markers to dissect the quantitative variation of complex characters by detecting QTL (van Stallen et al. 2005a, b).
Both published maps for chicory suffer from a lack of codominant and readily transferable markers, rendering their integration impossible, and thus limiting their use in the construction of a consensus genetic map for C. intybus L. In addition, as pointed out by Maliepaard et al. (1997), the use of dominant markers may cause problems with respect to the probability to detect linkage and the ability to distinguish linkage phase combinations. The development of large sets of transferable co-dominant markers would facilitate the process of constructing genetic maps and identifying major genes or QTL of interest, and, ultimately, could be valid for breeders in marker-assisted-selection schemes involving diversified genetic material.
SSR markers are considered as ideal co-dominant markers for genetic mapping in plants because of their neutral status, their abundance and polymorphism, their high information contents and their mostly monogenic inheritance (Morgante and Olivieri 1993). All these features have greatly facilitated SSR utilization in connecting maps from various mapping populations and the construction of consensus genetic maps (Cervera et al. 2001; Yu et al. 2003; Gonzalo et al. 2005). SSR markers were initially obtained after screening genomic libraries enriched with SSR motifs and selecting unique SSR sequences suitable for primer design (Zane et al. 2002; Squirrell et al. 2003). More recently, the availibility of large sets of EST resulted in genic sequence data allowing the search and mapping of SSR (EST-SSR or genic SSR, Varshney et al. 2005). In chicory, the number of available EST to derive EST-SSR was limited until recently. Consequently, our mapping project (CARTOCHIC) aimed at constructing a reference genetic map for C. intybus using a large number of SSR markers was based on the screening of SSR-enriched genomic libraries.
In this article, we first report the production and screening of 3 SSR-enriched libraries for identifying informative SSR markers used in the construction of three genetic maps in chicory; two derived from industrial chicory mapping populations, and one derived from a witloof mapping population. A total of 730 SSR sequences were surveyed to identify polymorphic markers that were subsequently used for the construction of genetic maps. In addition, a set of 81 STS was screened by SSCP, and polymorphic loci were added to the maps. Secondly, homologous loci between the three maps were used as bridge markers to construct a consensus genetic map of 472 markers covering 878 cM on 9 LG.
Materials and methods
Plant material
Three mapping populations were used. The first one, called K28K59, was an outbred population of 177 F ’1 individuals obtained after crossing the heterozygous and self-incompatible parental genotypes K28 and K59. Both parents are industrial chicories obtained from the improved Hungarian landrace population ‘Kospool’ (Florimond-Desprez, Cappelle-en-Pévèle, France). The second population, called Rubis118, was an F2 population of 96 plants obtained after selfing one individual of the hybrid cultivar ‘Rubis’, originated from crosses between the male sterile clone MS8 and the population variety ‘Cassel’ (Florimond-Desprez, Cappelle-en-Pévèle, France). The third mapping population, called BR, was an F2 population of 145 individuals originated from the cross between two witloof inbred lines: a white leaf-coloured line B and a red leaf-coloured line R (Hoquet Endives, Raillencourt, France). One F1 hybrid plant was self-pollinated to generate the mapping population.
Parents K28, K59, MS8 and the F1 Rubis118 were maintained through in vitro cloning procedures. The B and R inbred lines were propagated by selfing. The parental genotype ‘Cassel’ of the Rubis118 population was not maintained. The evaluation of polymorphism for this latter population was based on the comparison between MS8 and the F1 Rubis118 plant.
DNA extraction
Genomic DNA was extracted with the GenElute™ Plant Genomic DNA Miniprep Kit (Sigma–Adrich Chemie Gmbh, Germany) from 10 mg of dried leaves or 100 mg of fresh leaves. Yield and quality of extracted DNA were checked in a UV/vis spectrophotometer (Biophotometer, Eppendorf, Germany) and after electrophoresis in 0.8% agarose gels.
SSR markers
Three SSR-enriched libraries were used. The first one, named Lib1, was produced by the former company Agrogène (France) from DNA of the ‘855824’ industrial chicories (Florimond-Desprez, Cappelle-en-Pévèle, France). The library contained 420 enriched clones, divided in 161 clones with a (CA) n motif, 193 clones with a (CT) n motif, 33 clones with a (CTT) n motif and 33 clones with a (ATT) n motif. SSR markers obtained from this library were named A, B, C or D followed by a number (e.g., B42 or A149 in Table 1).
The second library (Lib2) was produced by the company Ecogenics GmbH (Zürich, Switzerland) from size selected genomic DNA of chicory (genotype K59) according to Armour et al. (1994), and enriched by magnetic bead selection with biotin-labelled (CA)13 and (GA)13 oligonucleotide repeats (Gautschi et al. 2000a, b). Of 2,185 colonies, 456 (111 with CA motif, 345 with GA motif) gave a positive signal after hybridisation. Positive recombinant clones were sequenced by the company Genoscreen (Lille, France). Markers were designed by the prefix sw followed by a series of numbers and letters (e.g., sw2H09.2 in Table 1).
A third library (Lib3) was produced in our laboratory using the enrichment protocol described by Glenn and Schable (2005). Using genomic DNA of K59 genotype, 2,228 recombinant clones were obtained after enrichment by magnetic bead selection with a mixture of 3′ biotinylated oligonucleotide repeats including (AG)12 and (TG)12. These clones, referred to as EU clones (e.g., EU0022 in Table 1), were sequenced by the company Genoscreen (Lille, France).
Both Lib2 and Lib3 sequences were trimmed for vector sequences in BioEdit v 4.8.10 and submitted to Sputnik (http://espressosoftware.com/sputnik/index.html) for the detection of SSR motifs. Local Blast was performed with Bioedit on a database including 2,390 sequences in order to search for inter- and intra-library redundancy. All groups of redundant clones were aligned and analysed in Bioedit (ClustalW Multiple Alignment) in order to define consensus sequences. Primers were designed with Primer3 v 0.3.0.
For each primer pair designed, one primer was labelled with the fluorescent dyes 6-Fam or Hex. The PCR assay was performed in 15 μl containing 1× PCR buffer with 2 mM MgCl2, 100 μM of each dNTP, 0.2 mg/ml BSA, 133 nM of each primer, 0.3 U Taq polymerase (Applied Biosystems) and 2 ng of template DNA. A touchdown (TD) procedure was applied : 5 min denaturation at 94°C, followed by (a) 5 cycles of 30 s at 94°C, TD 30 s at Tm (−1°C per cycle), 30 s at 72°C and (b) 35 cycles of 30 s at 94°C, 30 s at Tm −5°C, 30 s at 72°C, before 10 min of extension at 72°C.
