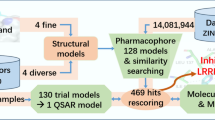
Abstract
The main study’s purpose is to detect novel natural products (NPs) that are potentially selective MAO-B inhibitors and, additionally, to computationally reposition the marketed drugs with a new therapeutic role for Parkinson’s disease. To reach the goals, 3D similarity search, docking, ADMETox, and drug repurposing approaches were employed. Thus, an unbiased benchmarking dataset was built including selective and nonselective inhibitors for MAO-B compliant with both ligand- and structure-based virtual screening approaches. A retrospective and prospective mining scenario was applied to SPECS NP and DrugBank databases to detect novel scaffolds with potential benefits for Parkinson’s disease patients. Out of the three best selected natural products, cardamomin showed excellently predicted drug-like properties, superior pharmacological profile, and specific interactions with MAO-B active site, indicating a potential selectivity over MAO-B. Two marketed drugs, fenamisal and monobenzone, were proposed as promising candidates repurposed for Parkinson’s disease. The application of shape, physicochemical, and electrostatic similarity searches protocol emerged as a plausible solution to explore MAO-B inhibitors selectivity. This protocol might serve as a rewarding tool in early drug discovery and can be extended to other protein targets.
Graphic abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Worldwide, over six million people suffer from Parkinson’s disease [1]. The monoamine oxidases (MAOs) are the enzymes responsible for the metabolism of monoamine neurotransmitters such as serotonin, norepinephrine, and dopamine, which regulate their concentrations in the central and peripheral tissues [2]. The MAOs exist as two isoforms, MAO-A, and MAO-B, which are the products of X-chromosomal gene Xp1 123, showing over 70% sequence similarity [3]. The two isoforms display distinct substrates and inhibitor specificities: MAO-A has a higher affinity for hydroxylated amines (noradrenaline and serotonin), whereas MAO-B interacts with non-hydroxylated amines (benzylamine and beta-phenylethylamine) [4]. Dopamine and tyramine show related affinities for both isoforms [5]. By their fundamental role in key physiological processes, both MAO isoforms are involved in the pathogenesis of various neuropsychiatric disorders, including mood disorders, depression, Alzheimer’s disease, Parkinson’s disease, etc. [6]. Although the administration of MAO inhibitors results in health improvement in the cases of certain psychiatric and neurological disorders, side effects and safety concerns co-occur [4]. Accordingly, MAOIs represent an alternative therapy when all other medications fail [1]. The identification of selective irreversible inhibitors of MAO-A and MAO-B elucidated partially these issues [4]. The relatively selective irreversible inhibitors of MAO-B, selegiline, and rasagiline, and the new effective, reversible MAO-B inhibitor, safinamide (SAG) were marketed for the treatment of Parkinson’s disease [4, 7]. Overcoming the main drawbacks of MAO inhibitors, irreversibility, and deficient selectivity requires continuous identification of new inhibitors that use the existing knowledge to elaborate rational design strategies.
Chemoinformatics tools are nowadays intensively used in the drug development process, and associated with experimental methods guide successfully the identification of novel drug candidates, ligand–target interactions [8], and concentrated potentially active ligands in targeted chemical libraries [8]. Additionally, in silico tools can predict off-target interactions, which require structural information about ligand and target proteins, as well as of pharmacokinetic properties [9,10,11] to prioritize the most promising molecules. However, a comprehensive theoretical investigation of selectivity is not substantial due to the absence of standard datasets adjusted on selectivity [12]. Nowadays the most frequently used virtual screening tools are similarity-based methods and docking. Ligand-based virtual screening (LBVS) relies on knowledge of the chemical structure of the compounds that bind to a protein target. They can provide predictive models suitable for hit/lead identification and optimization. Previously, MAO-A/B inhibitors were successfully developed using a ligand-based design strategy—3D-Quantitative Structure–Activity Relationship (3D-QSAR) [13]. These studies underlined that heterocyclic structure (oxadiazole, pyrrole, piperazine) defines a crucial pharmacophore feature to design MAO-B inhibitors [14]. Structure-based virtual screening (SBVS) is frequently used in hit/lead identification, by prioritizing a subset of compounds that have to be experimentally tested for biological activity [15]. Molecular docking involves the explicit prediction of the binding mode of each ligand inside the receptor active site and scoring to estimate the binding affinity [15].
The current analysis is aimed at identifying the shape pattern, physicochemical features, and specific ligand–receptor interactions that are correlated with selective and nonselective inhibition of MOA-B, respectively. The main goal is to detect novel natural products that are potentially selective MAO-B inhibitors, and repurposing of marketed drugs for the treatment of Parkinson’s disease, by using LBVS and validate them by docking and ADMETox properties prediction. The employment of high-quality benchmarking data sets, such as our novel selectivity oriented dataset for MAO-B, including experimentally validated selective and nonselective MAO-B inhibitors, are indispensable to retrospectively validate the virtual screening (VS) methodologies before applying it prospectively.
Methods
Datasets
Standard validation dataset for MAO-B
The standard validation dataset was built by collecting data points based on our previous work [16], which was adapted to the prerequisites of MAO-B selectivity problematic by setting the selectivity thresholds and topologically validated using BMF’s and MACCS keys. The bioactivity data for MAO-A and MAO-B were extracted from CHEMBL [17], retaining target type “single protein,” and standard relation “=”. In the case of multiple values for a compound-target pair, the minimum activity value was kept, followed by joining MAO-A with MAO-B biochemical data, identifying shared compounds, and removing duplicates. The salts were discarded, the charges and stereochemistry were systematized with the help of InstantJChem (Instant JChem, v.5.12.4, http://www.chemaxon.com). The non-drug-like ligands with RBNs > 20 or MW ≥ 600 were prefiltered according to the criteria adopted by Mysinger [18]. A preliminary physicochemical property filter to reduce “artificial enrichment,” which influences mainly SBVS, was applied [19]. Thus, the maximum and minimum values of drug-like descriptors registered for selective MAO-B inhibitors, calculated using FILTER (FILTER, OpenEye Scientific Software, Santa Fe, NM. http://www.eyesopen.com), which show better compliance to Lipinski’s rule of five (RO-5) [20] were enforced as a target-specific property filter: HBA = 0÷6, HBD = 0 ÷ 7, MW = 172.25 ÷ 570.76, RBN = 0 ÷ 17, XLogP = -0.27 ÷ 9.49, Polar Surface Area 2dPSA = 16.61 ÷ 106.38 (Fig. 1). The compounds showing IC50 > 20 μM against MOA-B were removed, resulting 501 compounds. The selectivity index (SI) for 501 compounds was calculated, based on potency ratios, i.e., half inhibitory concentrations (IC50), against the main target MAO-B and related target protein MAO-A: SI = IC50MAO-A/IC50MAO-B where IC50MAO-A stands for the affinity of the “ligand A” against MAO-A and IC50MAO-B is the affinity of the same ligand for MAO-B. The lowest threshold of SI for selective MAO-B inhibitors was set at 100, whereas the highest SI limit for nonselective inhibitors was established at 10. Based on this, we have: (1) 103 compounds showing IC50 < 1000 nM against MAO-B and SI > 100 designated as selective; and (2) 377 compounds presenting SI < 10 denoted as nonselective. The Bemis–Murcko scaffolds (BMF) [21] of MAO-A/B inhibitors were extracted with the help of InstantJChem (Table 1), resulting in 20 unique BMF for selective and 97 for nonselective MAO-B inhibitors (Figs. 2, 3). Since the number of unique BMF is low, we extended the number of ligands per framework such as the highest BMF/compound ratio is of 2 (Table 1) [18]. The binary selectivity sets (Table 1) were assembled according to selectivity class assignment, resulting in 33 selective and 150 nonselective MAO-B inhibitors. The distribution of biological activities for selective and nonselective MAO-B inhibitors is normal, showing the pIC50 maxima shifted toward higher values for selectives (pIC50 = 8) and to lower values (pIC50 = 5.5) for nonselectives (supplementary material). The SI gap between selective and nonselective MAO-B inhibitors extending 90 units guarantees the absence of boundary issues regarding the selectivity assignment (Table 1) [12]. However, several highly potent MAO-B inhibitors are not always selective, showing low nanomolar IC50 values (< 20 nM) for both MAO isoforms, e.g., CHEMBL2204752 IC50MAO-A = 6.6 nM, IC50MAO-B = 4.8 nM, CHEMBL3398531 IC50MAO-A = 14 nM, IC50MAO-B = 17 nM, and CHEMBL3398530 IC50MAO-A = 4 nM, IC50MAO-B = 20 nM (https://www.ebi.ac.uk/chembl/) [17]. The scaffolds of selective and nonselective MAO-B inhibitors are shown in Figs. 2 and 3.
Accurate validation datasets are necessary to assess virtual screening algorithms accurately, assuring the absence of the following problematic issues: (1) the lack of similarity of selectives and nonselectives regarding simple physicochemical descriptors which is denoted as “artificial enrichment” [19]; (2) low diversity of chemical space of selective class illustrated by a reduced number of chemotypes, the so-called analogue bias [22]; (3) “false negative” bias, which refers to the inclusion of active compounds into negative or decoy dataset [23]. The absence of “artificial enrichment” will be demonstrated by applying the mean (ROC-AUC) leave-one-out cross-validation with reference to low-dimensional physicochemical properties [19] (“Results and discussion” section). The extraction and multiplicity verification of BMF assures the omission of “analogue bias” in both selective and nonselective MAO-B subsets, while the “false negative” bias was excluded using experimentally validated nonselective MAO-B inhibitors. Considering the importance of the natural compounds for the development of many drugs, 400 natural products extracted from the SPECS NP database (their drug-like properties are shown in supplementary material) were used in prospective virtual screening investigation (https://www.specs.net, accessed in February 2020). Additionally, the approved DrugBank dataset was used for drug repurposing experiments to identify new possible indications of existing marketed drugs as modifying agents for Parkinson’s disease.
Assignment of ionization states and generation of tautomers
Thus, 400 natural compounds from SPECS NP and 2454 approved DrugBank dataset were prepared for docking by generating ionization states and tautomers at pH = 7.2 ± 0.2 employing LigPrep (LigPrep v.3.1, https://www.schrodinger.com/) from Schrödinger suite.
Conformer generation
Fast Rigid Exhaustive Docking (FRED), Rapid Overlay of Chemical Structures (ROCS), and Electrostatic Similarity for Lead Hopping (EON) use multiple rigid conformations to evaluate biding site complementarity and shape similarity, respectively. Therefore, the conformational space of each molecule has to be sampled before the screening. The conformers for MAO-B inhibitors included in the validation dataset, natural compounds from SPECS NP, and approved DrugBank were generated using OMEGA with default parameters (OMEGA v.2.5.1.4, OpenEye Scientific Software, Santa Fe, NM. www.eyesopen.com) [24]. Energy minimization of the conformers was done using the MMFF94 force field (http://www.eyesopen.com) [25].
3D Shape and electrostatic similarity search
Shape and chemical similarity coefficients were calculated with the help of ROCS software from OpenEye suite (ROCS v. 3.2.1.4, OpenEye Scientific Software, Santa Fe, NM. http://www.eyesopen.com) [26]. The basic concept of ROCS is that molecules are similar in shape if their volumes overlay well [26]. ROCS quantify and ranks the database molecules by means of Tanimoto-based coefficients, e.g., ShapeTanimoto (ShT), TanimotoCombo (TC), ComboScore (CS), ColorScore (CoS), ColorTanimoto (CoT), ScaledColor (SCo), Overlap (O) (Eq. 1) and Tversky-based indexes: FitTversky (FTv), FitColorTversky (FCoTv), FitTverskyCombo (FTvC), RefTversky (RTv), RefColorTversky (RCoTv), RefTverskyCombo (RTvC) (Eq. 2), which were used for further evaluation.
where I stand for the self-volume overlaps terms and the O terms denote the overlaps between molecules A and B, whereas β = 1 − α.