PCR products were analysed on a 3100 Avant Genetic Analyzer capillary sequencer (Applied Biosystems). Genotyping data were automatically collected and analysed (3100/3100 Avant Data Collection, Genescan® Analysis Software v 3.7.1, Genotyper v 3.7 NT). A subset of 18 monolocus and co-dominant SSR markers mapped on the 9 LG of the chicory map are provided in Table 1. A more complete set of SSR markers is available for academic research applications with a Materials Transfer Agreement. Applications should be sent to Dr Bruno Desprez, Florimond Desprez, BP41, 3 rue Florimond Desprez, 59242 Cappelle en Pévèle, France.
STS markers
STS sequences involved a set of 81 EST, cDNA, genes, and non-coding sequences of chicory listed in public databases or isolated in our laboratory. Table 2 summarizes the information related to the 46 STS mapped. SNP and InDel polymorphisms of STS were revealed by Capillary Electrophoresis—Single Strand Conformational Polymorphism (CE-SSCP) (Andersen et al. 2003) on an ABI 3100-Avant Genetic Analyser sequencer (Applied Biosystems).
Prior to primer design, sequence alignments between chicory cDNA or EST and Arabidopsis genes were searched with Blast tools on the NCBI site, Fasta 3 program in the EMBL-EBI site, and ClustalW Multiple Alignment in Bioedit v 4.8.10. Intron positions were determined and then compared to predictions by models for intron/exon boundaries determination with RNASPL tool, http://www.softberry.com/berry.phtml or GeneMark cDNA, http://opal.biology.gatech.edu/GeneMark/genemark_cDNA_all.cgi. For genes containing introns, primers were designed in regions surrounding putative intronic sequences, using the Primer3 v 0.3.0 software.
Each primer of the primer pair designed was labelled with a different fluorescent dye (6-Fam or Hex). The PCR assay was performed in 15 μl containing 1× PCR buffer with 2 mM MgCl2, 100 μM of each dNTP, 0.2 mg/ml BSA, 133 nM of each primer, 0.3 U Taq polymerase (Applied Biosystems) and 2 ng of template DNA. A TD procedure was applied: 5 min denaturation at 94°C, followed by (a) 5 cycles of 30 s at 94°C, TD 40 s at Tm (−1°C per cycle), 1 min at 72°C and (b) 35 cycles of 30 s at 94°C, 30 s at Tm −5°C, 1 min at 72°C, before 10 min of extension at 72°C.
Sample preparation, denaturation and CE-SSCP fractionation were performed according to User Bulletin ‘ABI PRISM® 3100 and 3100-Avant Genetic Analyzers: High Throughput Fluorescent SSCP Analysis’ (Applied Biosystems) with a minor modification in the concentration of Genescan™ polymer (3% instead of 7%). Data were collected (3100/3100 Avant Data Collection program) and normalised before analysis (Genescan® Analysis Software v 3.7.1 and Genotyper v 3.7 NT program).
Segregation analysis and linkage map construction
The segregation of each marker was evaluated by a χ2 test of goodness-of-fit to the expected Mendelian segregation ratio. For significantly skewed markers, the selection type (zygotic vs. gametic) was determined for co-dominant markers in F2 configuration type using two successive χ2 tests (Lorieux et al. 1995). Similarly, following the procedure described by Lorieux et al. (1995), χ2 tests for co-dominant markers with three and four alleles were derived. This procedure is available on request.
Linkage analysis and map calculations were performed using the program JoinMap v 3.0 (van Ooijen and Voorrips 2001). For each mapping population, linkage grouping was determined using a LOD threshold = 3. JoinMap setting parameters were: maximum recombination frequency = 0.45, LOD = 1.0, jump = 5. Pair-wise recombination frequencies were converted in map distances (cM) using the Kosambi mapping function (Kosambi 1944). Linkage analysis allowed to separate mapped markers between ‘map-specific markers’ and ‘bridge markers’ i.e., markers that share common alleles between mapping populations, thus identifying homologous loci. Homologous pair-wise marker data across two or three genetic maps were integrated with JoinMap (Map Integration option, van Ooijen and Voorrips 2001) before merging homologous groups and constructing the consensus or framework linkage map. The above-mentioned mapping parameters for generating the individual maps were also used for the consensus map. Subsequently, all homologous groups were drawn and aligned using the program MapChart v 2.1 (Voorrips 2002).
Genome coverage and marker clustering
Expected genome coverage G e was estimated by the expression proposed by Fishman et al. (2001): G e = G o + (2tG o)/n, where G o is the observed length in cM of the genetic map, t is the number of LG, and n is the number of marker intervals. We have also used the expression (4) of Chakravarti et al. (1991) generalized to all LG: G e = ΣG oi [(k i + 1)/(k i − 1)] where G oi is the observed length of the ith LG and k i is the number of mapped markers on the ith LG. Both expressions giving identical results, only the Fishman’s estimator is presented in Table 5. The proportion of genome coverage was then estimated by the ratio G o/G e. The probability that at least one marker is within d cM of a randomly chosen marker locus was estimated according to Lange and Boehnke (1982), as: P LB = 1 − e−2dk/Go, where k is the total number of loci.
Marker distribution between and within LG of each population was evaluated according to Remington et al. (1999). Inter-Linkage Groups Comparisons (ILGC analysis) were done under the hypothesis of a uniform marker distribution for all LG: the number k i in the ith LG followed a Poisson distribution with parameter λi = kG i /ΣG i , where G i is the estimated length of the ith LG given by the following expression: G i = G oi +2s (s is the average marker spacing). The cumulative probability function (cpf) was expressed for each LG by:
Poisson probabilities [P(λ i ≥ k i ) or P(λ i ≤ k i )] were significant when the cpf value or one minus the cpf value were less than α/2 (two-tailed Poisson test under cumulative distribution, α = 0.05).
Within-Linkage Groups Comparisons (WLGC analysis) for marker clustering were done after dividing each ith LG in windows of width W i:
-
1.
W i was fixed for all LG (W i = 10 cM)
-
2.
W ij was defined after selecting a subset of markers.
The criteria of choice for selected markers were: uniform distribution on the LG, co-dominance, minimum number of missing data, and low impact of segregation distortion. Intervals between markers j in the ith LG (W ij ) were then variable.
In both cases, if markers are randomly distributed, the number of mapped markers in the ith LG (k i ) followed a Poisson distribution with parameter λ i = k i W i /G i (W i fixed) or λ ij = k i W ij /G i (W ij variable). Poisson probabilities, calculated for each interval of the ith LG, was significant to a α/2 threshold. Intervals with significant Poisson probabilities P(λ i ≤ k i ) revealed significant clustering of markers.