Electrostatic Similarity for Lead Hopping (EON) measures the electrostatic similarity of small molecules using a field-based algorithm which accounts for electrostatic potential in addition to shape (EON v. 2.2.0.5, OpenEye Scientific Software, Santa Fe, NM. http://www.eyesopen.com). EON generates two 3D electrostatic Tanimoto coefficients (ET), ET_coulomb (EON_ET_coul), and ET Poisson Boltzmann (EON_ET_pb) and a combined one ET-Combo (EON_ET_C), which sum Shape Tanimoto (EON_ET_ShT) and Poisson Boltzmann term (EON_ET_pb).
The reference molecules were chosen to include essential information, i.e., shape, structural features, related to selective interaction with MAO-B: (1) safinamide a selective MAO-B inhibitor (SAG) ((S)-(+)-2-[4-(fluorobenzyloxy-benzylamino)propionamide) (SI > 13,038, IC50MAO-B = 0.00767 μM), denoted query 1 (q1); and (2) the lowest energy conformer of selective inhibitor (2E)-2-[(5-bromofuran-2-yl)methylidene]-5-methoxy-2,3-dihydro-1H-inden-1-one (SI = 42, IC50MAO-B = 0.0044 μM) denoted query 2 (q2) [27] (Fig. 4). 3D Similarity search was applied in a retrospective manner to our in-house benchmarking validation dataset for MAO-B to identify the coefficients which yield the best AUC values, and to calculate similarity thresholds. Afterward, a 3D similarity search was applied prospectively to SPECS NP and approved DrugBank datasets to identify similar compounds that may display potentially selective inhibition against MAO-B. The workflow diagram shown in Fig. 5 compiles the stages completed in this section.
Protein preparation
The X-ray structures of MAO-A cocrystallized with harmine (HRM) (PDB ID: 2z5x) and MAO-B complex with SAG (PDB ID: 2v5z) downloaded from Protein Data Bank (https://www.rcsb.org/) were prepared for docking using MakeReceptor (MakeReceptor v.3.2.0.2, OpenEye Scientific Software, Santa Fe, NM. http://www.eyesopen.com): (i) active site boxes: 7622 Å3 and 5041 Å3, (ii) inner/outer contours: 750 Å3 and 585 Å3. In the absence of any constraints, the default parameters were involved. Water molecules of the active site that exceeded 5Å from cocrystallized ligands were deleted, considering only water molecules that may provide realistic energetic stability to the resulting protein–ligand complex [28].
Docking
To estimate the binding affinity of the investigated molecules, docking analysis was performed employing FRED software (FRED v.3.2.0.2, OpenEye Scientific Software, Santa Fe, NM. www.eyesopen.com) [29, 30]. The conformationally sampled ligands and target proteins were treated as rigid structures during the docking process. The lowest energy ten docking poses were retained for each ligand. The generated ligand–protein complexes were visually inspected by comparing the atomic coordinates of each docked ligand with the X-ray coordinates of the reference ligands, SAG, and HRM. The Chemgauss 4 (CG4) scoring function [29, 30] was used to score ligand poses placement inside the active site. The most likely binding conformations were selected based on the binding mode, significant interactions with key binding site residues, and the values of the CG4 score.
The performance of the docking experiment was checked by redocking cocrystallized ligands SAG and HRM into active sites MAO-B (PDB ID: 2v5z), and MAO-A (PDB ID: 2z5x), respectively. The root-mean-square deviation (RMSD) values between the coordinates of the atoms of cocrystallized ligands and docked poses were calculated. After acknowledging the docking accuracy, the prioritized SPECS NP and approved DrugBank datasets were docked into MAO-A and MAO-B active site. A single docked structure for each molecule was retained.
Evaluation methods
The ROC-AUC method is recurrently used to assess the effectiveness of a diagnostic test. The ROC curve is represented based on the sensitivity (Se, Eq. 3), which designates the fraction of actives predicted as actives, as a function of (1 − Sp) where Sp represents the specificity or true negative rate (the fraction of inactives predicted as inactives, Eq. 4), at any threshold [31],
where TP (true positives) designates the number of correctly predicted actives; FN (false negatives) is the number of incorrectly predicted inactives; TPR (true positive rate) represents the fraction of correctly predicted actives divided by the total number of inactives when the ith active in the ranking list is recovered. The ROC curve defining optimal distributions shows no overlap between the Se and (1 − Sp) scores for actives and, respectively, inactives. The greater the value of the AUC (Eq. 5), the better classification is achieved by the virtual screening method regarding the identification of selective compounds. A special case represents the ROC curve for a data set of actives and inactives displaying randomly distributed classes, where Se = 1 − Sp (AUC = 0.5), indicating the random assignment of the compounds.
Validation
The validation of custom made selectivity focused dataset for MAO-B was performed using the arithmetic mean of the area-under-curve (AUC) of receiver operating curve (ROC) denoted mean (ROC AUCs), by applying Leave-One-Out (LOO) Cross-Validation (CV) [32], which have to show values close to 0.5. The topological similarity between selectives and nonselectives was demonstrated by employing MACCS structural keys (https://www.knime.com/). The absence of “artificial enrichment” is proved by mean (ROC AUCs) values close to 0.5 using six basic drug-like physicochemical descriptors HBA, HBD, MW, RBN, XlogP, 2DPSA. During individual AUC values calculation, the reference molecule was eliminated from the vector containing the similarity data. This validation was necessary to demonstrate the effectiveness of the methodology used to lower the analogue biases in the course of validation dataset construction. Virtual screening experiments were validated by calculating AUCs [31] (Eqs. 5 and 6) using in-house developed software, ETICI (Evaluation Tool In ChemInformatics) [33].
ADME and toxicity related risk profiles
Although significant progress has been made in the last years, accurate prediction of several essential pharmaceutical descriptors is still a challenge, because of complex fundamental physiological mechanisms [34]. The chronic character of Parkinson’s disease demands the treatments to be lifelong and therefore the drugs have to be safe and well tolerated for long periods [1]. Therefore, ADME (absorption, distribution, metabolism, excretion) properties and the toxicity risk profile of the prioritized natural compounds from the SPECS NP database were manifold estimated using the QikProp module of Schrödinger (QikProp v. 5.2, https://www.schrodinger.com/), SwissADME [35] and Osiris Property Explorer (Osiris, http://www.organic-chemistry.org/prog/peo/). The wide variety of predicted drug-like properties are given in the Results section.
Drug repositioning
In silico approaches such as 3D similarity, pharmacophore modeling, docking [11], whether used conjointly with high-throughput screening (HTS) or independently, reported several approved drugs interacting with new multifunctional protein targets, which simplified the repositioning for other diseases [36]. Advances in rational drug repositioning for Parkinson’s disease with potential clinical uses provide a possible expeditious direction to drug discovery [1]. In the current paper, we investigate potential inhibitory activity toward MAO-B of the computationally repurposed candidates as potential disease-modifying agents for Parkinson’s disease. Drug safinamide (Fig. 4), a selective MAO-B inhibitor, used for the treatment of Parkinson’s disease since February 2015 in Europe and March 2017 in the USA, was employed as a reference molecule for the repurposing of approved DrugBank dataset.
Results and discussion
Workflow
The workflow scheme (Fig. 5) adopted in this paper includes (1) construction of selectivity-based MAO-B dataset; (2) retrospective 3D similarity search (shape, physicochemical, and electrostatics) involving our custom made selectivity oriented dataset for MAO-B; (3) identification of 3D similarity coefficients with significant AUC values; (4) calculation of similarity thresholds; (5) prospective 3D similarity search of SPECS NP and DrugBank datasets to identify novel potentially selective MAO-B inhibitors; (6) docking experiments carried out to identify specific ligand–MAO-B interactions; (7) prediction of ADMETox profile of the hits.
Validation of selectivity oriented dataset
In order to validate the benchmarking dataset, the mean (ROC AUCs) parameter, based on physicochemical descriptors and MACCS fingerprints, was derived. In the case of physicochemical descriptors, the mean (ROC AUCs) value resulted from the LOO CV procedure is 0.653. Although it cannot be evaluated as ideal, it does not provide substantial artificial enrichment and similar values can be found in the literature [32]. The 70% of BMFs identified as shared between selective and nonselective MAO-B inhibitors meet the criteria used to design the benchmark data sets for virtual screening [37]. Furthermore, we used MACCS keys to calculate the mean (ROC AUCs), which give indications about the topological similarity between selective and nonselective MAO-B inhibitors, which is satisfactory (0.604). “False negative” bias was excluded by using confirmed experimental nonselectives. Thus, the selectivity oriented benchmarking dataset for MAO-B, developed by us, is free of VS problematic biases including “analogue bias“, “artificial enrichment,” and “false negative” bias and can be employed further for evaluation of various LBVS and SBVS methods. Further, we used MAO-B selectivity focused dataset in retrospective VS experiments involving shape, physicochemical, and electrostatic 3D similarity methods.
Retrospective 3D similarity
Regardless of the origin of the 3D reference molecule structure, i.e., bioactive conformation (SAG, q1) and the lowest energy conformer ((2E)-2-[(5-bromofuran-2-yl)methylidene]-5-methoxy-2,3-dihydro-1H-inden-1-one, q2), the quality of alignments and implicitly overall enrichments yielded by ROCS are satisfactory. As can be seen in Fig. 6, the MAO-B selective inhibitors arrange more systematically on reference molecules displaying slight structural variability localized at position 3 of ring B belonging to q1 and a larger structural fluctuation of the substituent at position 4, which is responsible for the affinity for MAO-B. Similarly, the overlays on q2 show larger structural irregularities on the linker and furan ring for nonselective MAO-B inhibitors related to selective ones. The structural diversity of our custom made selectivity oriented dataset for MAO-B is higher; therefore, the overlays look more evenly than one can expect. However, the overlays on q1 are of better quality than those observed for q2.
ROCS overly of the top ten selective and nonselective MAO-B inhibitors from selectivity focused dataset against query q1 (a) and (c); against query q2 (b) and (d), ranked by TC (BIOVIA Discovery Studio, www.3dsbiovia.com)
The 3D similarity algorithm retrieves hits with diverse topologies; however, ShT values higher than 0.8 (against both q1 and q2) observed for selectives are higher (18.18%) than those observed for nonselectives (11.33%) (Fig. 7). Still, their SI values register a large variation, since the geometrical distribution of crucial molecular features gives, to a certain degree, information regarding size and shape requirements to support ligand–receptor interactions [38].