When both analyses (fixed W i , variable W ij ) were significant, or one was significant and the other close to the α/2 level, the corresponding regions were represented by dark-grey patches in genetic maps (Figs. 2, 3). When only one analysis was significant or close to the α/2 level, the corresponding regions were represented by pale-grey patches (Figs. 2, 3).
Results
SSR screening
After sequencing inserts from clones of the three SSR-enriched libraries, a database of 2,390 sequences was generated containing 420 sequences from the Lib1 library, 719 sequences from the Lib2 library, and 1,251 sequences from the Lib3 library. The level of enrichment in SSR motifs ranged from 75 to 85%. Unique sequences containing a SSR repeat varied from 42% for the Lib1 library to 53 and 57% for the Lib3 and Lib2 library, respectively. Inter-library redundant sequences represented only 10–20%, indicating that the three libraries represented three complementary sources of SSR sequences.
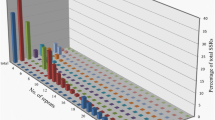
The distribution of sequences into different fractions was available for the Lib3 library, as it was completely produced and screened in our laboratory, and is illustrated in Fig. 1. Among the 1,251 Lib3 sequences, 332 sequences (26%) were removed according to several quality criteria: poor sequence quality (1%), lack of SSR (9%), SSR number or repeats <10 (8%), insufficient number of bases flanking the SSR to design primers (4%), the presence of repetitive sequences other than SSR (4%). The latter sequences showed similarities to the minisatellites identified in Beta species by Schmidt et al. (1991). Alignments of 608 redundant sequences containing SSR repeats allowed the constitution of contigs from which 188 (15%) consensus SSR sequences were derived (Fig. 1). By adding 311 (25%) unique SSR sequences available for primer design, the complete set of informative SSR was 499 SSR or 40% of the sequences from the Lib3 library.
Distribution of the 1,251 sequences derived from the Lib3 SSR-enriched-library. SSR redundant sequences were associated in contigs from which unique SSR consensus sequences were extracted, the remaining sequences being noted ‘copy sequences’. The two fractions containing informative SSR sequences for genetic analysis are the SSR (consensus) and SSR (unique) fractions
The level of putatively useful SSR from the Lib1 library was similar to the Lib3 library (155 SSR or 37%). This level was reduced in the Lib2 library (208 SSR or 28%), mainly because the mean size of the inserts containing the SSR was significantly smaller (246 bp in the Lib2 library vs. 500 bp in Lib3 or Lib1 libraries) thus increasing the number of SSR truncated sequences or SSR sequences with insufficient flanking sequences to design primers.
The 5 parental genotypes (K28, K59, MS8, B and R) and the F1 Rubis118 were genotyped for polymorphism survey using 367 Lib3 SSR from the 499 possible informative SSR. Thereof 258 (70%) turned out to be polymorphic, and 187 (51%), 153 (41.6%), and 98 (26.7%) could be used as markers in the K28K59, Rubis118, and BR mapping populations, respectively. Similarly, from the 155 available Lib1 SSR 114 (73%) were polymorphic, and 79 (50.9%), 64 (41.2%), and 55 (35.4%) could be used as markers in K28K59, Rubis118 and BR, respectively. The level of polymorphisms was lower for the Lib2 SSR (135 markers, i.e., 65%), and the number of informative markers was 97 (46.6%) for K28K59, 79 (37.9%) for Rubis118 and 67 (32.2%) for BR.
In total, 730 SSR were surveyed for polymorphism. The K28K59 population represented the most informative mapping population (363 markers), most likely due to its F1 outbred structure, followed by Rubis118 (296 markers). In comparison, the BR witloof population revealed less polymorphic markers (220 markers), in accordance with the reduced genetic base of this cultigroup.
STS screening
An SSCP analysis of 81 STS on the parental genotypes revealed that 46 (57%) were polymorphic in at least one of the 3 mapping populations. A strong divergence in terms of polymorphic loci between the industrial chicory populations and the witloof population was apparent: 28 (34.5%) and 33 (40.7%) STS were polymorphic in K28K59 and Rubis118, whereas only 8 (9.8%) STS were polymorphic in BR. Table 2 lists the assignments of these 46 STS markers to the 9 LG of the chicory map. Except for LG7 with only one STS marker, the distribution of STS was dispersed throughout the genome, ranging from 3 (LG5) to 9 markers (LG8) per LG.
Genetic maps for industrial and witloof chicory
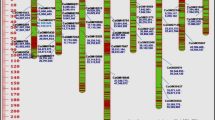
The 177 progenies of the K28K59 population could be genotyped for 291 marker loci (Fig. 2). Seventeen markers were excluded because their complex profiles made reliable identification of alleles impossible. The majority of markers (55%) were informative in only one parent (backcross configuration). The remaining 45% markers were informative in both parents (F2 configuration, configurations with three or four alleles) and allowed the integration of the genetic maps of both parents. The genetic map included 274 mapped markers and covered 749.8 cM with an average marker spacing of 2.7 cM. Loci were distributed over 9 LG and were represented by 68 Lib1 SSR, 37 Lib2 SSR, 143 Lib3 SSR, and 26 STS (Fig. 2). LG9, LG7, and LG4 had the lowest number of markers k i (19 < k i < 21). LG7, LG4, and LG2 had the smallest coverage (observed genome coverage G oi < 66 cM). Conversely, LG1, LG3, LG6, and LG8 were top groups for mapped markers (31 < k i < 48), with G oi ranging from 89.2 cM (LG6) to 106.6 (LG1).
Genetic maps of 3 chicory populations (K28K59, Rubis118, BR). Bridge markers are underlined. Skewed markers are followed by ‘*’ representing the probability associated to χ2 tests of goodness-of-fit (*P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.0001). Marker clustering (WLGC) are visualized through grey patches inside LG and rectangles surrounding significant markers (dark grey: significant WLGC test, pale grey: WLGC test just below the threshold)
The 96 progenies of the Rubis118 population were genotyped for 302 polymorphic markers. Twenty complex markers were excluded from the mapping procedure. As showed in Fig. 2, the genetic map included 282 markers and covered 806.1 cM with an average marker spacing of 2.8 cM. These loci were represented by 59 Lib1 SSR, 65 Lib2 SSR, 130 Lib3 SSR, and 28 STS. LG2, LG4, and LG7 were characterized by the smallest number of markers (19 < k i < 25). LG2 and LG4 had also the smallest genetic length (G oi < 64 cM). LG1, LG3, LG6, and LG8 had the largest genetic lengths (G oi > 100 cM) and were constituted by the largest number of markers (35 < k i < 43). In the Rubis118 map 80% of the markers were co-dominant, ranging from 63% for LG8 to 91% for LG4.