The analysis of AUC values (Fig. 8) emerging from 3D similarity searches was performed for our custom made selectivity focused MAO-B dataset in addition to queries q1 and q2. The AUCs are shown in Fig. 8, while their standard deviations are included in the supplementary material. The global discriminative performance of AUC for the selectivity focused dataset is satisfactory, showing values greater than 0.7 for FTv, FTvC, and TC (q1) and FTvC, FTv, TC, ST, CS, and FCoTv (q2). ShT enrichment is satisfactory, making differences among selective and nonselective MAO-B inhibitors, but shape similar compounds show various physicochemical properties and pharmacophore arrangements, i.e., the AUCs for CS and CoT which include “color features” are lower 0.666 and 0.582 for q1, whereas against q2 the AUCs are of 0.610 and 0.705 (Fig. 8). Although both queries q1 and q2 are selective against MAO-B (Fig. 4), they didn’t show a noticeable similarity between themselves (TC = 0.959, ShT = 0.712, CoT = 0.247). In terms of AUC q2 performs slightly better than q1, on regular basis, e.g., the highest AUC value provided by query q2 is of 0.751 (FTvC), which in the case of query q1 is of 0.741 (FTv) (supplementary material). The 3D coefficients, which yields the best enrichments for selectivity oriented dataset, include both shape and color features. This is consistent with the previous observation, which states that selectivity is determined by both shape, and pharmacophore features [38]. Excepting shape-based coefficients ShT and FTv, the coefficients which include color field, i.e., CoS, TC, FCTv, FTvC, and CS furnish more accurate rankings (Fig. 8). Tversky scores involve asymmetric comparison, which depends on the molecular size of the query MWq1= 302.349 and MWq2= 333.181 and database molecules MWaverage = 293.012 for selectives and MWaverage = 349.074 for nonselectives. Wherefore, RTv, and FTv scores fluctuate more widely than Tanimoto-based scores. FTv displayed higher AUC values compared to RTvC. The AUC corresponding to Tversky coefficients ranges 0.437 ÷ 0.741 for q1, and 0.509 ÷ 0.750 for q2. FTv-based coefficients fluctuate in the range 0.650 ÷ 0741 (q1), and 0.702 ÷ 0.751 (q2), exceeding the AUC values of RTv coefficients which range 0.437 ÷ 0.597 (q1), and 0.510 ÷ 0.617 (q2). Hence, it results that MAO-B selective inhibitors are prioritized more appropriately at the top of the ranking list by the FTv-based coefficients by applying q2.
The highest AUC values for EON electrostatic coefficients were recorded for EON-ST 0.747 (q2), 0.738 (q1), EON_ET_C 0.738 (q1), and EON_ET_pb 0.702 (q1). In the case of query q2, the maximal AUC value is only 0.683 for EON_ET_C. Further, we used shape, physicochemical and electrostatic similarity coefficients whose AUCs are higher than 0.7: TC (q1, q2), CS (q2), ShT (q2), FCTv (q2), FTv (q1, q2), FTvC (q1, q2), EON_ET_C (q1), EON_ET_pb (q1), EON_ET_ST (query q1, q2) in a prospective experiment to prioritize novel natural compounds from SPECS NP database with potential ability to inhibit selectively MAO-B. Apart from AUC calculation, we estimated the average values of 3D similarity coefficients for selective and nonselective compounds to establish the thresholds for further prospective virtual screening (supplementary material). These threshold values which register a relevant difference between selectives and nonselectives were used to select natural compounds from SPECS NP and DrugBank, whose similarity values exceed explicitly the average value for selective MAO-B inhibitors (Table 2). A total number of 310 3D similarity values from the SPECS NP database (Table 2), which satisfy the thresholds of any of six 3D coefficients relative to q1, were recorded which corresponds to 182 unique compounds. Among these 182 compounds, one compound (AI-372/20970054; 1-(2,4-dihydroxyphenyl)-2-phenylethanone) comply with all six 3D coefficient thresholds. In the case of query q2, 208 similarity values exceed at least one of the seven 3D coefficient thresholds enforced, are associated with 110 compounds (Table 2), of which two compounds, AA-504/20834003 (cardamomin, (E)-1-(2,4-dihydroxy-6-methoxyphenyl)-3-phenylprop-2-en-1-one) and AC-776/25122088 ((E)-1-(2-hydroxy-4-methoxyphenyl)-3-phenylprop-2-en-1-one), complies with all seven thresholds, including the compound which obeys the conditions for q1. Similar treatment of DrugBank database prioritized two drugs fenamisal (DB06807) and monobenzone (DB00600) which satisfy all seven thresholds in favor of q1, whereas two drugs thiohexam (DB14200), benzylparaben (DB14176) whose similarity coefficients values obey the thresholds corresponding to q2 were excluded from the study since they are allergens and show other potential adverse effects such as endocrine disruption, and oxidative DNA damage [39].
Docking outcomes
In the first step, the ability of FRED to reproduce the interactions observed in the X-ray co-crystals of SAG with MAO-B (PDB ID: 2v5z) and of HRM with MAO-A (PDB ID: 2z5x) was checked. The crystallographic structures of the ligands were extracted and redocked into the 2v5z and 2z5x binding sites. The reproduction of the conventional hydrogen bonding interaction and π-donor hydrogen bond of SAG with Gln206, and carbon–hydrogen bond with Ile199 detected in the X-ray co-crystal 2v5z was noticed. The hydrophobic interactions observed between SAG and MAO-B binding site residues occurs with Leu171 (π-sigma), Ile199, Ile316, Phe343 and Tyr398 (π-alkyl), Tyr326 (π-π T-shaped), and with Cys172 (π-sulfur) (Fig. 9). Regarding the HRM redocking, significant hydrophobic interactions between HRM and MAO-A active site residues appear with Tyr444, Tyr407, Leu337, Ile180 and Ile335 (π-alkyl), Tyr407 (π–π stacked), Phe208 and Il335 (π-sigma), and with Cys323 (π-sulfur) (supplementary material). For both redocked ligands, the specific water hydrogen bonds were reproduced. These good results were also confirmed by the very low RMSD values of 0.589 Å and 0.564 Å, respectively, calculated between the best docked pose of SAG (Fig. 9a) and HRM (supplementary material) and of their X-ray coordinates. These arguments attest that the docking procedure used in the current investigation is appropriate and can be further applied to the compounds selected by the 3D similarity method from SPECS NP and DrugBank databases.
The three compounds from the SPECS NP database AA-504/20834003, AC-776/2512208, and AI-372/20970054 which adhere to all 3D coefficient thresholds (Table 2, Fig. 10) were subjected to docking in the binding sites of MAO-B and MAO-A receptors to investigate their presumable specific interactions with MAO-B.
According to the CG4 score results, all compounds showed higher binding affinity (CG4AA-504/20834003 = − 15.149, CG4AC-776/2512208 = − 14.237, and CG4AI-372/20970054 = − 13.655) for MAO-B than for MAO-A (CG4AA-504/20834003 = − 13.019, CG4AC-776/2512208 = − 13.287, and CG4AI-372/20970054 = − 12.390). The visual analysis of the docking outcomes indicated that the orientation of the bound possesses of these three compounds (Fig. 11) closely mimic the orientations of the native ligands in the X-ray crystal structures of MAO-A (supplementary material) and MAO-B (Fig. 9).
Hence, AA-504/20834003 and AC-776/2512208 tightly bind to MAO-B by forming H-bonding interactions (1) their carbonyl group with Tyr326 and Gln206, and (2) 4-OH group of AA-504/20834003 and a 4-OCH3 group of AC-776/2512208 with Tyr435. The interaction with Tyr326 residue of MAO-B is known to be essential for the substrate/inhibitor selectivity [40]. Moreover, the Ile199 and Ile316, which are also known as critical residues for selective interaction with MAO-B [41], showed strong hydrophobic interaction with these natural compounds. The unsubstituted aromatic rings of both ligands participated in hydrophobic π-alkyl interactions with Leu164, Ile316, Ile199, and supplementary π–π-T-shaped interaction with Phe168 for AA-504/20834003. The substituted aromatic rings of both ligands are involved in π-alkyl interactions with Leu171 and Ile198. We also observed direct π-alkyl interaction (not water-mediated) with Cys172. The 4-methoxy moiety of AC-776/2512208 substituted aromatic ring made п-sigma type contacts with Tyr398 and Tyr435. Additionally, the conserved water molecules within the receptor active site were important for enhancing the interaction of these compounds with MAO-B. In this light, two water molecules mediated hydrogen bonds between both AC-776/2512208 and AA-504/20834003 and the MAO-B receptor as follow: (1) HOH1170 bridged the substituted aromatic ring of compounds with Tyr435, Ile198, Gly205, and Gln206 residues, and (2) HOH1229 bridged the carbonyl unit with Tyr326, Gln206, and Ile199. We also observed water-mediated H-bonding (HOH1155) with Cys172 and the 4-OH unit of the substituted aromatic ring of AA-504/20834003. The water-mediated hydrogen bonding interactions were similar to that of the SAG ligand.
AI-372/20970054 binds to MAO-B by forming H-bonding interactions as follows: (1) the carbonyl unit with Tyr398, and (2) the 2-OH and 4-OH of the substituted aromatic ring with Tyr435, Cys172, and Gly434, respectively. Moreover, the substituted aromatic ring of the ligand is involved in п-п-stacked interactions with Tyr398, Tyr435, and FAD cofactor. By contrast with the other two ligands, the substituted aromatic rings make two п-alkyl interactions with Leu171 and Ile198 and one п-sulfur with Cys172. The compound orientation mimics the placement of the native ligand into the active site but it is slightly displaced to FAD compared to the positioning of the other two compounds. This could be assigned to the lack of the ethylene unit and the methoxy group. Furthermore, the less prominent interactions with Tyr444, Tyr407, and Ile335 are considered to play an important role in MAO-A catalytic activity (supplementary material), allowed an increased selectivity of all three ligands to MAO-B. Likewise, Geha and co-workers [42] suggested that Tyr407 and Tyr444 in MAO-A and the corresponding Tyr398 and Tyr435 in MAO-B may form an aromatic sandwich that consolidates the substrate binding. All three ligands were situated into the aromatic cage framed by FAD aromatic ring, Tyr326, Phe343, Tyr398, and Tyr435 residues of MAO-B active site. Taken together, these results predicted all three natural compounds as selective MAO-B inhibitors. The outcomes are also in line with the continuous need of developing specific MAO-B inhibitors since they, used mainly for the treatment of Parkinson’s disease, have the advantage of less severe side effects compared to the drugs which are MAO-A inhibitors [6].
ADME and toxicity risk profiles
The SPECS NP top hits were passed through the ADME and toxicity related risks filters to evaluate their drug-like properties. Thus, passive gastrointestinal absorption (HIA) and brain permeability (BBB) properties were predicted simultaneously with SwissADME software (Fig. 12) [35]. The AC-776/25122088, AA-504/20834003, and AI-7372/20970054 are predicted as passively crossing the BBB (in the yellow region), being not subject to active efflux (red dots).
Also, six physicochemical properties such as lipophilicity, size, polarity, solubility, flexibility, and saturation are considered for Bioavailability Radar construction. A pool of descriptors defining the pink area on the radar plot of the molecule has to fall entirely in the delimited area to be considered drug-like (supplementary material). The pink area denotes the optimal range for each property (polarity: TPSA between 20 and 130 Å2, lipophilicity: XLOGP3 between − 0.7 and + 5.0, solubility: log S not higher than 6, saturation: the fraction of carbons in the sp3 hybridization not less than 0.25, size: MW between 150 and 500 g/mol, and flexibility: no more than 9 rotatable bonds. The SwissADME parameters values from Table 3 and supplementary material indicate that all compounds obey the mentioned criteria, as follows: (1) have good oral bioavailability, (2) are completely contained in the pink area, and (3) exhibit drug-like properties.
The ADME properties values predicted by QikProp software from the Schrödinger suite (Table 4) fall into the recommended ranges, indicating that all three SPECS NP have an excellent pharmacokinetic profile with high bioavailability, good water solubility and good predicted BBB permeability. Hence, we can state that the molecules investigated in the current paper exhibit drug-like properties.