The 145 progenies of the BR population were genotyped for 190 markers. After excluding 12 complex SSR markers, the remaining loci were distributed over 9 LG (37 Lib1 SSR, 61 Lib2 SSR, 73 Lib3 SSR, and 7 STS). The genetic map included 178 markers and covered 891.4 cM with an average marker spacing of 5 cM (Fig. 2). LG2 was the smallest group (46.7 cM, 11 markers). The number of markers was relatively stable for six LG (LG1, LG3, LG4, LG7, LG8, and LG9; 18 < k i < 23). The highest value of k i was found for LG6 (33 markers), also characterised by a large size (121.9 cM). In the BR map 72% of the markers were co-dominant, ranging from 60% for LG6 to 80% for LG5 and LG8.
The level of recombination seemed higher in the BR witloof chicory map when compared to the industrial chicory maps, as revealed by the differences in LG lengths. Five LG had larger sizes in the BR map. The remaining LG had similar sizes between maps (LG9, LG7) or were less well covered in the BR map (LG2, LG3). The number k i was high (33 < k i < 43) in LG6 of the three maps. This number was also remarkably homogeneous for LG4 and LG7 (19 < k i < 22).
Bridge markers to construct the consensus map
Bridge markers or homologous loci mapped in at least 2 populations were used to identify co-linear regions between the 3 maps (Table 3). The numbers of bridge markers mapped in each of the 3 populations are presented in Table 3a. As a consequence of the lower number of markers mapped in the BR population, bridge markers were less numerous in the in the BR map than in the K28K59 and Rubis118 maps (96 vs. 154 and 161, respectively). Nonetheless, the proportion of bridge markers in the whole set of mapped markers was similar for all maps (53.9–57.1%), even though the proportion of bridge markers per homologous LG varied (e.g., LG7 and LG9).
Two by two map comparisons revealed a total of 193 connections between the three maps. The highest number of connections (97) consisted of bridge markers common to the 9 LG of the K28K59 and the Rubis118 maps (Table 3b). The numbers of bridge markers connecting the industrial chicory maps with the witloof map were relatively close (31–38, 16–19%). Triple-map connections K28K59-Rubis118-BR were the least numerous but present in all LG, ranging from one (LG2, LG9) to five connections (LG5, LG6).
Co-linear intervals flanked by bridge markers in the three individual maps were integrated to construct the consensus LG. The consensus map thus obtained contained 472 loci dispersed over 9 LG (Fig. 3), and covered 878 cM (average marker spacing of 1.8 cM). Markers were represented by 105 Lib1 SSR, 118 Lib2 SSR, 208 Lib3 SSR, and 41 STS. Five LG (LG1, LG3, LG6, LG8, and LG9) had observed genetic lengths G oi > 100 cM, and were also the richest LG for the number of markers (48 < k i < 75). The remaining 4 LG (LG2, LG4, LG5, and LG7) contained less markers (35 < k i < 43) and had reduced lengths (G oi < 87 cM). LG7 had the lowest number of mapped markers as most of them were bridge markers in the three individual maps.
Segregation distortion
The calculation of a χ2 test of goodness-of-fit for all markers revealed that the K28K59 and BR maps shared similar proportions of skewed markers (24 markers or 8.7% in K28K59, 15 markers or 8.4% in BR), whereas the Rubis118 map was characterized by a relative high proportion of skewed markers (55 markers or 19.5%). In the latter map, 51 distorted markers were concentrated in LG4, LG7, LG8, and LG9 (Fig. 2). They appeared clustered in specific regions (LG4: 7 markers; LG9: 11 markers) or represented the majority of the markers (LG7: 15 markers; LG8: 18 markers). For each of the four putative SDR, the source of the distortion was identified: zygotic selection factors seemed responsible for the SDR on LG4 and LG8, whereas the SDR on LG7 and LG9 most likely resulted from gametic selection factors.
Small clusters of distorted markers existed also in the K28K59 map (LG1, LG2, LG4, LG8 and LG9). Zygotic selection factors might explain the distorted regions in LG4 and LG8, and gametic selection factor(s) the SDR in LG9. This latter region was homologous to the SDR identified in LG9 of the Rubis118 map, suggesting the presence of the same loci affecting the viability of gametes. Distorted markers on LG1 and LG2 segregated according to a backcross-type configuration; in this case, the type of selection could not be determined.
Most of the distorted markers were dispersed in the BR map. Gametic selection factor(s) could explain the two distorted markers in LG7, whereas zygotic selection factor(s) could be associated to the four distorted markers in LG8.
Marker distribution
Marker distribution among LG (ILGC analysis) was evaluated for the 4 maps by comparing the observed number of markers per LG to an expected number calculated after re-estimating the size of each LG (Remington et al. 1999). The significant cpf values (Table 4) ranged from 0.975 to 0.99 for LG containing more markers than expected and from 0.025 to 0.0007 for LG containing fewer markers than expected under random distribution. The analysis revealed a balanced distribution of markers, even though map-specific situations existed, such as the lack of markers for LG9 of K28K59, LG7 of Rubis118, and LG8 of BR, or the high accumulation of markers in LG2 and LG8 of K28K59 and in LG3 of Rubis118. The reference map reflected the balanced and specific situations observed in the 3 individual maps (Table 4). Only LG6 had significantly more markers than expected, a consequence of the accumulation of markers in this particular LG across all maps (significant for BR, close to significant cpf values in K29K59 and Rubis118).
A second level of marker distribution was analysed by dividing each LG in segments or windows to assess clustering in specific areas (WLGC analysis). The analysis revealed 65 regions characterised by clustered markers (Figs. 2, 3), and all LG from all maps contained at least one clustered region. Six LG (LG2, LG3, LG4, LG6, LG7, and LG9) had homologous clustered regions in all maps, whereas 6 LG (LG1, LG2, LG3, LG5, LG6 and LG8) possessed homologous clustered regions in 3 maps. Clustered regions formed a significant proportion of mapped markers in several LG (LG2, LG4, LG7, LG8 for K28K59, LG4, LG9 for Rubis118, and LG3 for BR). Clustered regions of the consensus map reflected the most significant regions observed in the 3 mapping populations (Fig. 3), i.e., 4 LG (LG1 LG2, LG3 and LG6) with two clustered regions, the first one in the distal part close to the end of the group, the second one close to the middle of the group, and 5 LG (LG4, LG5, LG7, LG8 and LG9) with only one clustered region.