OSIRIS calculations are used generally to estimate the risks of side effects as well as drug relevant properties of the compounds. Thus, AA-504/20834003 has no mutagenic, tumorigenic, irritant, or reproductive effective potential. Probably, the supplementary hydroxyl group and the migration of the methoxy unit at the 6-substitution position of the AA-504/20834003 aromatic ring compared with AC-776/25122088 reduced completely the side effects risk. AC-776/25122088 displayed only the reproductive effect risk. Compound AI-372/20970054 shows potential high risks for undesired effects like mutagenicity and irritant. Conclusively, ADME and toxicity risk profiles suggest AA-504/20834003, in comparison with the other two compounds, display excellent predicted drug-like properties and superior pharmacological profiles.
Drug repositioning of DrugBank database
To expand the chemical space of MAO-B inhibitors by repositioning of approved drugs from DrugBank [43], the same 3D similarity search scenario applied to the SPECS NP database was used. The top hits retrieved by both queries q1 and q2 include drugs used to treat several health issues such as depigmentation monobenzone (DB00600), and tuberculosis fenamisal (DB06807).
The two approved drugs (Fig. 13) prioritized by all 3D coefficients (Table 2) were further docked in the active site of MAO-B (2v5z). Ten lowest energy drug poses were analyzed to identify the pose with the lowest docking score and significant interactions with binding site residues, which can illustrate their possible activity on MAO-B.
DB06807 (fenamisal) compared with DB00600 (monobenzone) was found to be the best molecule based on its lowest docking score (CG4DB06807 = − 13.039 and CG4DB00600 = − 12.243). Despite the structural differences that contribute to slightly different placement in the receptor active site, the approved drugs displayed interesting molecular interactions with the MAO-B receptor (Fig. 14) suggesting a potential role of their skeletal structure regarding the presumable activity against MAO-B. The fenamisal orientation mimics the arrangement of the SAG in the active site but it is slightly displaced toward FAD compared to the positioning of the monobenzone which simulates perfectly the SAG alignment. In terms of structural and MAO-B interactions, fenamisal behaves similarly to AI-372/20970054. Both approved drugs make π–π T-shaped interaction with Tyr326 residue which is essential for MAO-B selectivity [40]. Likewise, strong hydrophobic interactions of π-alkyl type were achieved between the unsubstituted ring of monobenzone with Ile199, Leu164, and Ile316 residues, also known to be critical for MAO-B selectivity [41]. Additional π–π-stacked interactions with Tyr398 and Tyr435 and donor H-bond with Gln206 were observed for fenamisal. The latter was also made by monobenzone. Moreover, fenamisal binds to MAO-B by forming H-bond interactions (1) at carbonyl unit with Tyr398 and (2) at the 2-OH unit with Tyr435. We observed that HOH1155 connected Cys172 and Tyr435 residues to the 2-OH unit of fenamisal substituted aromatic ring. Contacts of π-sigma type with Leu171 and of π-sulfur type with Cys172 were realized by both drugs.
In the absence of experimental values for ADME and BBB permeation fenamisal and monobenzone have been subjected to ADME and toxicity risk filters as the hits originating from the SPECS NP database to allow a direct comparison of predicted properties. The passive gastrointestinal absorption (HIA) and brain permeability (BBB) computed simultaneously with SwissADME (Fig. 12) indicated both approved drugs to passively permeate the BBB (yellow region) and not to be effluated from the CNS (red dot). The lipophilicity, size, polarity, solubility, flexibility, and saturation used for Bioavailability Radar construction were located in the delimited pink area on the radar plot, pointing that both approved drugs respect the drug-like properties (Table 3 and supplementary material). As for SPECS NP, the OSIRIS Property Explorer was used to check the potential toxicity of these candidates. Toxicity screening results presented in Table 3 showed that both drugs pose a high risk of side effects like tumorigenicity. Furthermore, fenamisal presents a high risk of mutagenicity while monobenzone poses a high irritant effect.
In short, our drug repurposing experiment is able to identify two drugs that can be repositioned for Parkinson’s disease. Because these drugs have indications for other diseases, in the first step they have to be assessed for BBB penetration in vivo [1] and if proved so, then clinically tested for their efficacy in Parkinson’s disease treatment. Athauda et al. reviewed the safety and potential efficacy of several promising candidates repurposed for Parkinson’s disease which undergoes the latter stages of clinical testing. They pointed out that drugs with no indication for the central nervous system, e.g., ambroxol, deferiprone, etc. cross the blood–brain barrier [1]. However, fenamisal and monobenzone have low molecular weight, low hydrogen bond donors and acceptors, and high lipid solubility, which facilitates the passage of the lipid membrane [20].
Conclusions
In this study, 3D similarity methods including shape, physicochemical, and electrostatics were engaged in a retrospective calibration experiment to ascertain the most appropriate similarity coefficients and estimate their associated thresholds for prospective screening. A novel standard validation dataset for MAO-B focused on selectivity, validated by mean (ROC-AUC) cross-validation procedure based on MACCS keys and physicochemical descriptors, was generated. Nine 3D similarity coefficients showing AUC > 0.7 in retrospective virtual screening experiment, containing various levels of structural information, were further involved in a prospective screening experiment on SPECS NP and Drug Bank databases using a consensus scheme. The interactions of the prioritized hits including three natural compounds and two approved drugs with MAO-B were confirmed by docking as potentially selective inhibitors for MAO-B. From these natural compounds, AA-504/20834003 exhibited great predicted drug-like properties, an excellent pharmacological profile, and specific interactions with the MAO-B binding site. Moreover, drug reposition was used to explore the efficacy of already marketed drugs fenamisal, and monobenzone that could potentially be repurposed for treating Parkinson’s disease. We assume that onward development of this kind of investigation for prioritization of potentially selective inhibitors for closely related enzymes represents an advantage for the drug discovery process.