Genome coverage
Parameters G e, G o/G e and P LB have to be calculated under the hypothesis that marker loci are evenly distributed along the genome (Chakravarti et al. 1991; Lange and Boehnke 1982). As marker distribution analysis had revealed that clustered regions were present in all maps, a set of framework markers for each map was selected to reduce biased estimations of genome coverage parameters (Cervera et al. 2001; Fishman et al. 2001). The framework markers, selected for uniform distribution, co-dominance, low missing data, and low segregation distortion, were used to compute 4 framework maps (data not shown). The genome coverage parameters for the framework maps were subsequently compared with those for the complete set of markers.
Ge values estimated from the framework set of markers ranged from 932 to 1,126 cM, depending on the map (Table 5). Reduced Ge values were obtained when the complete sets of markers were used (range 801–986 cM), in agreement with the presence of clustered markers in all maps. As a consequence, genome coverages (Go/Ge) estimated from framework sets of markers were lower (range 81–83%) than those calculated from the complete sets of markers (range 90–94%).
To test the homogeneity of the repartitioning of markers inside LG, the probability P LB was calculated for 2 conditions d = 5 cM and d = 10 cM (Table 5). Considering the total set of markers, P LB (d = 10 cM) values were close to 1 (with a minimum of 0.97 for BR), whereas P LB (d = 5 cM) values were superior to 0.96 for K28K59 and Rubis118, and only 0.83 for BR, reflecting the lower number of polymorphic markers in this population. The selection of framework markers gave similar genome coverage values for the individual maps (0.77 < P LB (d = 10 cM) < 0.84) indicating that a restricted set of informative markers with uniform level of genome coverage can be used for breeding applications (e.g., diversity analysis, QTL identification, markers-assisted selection).
The above analyses indicated good and homogenous levels of genome coverage for all maps, especially for the industrial chicories, and suggested the absence of large non-covered regions in the LG. As for the BR map, the genome coverage was comparable to the other maps albeit that the density per LG was lower. These results corroborated with the fact that all markers, except for the few SSR markers with complex profiles that rendered allele identification ambiguous, could be associated with LG of the 3 maps.
Discussion
Merging genetic maps to construct a reference map has been described for several species (e.g., Jeuken et al. 2001; Cervera et al. 2001; Yu et al. 2003; Yan et al. 2005; Gonzalo et al. 2005). Two genetic maps have been published for chicory based on AFLP and RAPD markers, respectively (de Simone et al. 1997; van Stallen et al. 2003), but the lack of common co-dominant markers renders their merging impossible. We have constructed a consensus genetic map in chicory based on genetic data from 3 progenies using a large set of SSR markers from three SSR-enriched libraries. In addition, a subset of STS (non-coding sequences, cDNA, genes) of chicory was added to the maps following an SSCP-based protocol (Aubert et al. 2006). The consensus map was constructed by integrating the maps of two progenies from crosses between industrial chicories and one from a cross between witloof chicories. The industrial chicory parental genotypes showed high levels of polymorphisms for both SSR and STS markers in comparison to the witloof chicories, reflecting the narrow genetic base of the latter. The consensus map measures 878 cM and contains 472 molecular markers distributed over 9 LG, which is in agreement with the haploid chromosome number of the genus Cichorium (Dujardin et al. 1979). Marker distribution analyses indicated a good coverage of the genome, and suggested the absence of large non-covered regions.
Systematic comparisons of homologous regions in our maps revealed the presence of common SDR as well as of homologous regions with localized clustering of markers. Unlike Cervera et al. (2001), who rejected high distorted loci from the mapping process, we introduced all markers and compared the resulting orders among the three maps (Cloutier et al. 1997; Lashermes et al. 2001; Song et al. 2006). Sequential χ2 tests described in Lorieux et al. (1995) allowed the distinction between zygotic and gametic selection in our mapping population, each explaining about half of the segregation distortions observed. Markers affected by zygotic selection, as revealed on SDR of LG4 and LG8, were characterised by an excess of heterozygotes. Similar results were found in inbred species like lettuce (Kesseli et al. 1994), or outbred species like coffee (Lashermes et al. 2001), alfalfa (Kalo et al. 2000) and white clover (Jones et al. 2003).
Segregations affected by gametic selection were observed for most markers in LG7 of the Rubis118 map, and for markers on LG9 of both Rubis118 and K28K59 maps. Most of the markers on LG7 in Rubis118 showed an excess of one homozygous class in comparison to the other homozygous class, whereas the frequency of the heterozygous class remained close to 0.50. This observation suggests one or more gametophytic factors on LG7 causing a deficiency in the viability and/or transmission of male or female gametes. Similar distorted segregations have been reported for F2 progenies of maize (Lu et al. 2002). The distorted segregation of markers in the SDR of LG9—a deficiency for the heterozygous class and one homozygous class, and an excess for the other homozygous class—may be the result of gametic selection in both parents.
In chicory self-incompatibility and pollen competition were reported (Eenink 1982; Desprez et al. 1994), but numerous other genetic factors such as meiotic drive, selective germination or seedling death (Korbecka et al. 2002), negative epistatic interactions (Torjek et al. 2006) could also explain the presence of SDR (see Song et al. 2006 for review). The higher level of distortion for the Rubis118 population, when compared to K28K59 and BR, may be associated with constraints in the fitness of F1 gametes and/or F2 genotypes originating from parental material initially selected for their out-crossing performance and subsequently submitted to forced selfing to generate the mapping population. In contrast, K28 and K59 were compatible genotypes chosen from an open-pollinated pre-breeding population and thus had encountered less selection pressure. Similarly, the witloof chicory parents of the BR population had been obtained after selection for inbreeding performance.
We have adapted the method described by Remington et al. (1999) to asses the level of marker distribution between the LG of the maps (ILGC tests) and inside each LG (WLGC tests). ILGC tests showed significant levels of marker concentrations or deficiencies in a limited number of LG per map, whereas WLGC tests revealed a large number of regions with clustered markers, in particular 13 clustered homologous regions dispersed over the 9 LG of at least 3 maps. Clustering of markers in genetic maps have been reported in species like lettuce (Jeuken et al. 2001), tomato (Areshchenkova and Ganal 1999) or barley (Ramsay et al. 2000), but seemed less evident in sunflower (Tang et al. 2002), melon (Gonzalo et al. 2005), or rose (Yan et al. 2005).
Non random distribution of SSR from enriched-libraries in comparison to SSR motifs present in EST sequences was recently reported in sugar beet (Laurent et al. 2007). In barley SSR from enriched-libraries tended to be concentrated in retrotransposons and repetitive elements-rich regions (Ramsay et al. 1999) and were associated to centromeric regions in barley and tomato (Areshchenkova and Ganal 1999; Ramsay et al. 2000), where inhibition of recombination occurs (Copenhaver et al. 1999). An association of markers with centromeric regions could be suggested to exist in chicory, particularly for LG2, LG3, LG4, LG6, LG7, and LG9 that shared chromosomal regions with homologous clustered markers in all maps. This hypothesis can be tested once centromere-specific markers for chicory or other members of the Asteraceae family become available, as reported by Pouilly et al. (2008) for oilseed rape and by Luce et al. (2006) for maize.
The reference map of chicory opens new perspectives in different directions such as, gene tagging, genetic analysis of quantitative-inherited traits, marker-assisted selection, or comparative genomics with other Asteraceae. To further improve the map, it is envisaged to integrate large numbers of gene loci. While our mapping project was in progress, numerous EST for chicory have been published (3,348 EST from embryogenic and non-embryogenic cDNA libraries—Legrand et al. 2007; 12,226 EST from root, leaf and nodules cDNA libraries—Dauchot et al. 2009) and released by the Genome Mapping Project for the Compositae (Asteraceae) at UC Davis Genome Centre (38,323 EST or 22,291 unigenes for C. intybus, as well as 30,171 EST or 19,065 unigenes for its close relative C. endivia: http://cgpdb.ucdavis.edu/cgpdb2/est_info_library.php). In addition, the availability of the EST offers the possibility to find additional SSR markers. The screening of EST-SSR is an efficient alternative to genomic library construction to identify EST-SSR markers (Varshney et al. 2005). In most of the plant species, SSR with a minimum repeat length of 20 bp are present in ~5% EST, with trinucleotides as preponderant motifs followed by dinucleotides or tetranucleotides (Varshney et al. 2005). Although the level of polymorphism was usually thought lower in EST-SSR than in SSR derived from enriched libraries (Eujayl et al. 2001; Rungis et al. 2004), a comparison in sugar beet showed more polymorphism for EST-SSR and emphasised their utility in genetic mapping (Laurent et al. 2007). Successful PCR amplification of Conserved Orthologous Set (COS) of EST from lettuce and sunflower in several other members in the Asteraceae family, including chicory, has been reported (Chapman et al. 2007). This suggests that the transfer and mapping of EST from related species is possible, even though the transferability of sunflower EST-SSR and INDELs was limited to only 18% when tested in lettuce or prickly lettuce (Heesacker et al. 2008). For chicory, transferable EST may be expected to be found especially in species belonging to same Cichoriodeae subfamily, such as lettuce.
Finally, in order to link the presented map with the chromosomes of chicory, next mapping steps will consist in finding of the ends of each LG by linking terminal markers with telomere-specific sequences (Hu 2006), and identifying the centromeric regions. Furthermore, through the development of BAC libraries, an assignment of the 9 LG to the corresponding chromosomes in chicory could be envisaged by BAC landed—Fluorescence In Situ Hybridisation protocols (Dong et al. 2000; Kim et al. 2002; Wang et al. 2007). This approach would be the first step to integrate genetic data with physical regions of the chicory genome.
References
Andersen PS, Jespersgaard C, Vuust J, Christiansen M, Larsen LA (2003) High-throughput single strand conformation polymorphism mutation detection by automated capillary array electrophoresis: validation of the method. Hum Mutat 21:116–122
Areshchenkova T, Ganal MW (1999) Long tomato microsatellites are predominantly associated with centromeric regions. Genome 42:536–544
Armour JAL, Neumann R, Gobert S, Jeffreys AJ (1994) Isolation of human simple sequence repeat loci by hybridisation selection. Hum Mol Genet 3:599–605
Aubert G, Morin J, Jacquin F, Loridon K, Quillet M-C, Petit A, Rameau C, Lejeune-Hénaut I, Huguet T, Burstin J (2006) Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor Appl Genet 112:1024–1041
Bannerot H, de Coninck B (1970) L’utilisation des hybrides F1: nouvelle méthode d’amélioration de la chicorée de Bruxelles. Symp. Int. Section Horticole d’Eucarpia. La chicorée de Bruxelles. Gembloux, Belgique, pp 99–118
Barcaccia G, Pallottini L, Soattin M, Lazzarin R, Parrini P, Lucchin M (2003) Genomic DNA fingerprints as a tool for identifying cultivated types of radicchio (Cichorium intybus L.) from Veneto, Italy. Plant Breed 122:178–183
Bellamy A, Vedel F, Bannerot H (1996) Varietal identification in Cichorium intybus L. and determination of genetic purity of F1 hybrid seed samples, based on RAPD markers. Plant Breed 115:128–132
Bremer K (1994) Asteraceae: cladistics and classification. Timber Press, Portland
Cervera M-T, Storme V, Ivens B, Gusmao J, Liu BH, Hostyn V, van Slycken J, van Montagu M, Boerjan W (2001) Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra and P. trichocarpa) based on AFLP and microsatellite markers. Genetics 158:787–809
Chakravarti A, Lasher LK, Reefer JE (1991) A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128:175–182
Chapman M, Chang J, Weisman D, Kesseli R, Burke J (2007) Universal markers for comparative mapping and phylogenetic analysis in the Asteraceae (Compositae). Theor Appl Genet 115:747–755
Cloutier S, Cappadocia M, Landry BS (1997) Analysis of RFLP mapping inaccuracy in Brassica napus L. Theor Appl Genet 95:83–91
Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD, McCombie WR, Martienssen RA, Marra M, Preuss D (1999) Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286:2468–2474
Dauchot N, Mingeot D, Purnelle B, Muys C, Watillon B, Boutry M, Van Cutsem P (2009) Construction of 12 EST libraries and characterization of a 12, 226 EST dataset for chicory (Cichorium intybus) root, leaves and nodules in the context of carbohydrate metabolism investigation. BMC Plant Biol 9:14
de Simone M, Morgante M, Lucchin M, Parrini P, Marocco A (1997) A first linkage map of Cichorium intybus L. using a one-way pseudo-testcross and PCR-derived markers. Mol Breed 3:415–425
Desprez FF, Bannerot H (1980) A study of pollen tube growth in witloof chicory. In: Maxon Smith JW, Langton A (eds) Proceedings Eucarpia meeting on leafy vegetables, Littlehampton, pp 47–52
Desprez B, Delesalle L, Dhellemmes C, Desprez M (1994) Genetics and breeding of industrial chicory. C R Acad Agric Fr 80:47–62
Dong F, Song J, Naess SK, Helgeson JP, Gebhardt C, Jiang J (2000) Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor Appl Genet 101:1001–1007
Dujardin M, Louant BP, Tilquin JP (1979) Determining the caryogram of Cichorium intybus L. Ann Amélior Plantes 29:305–310
Eenink AH (1980) Breeding research on witloof chicory for the production of inbred lines and hybrids. In: Maxon Smith JW, Langton A (eds) Proceedings Eucarpia meeting on leafy vegetables, Littlehampton, pp 5–11
Eenink AH (1981) Compatibility and incompatibility in witloof-chicory (Cichorium intybus L.). 2. The incompatibility system. Euphytica 30:77–85
Eenink AH (1982) Compatibility and incompatibility in witloof-chicory (Cichorium intybus L.). 3. Gametic competition after mixed pollinations and double pollinations. Euphytica 31:773–786
Eujayl I, Sorrels M, Baum M, Wolters P, Powell W (2001) Assessment of genotypic variation among cultivated durum wheat based on EST-SSRs and genomic SSRs. Euphytica 119:39–43
Fishman L, Kelly AJ, Morgan E, Willis JH (2001) A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159:1701–1716
Gautschi B, Tenzer I, Muller JP, Schmid B (2000a) Isolation and characterization of microsatellite loci in the bearded vulture (Gypaetus barbatus) and cross-amplification in three Old World vulture species. Mol Ecol 9:2193–2195
Gautschi B, Widmer A, Koella J (2000b) Isolation and characterization of microsatellite loci in the dice snake (Natrix tessellata). Mol Ecol 9:2192–2193
Glenn TC, Schable NA (2005) Isolating microsatellite DNA loci. Methods Enzymol 395:202–222
Gonzalo M, Oliver M, Garcia-Mas J, Monfort A, Dolcet-Sanjuan R, Katzir N, Arùs P, Monforte A (2005) Simple-sequence repeat markers used in merging linkage maps of melon (Cucumis melo L.). Theor Appl Genet 110:802–811
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Heesacker A, Kishore VK, Gao W, Tang S, Kolkman JM, Gingle A, Matvienko M, Kozik A, Michelmore RM, Lai Z, Rieseberg LH, Knapp SJ (2008) SSRs and INDELs mined from the sunflower EST database: abundance, polymorphisms, and cross-taxa utility. Theor Appl Genet 117:1021–1029
Hendriks T, Scheer I, Quillet M-C, Randoux B, Delbreil B, Vasseur J, Hilbert J-L (1998) A nonsymbiotic hemoglobin gene is expressed during somatic embryogenesis in Cichorium. Biochim Biophys Acta 1443:193–197
Hu J (2006) Defining the sunflower (Helianthus annuus L.) linkage group ends with the Arabidopsis-type telomere sequence repeat-derived markers. Chromosome Res 14:535–548
Jeuken M, van Wijk R, Peleman J, Lindhout P (2001) An integrated interspecific AFLP map of lettuce (Lactuca) based on two L. sativa × L. saligna F2 populations. Theor Appl Genet 103:638–647
Jones ES, Hughes LJ, Drayton MC, Abberton MT, Michaelson-Yeates TPT, Bowen C, Forster JW (2003) An SSR and AFLP molecular marker-based genetic map of white clover (Trifolium repens L.). Plant Sci 165:531–539
Kalo P, Endre G, Zimanyi L, Csanadi G, Kiss GB (2000) Construction of an improved linkage map of diploid alfalfa (Medicago sativa). Theor Appl Genet 100:641–657
Kaur N, Gupta AK (2002) Applications of inulin and oligofructose in health and nutrition. J Biosci 27:703–714
Kesseli RV, Paran I, Michelmore RW (1994) Analysis of a detailed genetic linkage map of Lactuca sativa (lettuce) constructed from RFLP and RAPD markers. Genetics 136:1435–1446
Kiers AM, Mes THM, vanderMeijden R, Bachmann K (2000) A search for diagnostic AFLP markers in Cichorium species with emphasis on endive and chicory cultivar groups. Genome 43:470–476
Kim J-S, Childs KL, Islam-Faridi MN, Menz MA, Klein RR, Klein PE, Price HJ, Mullet JE, Stelly DM (2002) Integrating karyotyping of sorghum by in situ hybridization of landed BACs. Genome 45:402–412
Koch G, Jung C (1997) Phylogenetic relationships of industrial chicory varieties revealed by RAPDs and AFLPs. Agronomie 17:323–333
Korbecka G, Klinkhamer PGL, Vrieling K (2002) Selective embryo abortion hypothesis revisited – a molecular approach. Plant Biol 4:298–310
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eug 12:172–175
Lange K, Boehnke M (1982) How many polymorphic genes will it take to span the human genome? Am J Hum Genet 34:842–845
Lashermes P, Combes M-C, Prakash NS, Trouslot P, Lorieux M, Charrier A (2001) Genetic linkage map of Coffea canephora: effect of segregation distortion and analysis of recombination rate in male and female meioses. Genome 44:589–596
Laurent V, Devaux P, Thiel T, Viard F, Mielordt S, Touzet P, Quillet M-C (2007) Comparative effectiveness of sugar beet microsatellite markers isolated from genomic libraries and GenBank ESTs to map the sugar beet genome. Theor Appl Genet 115:793–805
Legrand S, Hendriks T, Hilbert J-L, Quillet M-C (2007) Characterization of expressed sequence tags obtained by SSH during somatic embryogenesis in Cichorium intybus L. BMC Plant Biol 7:27
Li G, Kemp PD (2005) Forage chicory (Cichorium intybus L.): a review of its agronomy and animal production. In: Sparks DL (ed) Advances in agronomy, vol 88. Academic Press, London, pp 187–222
Lorieux M, Perrier X, Goffinet B, Lanaud C, Gonzales de Leon D (1995) Maximum-likelihood models for mapping genetic markers showing segregation distortion. 2. F2 populations. Theor Appl Genet 90:81–89
Lu H, Romero-Severson J, Bernardo R (2002) Chromosomal regions associated with segregation distortion in maize. Theor Appl Genet 105:622–628
Lucchin M, Varotto S, Barcaccia G, Parrini P (2008) Chicory and endive. In: Prohen J, Nuez F (eds) Vegetables I. Springer, New York, pp 3–48
Luce AC, Sharma A, Mollere OSB, Wolfgruber TK, Nagaki K, Jiang J, Presting GG, Dawe RK (2006) Precise centromere mapping using a combination of repeat junction markers and chromatin immunoprecipitation—polymerase chain reaction. Genetics 174:1057–1061
Maliepaard C, Jansen J, van Ooijen JW (1997) Linkage analysis in a full-sib family of an outbreeding plant species: overview and consequences for applications. Genet Res 70:237–250
Morgante M, Olivieri A (1993) PCR-amplified microsatellites as markers in plant genetics. Plant J 3:175–182
Panero JL, Funk VA (2002) Toward a phylogenetic subfamilial classification for the Compositae. Proc Biol Soc Wash 115:909–922
Pécaut P (1962) Etude sur le système de reproduction de l’endive (Cichorium intybus L.). Ann Amélior Plantes 12:265–296
Pool-Zobel BL (2005) Inulin-type fructans and reduction in colon cancer risk: review of experimental and human data. Br J Nutr 93:S73–S90
Pouilly N, Delourme R, Alix K, Jenczewski E (2008) Repetitive sequence-derived markers tag centromeres and telomeres and provide insights into chromosome evolution in Brassica napus. Chromosome Res 16:683–700
Rambaud C, Dubois J, Vasseur J (1993) Male-sterile chicory cybrids obtained by intergeneric protoplast fusion. Theor Appl Genet 87:347–352
Ramsay L, Macaulay M, Cardle L, Morgante M, Ivanissevich SD, Maestri E, Powell W, Waugh R (1999) Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant J 17:415–425
Ramsay L, Macaulay M, Ivanissevich SD, MacLean K, Cardle L, Fuller J, Edwards KJ, Tuvesson S, Morgante M, Massari A, Maestri E, Marmiroli N, Sjakste T, Ganal M, Powell W, Waugh R (2000) A simple sequence repeat-based linkage map of barley. Genetics 156:1997–2005
Remington DL, Whetten RW, Liu B-H, O’Malley DM (1999) Construction of an AFLP genetic map with nearly complete genome coverage in Pinus taeda. Theor Appl Genet 98:1279–1292
Rick CM (1953) Hybridization between chicory and endive. J Am Soc Hort Sci 61:459–466
Rungis D, Bérubé Y, Zhang J, Ralph S, Ritland CE, Ellis BE, Douglas C, Bohlmann J, Ritland K (2004) Robust simple sequence repeat markers for spruce (Picea spp.) from expressed sequence tags. Theor Appl Genet 109:1283–1294
Schmidt T, Jung C, Metzlaff M (1991) Distribution and evolution of two satellite DNAs in the genus Beta. Theor Appl Genet 82:793–799
Song X-L, Sun X-Z, Zhang T-Z (2006) Segregation distortion and its effect on genetic mapping in plants. Chin J Agric Biotechnol 3:163–169
Squirrell J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, Gibby M, Powell W (2003) How much effort is required to isolate nuclear microsatellites from plants? Mol Ecol 12:1339–1348
Tang S, Yu J-K, Slabaugh M, Shintani D, Knapp S (2002) Simple sequence repeat map of the sunflower genome. Theor Appl Genet 105:1124–1136
Torjek O, Witucka-Wall H, Meyer R, von Korff M, Kusterer B, Rautengarten C, Altmann T (2006) Segregation distortion in Arabidopsis C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interaction of two loci. Theor Appl Genet 113:1551–1561
van Cutsem P, du Jardin P, Boutte C, Beauwens T, Jacqmin S, Vekemans X (2003) Distinction between cultivated and wild chicory gene pools using AFLP markers. Theor Appl Genet 107:713–718
van Ooijen JW, Voorrips RE (2001) JoinMap 3.0, software for the calculation of genetic linkage maps. Plant Research International, Wageningen
van Stallen N, Noten V, Demeulemeester M, de Proft M (2000) Identification of commercial chicory cultivars for hydroponic forcing and their phenetic relationships revealed by random amplified polymorphic DNAs and amplified fragment length polymorphisms. Plant Breed 119:265–270
van Stallen N, Vandenbussche B, Verdoodt V, de Proft M (2003) Construction of a genetic linkage map for witloof (Cichorium intybus L. var. foliosum Hegi). Plant Breed 122:521–525
van Stallen N, Vandenbussche B, Londers E, Noten V, de Proft M (2005a) QTL analysis of important pith characteristics in a cross between two inbred lines of chicory (Cichorium intybus var. foliosum): a preliminary study. Plant Breed 124:54–58
van Stallen N, Vandenbussche B, Londers E, Noten V, de Proft M (2005b) QTL analysis of production and taste characteristics of chicory (Cichorium intybus var. foliosum). Plant Breed 124:59–62
Varotto S, Pizzoli L, Lucchin M, Parrini P (1995) The incompatibility system in Italian red chicory (Cichorium intybus L.). Plant Breed 114:535–538
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang K, Guo W, Zhang T (2007) Development of one set of chromosome-specific microsatellite-containing BACs and their physical mapping in Gossypium hirsutum L. Theor Appl Genet 115:675–682
Wittop Koning DA, Leroux A (1972) La chicorée dans l’histoire de la médecine et dans la céramique pharmaceutique. Rev Hist Pharm (Suppl) 215: 28 p
Yan Z, Denneboom C, Hattendorf A, Dolstra O, Debener T, Stam P, Visser PB (2005) Construction of an integrated map of rose with AFLP, SSR, PK, RGA, RFLP, SCAR and morphological markers. Theor Appl Genet 110:766–777
Yu J-K, Tang S, Slabaugh MB, Heesacker A, Cole G, Herring M, Soper J, Han F, Chu W-C, Webb DM, Thompson L, Edwards KJ, Berry S, Leon AJ, Grondona M, Olungu C, Maes N, Knapp SJ (2003) Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci 43:367–387
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: a review. Mol Ecol 11:1–16
Acknowledgments
The present work was financed by a public–private partnership ‘Groupement d’Intérêt Scientifique’ CARTOCHIC involving UMR 1281 SADV, Université de Lille 1, Florimond Desprez Veuve and Fils SAS, Leroux SAS, and SARL Hoquet Endives. We would like to thank Louis Delesalle, Jean-Christophe Lepeltier, Pierre Devaux (Florimond-Desprez) and Pascal Lévêque (Hoquet Endives) who have generated and maintained the Rubis118 and BR populations. We also acknowledge the SREL laboratory of Dr Travis Glenn (Travis Glenn, Cris Hagen, Lucy Dueck, SREL, Aitken, SC, USA) for the training at the first authors in the SSR enrichment protocol, and Aude Darracq (UMR 8016 GEPV, Université de Lille 1) for the development of a program devoted to the identification of unique SSR sequences. Helpful comments and critical reading of the manuscript by Bruno Desprez and Pierre Devaux have also been greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Thierry Cadalen and Monika Mörchen contributed equally to this work.
Rights and permissions
About this article
Cite this article
Cadalen, T., Mörchen, M., Blassiau, C. et al. Development of SSR markers and construction of a consensus genetic map for chicory (Cichorium intybus L.). Mol Breeding 25, 699–722 (2010). https://doi.org/10.1007/s11032-009-9369-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-009-9369-5