Abbreviations
- AUC:
-
Area under the curve
- BMF:
-
Bemis–Murcko framework
- CG4:
-
Chemgauss4
- CS:
-
ComboScore
- CoS:
-
ColorScore
- CoT:
-
ColorTanimoto
- FCoTv:
-
FitColorTversky
- FTv:
-
FitTversky
- FTvC:
-
FitTverskyCombo
- FRED:
-
Flexible ligand–rigid protein docking
- HBA:
-
Hydrogen bond acceptor
- HBD:
-
Hydrogen bond donor
- HRM:
-
Harmine
- LB:
-
Ligand based
- MAO-A:
-
Monoamine oxidase A
- MAO-B:
-
Monoamine oxidase B
- MAOIs:
-
Monoamine oxidase inhibitors
- SPECS NP:
-
SPECS natural products
- O:
-
Overlap
- PDB:
-
Protein Data Bank
- q1 :
-
Query 1
- q2 :
-
Query 2
- RBN:
-
Rotatable bond
- RMSD:
-
Root mean squared deviation
- ROC:
-
Receiver operating characteristic
- ROCS:
-
Rapid overlay of chemical structures
- RCoTv:
-
RefColorTversky
- RTv:
-
RefTversky
- RTvC:
-
RefTverskyCombo
- SB:
-
Structure based
- SAG:
-
Safinamide
- ShT:
-
ShapeTanimoto
- SCo:
-
ScaledColor
- TC:
-
TanimotoCombo
- VS:
-
Virtual screening
References
Athauda D, Foltynie T (2018) Drug repurposing in Parkinson’s disease. CNS Drugs 32(8):747–761. https://doi.org/10.1007/s40263-018-0548-y
Mittal R, Debs LH, Patel AP, Nguyen D, Patel K, O’Connor G, Grati M, Mittal J, Yan D, Eshraghi AA, Deo SK, Daunert S, Liu XZ (2017) Neurotransmitters: the critical modulators regulating gut-brain axis. J Cell Physiol 232:2359–2372. https://doi.org/10.1002/jcp.25518
Tripathi RKP, Ayyannan SR (2019) Monoamine oxidase-B inhibitors as potential neurotherapeutic agents: an overview and update. Med Res Rev. https://doi.org/10.1002/med.21561
Ugun-Klusek A, Theodosi TS, Fitzgerald JC, Burté F, Ufer C, Boocock DJ, Yu-Wai-Man P, Bedford L, Billet EE (2019) Monoamine oxidase-A promotes protective autophagy in human SH-SY5Y neuroblastoma cells through Bcl-2 phosphorylation. Redox Biol 20:167–181. https://doi.org/10.1016/j.redox.2018.10.003
Yeung AWK, Georgieva MG, Atanasov AG, Tzvetkov NT (2019) Monoamine oxidases (MAOs) as privileged molecular targets in neuroscience: research literature analysis. Front Mol Neurosci 12:143. https://doi.org/10.3389/fnmol.2019.00143
Finberg JPM, Rabey JM (2016) Inhibitors of MAO-A and MAO-B in psychiatry and neurology. Front Pharmacol 7:340. https://doi.org/10.3389/fphar.2016.00340
Bette S, Shpiner DS, Singer C, Moore H (2018) Safinamide in the management of patients with Parkinson’s disease not stabilized on levodopa: a review of the current clinical evidence. Ther Clin Risk Manag 14:1737–1745. https://doi.org/10.2147/TCRM.S139545
Tanrikulu Y, Krüger B, Proschak E (2013) The holistic integration of virtual screening in drug discovery. Drug Discov Today 18(7–8):358–364. https://doi.org/10.1016/j.drudis.2013.01.007
Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, Lavan P, Weber E, Doak AK, Côté S, Shoichet BK, Urban L (2012) Large-scale prediction and testing of drug activity on side-effect targets. Nature 486(7403):361–367. https://doi.org/10.1038/nature11159
Crisan L, Pacureanu L, Bora A, Avram S, Kurunczi L, Simon Z (2013) QSAR study and molecular docking on indirubin inhibitors of Glycogen Synthase Kinase-3. Cent Eur J Chem 11(1):63–77. https://doi.org/10.2478/s11532-012-0133-z
Crisan L, Avram S, Pacureanu L (2017) Pharmacophore-based screening and drug repurposing exemplified on glycogen synthase kinase-3 inhibitors. Mol Divers 21(2):385–405. https://doi.org/10.1007/s11030-016-9724-5
Stumpfe D, Lounkine E, Bajorath J (2011) Molecular test systems for computational selectivity studies and systematic analysis of compound selectivity profiles. Methods Mol Biol 672:503–516. https://doi.org/10.1007/978-1-60761-839-3_20
Bautista-Aguiler OM, Esteban G, Bolea I, Nikolic K, Agbaba D, Moraleda I, Iriepa I, Samadi A, Soriano E, Unzeta M, Marco-Contelles J (2014) Design, synthesis, pharmacological evaluation, QSAR analysis, molecular modeling and ADMET of novel donepezil-indolyl hybrids as multipotent cholinesterase/monoamine oxidase inhibitors for the potential treatment of Alzheimer’s disease. Eur J Med Chem 75:82–95. https://doi.org/10.1016/j.ejmech.2013.12.028
Ogunrombi MO, Malan SF, Terre’Blanche G, Castagnoli N Jr, Bergh JJ, Petzer JP (2008) Structure-activity relationships in the inhibition of monoamine oxidase B by 1-methyl-3-phenylpyrroles. Bioorg Med Chem 16(5):2463–2472. https://doi.org/10.1016/j.bmc.2007.11.059
Ferreira LLG, Santos RN, Oliva G, Andricopulo AD (2015) Molecular docking and structure-based drug design strategies. Molecules 20:13384–13421. https://doi.org/10.3390/molecules200713384
Pacureanu L, Avram S, Crisan L (2020) Comprehensive investigation of selectivity landscape of glycogen synthase kinase-3 inhibitors. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1747544
Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Kruger FA, Light Y, Mak L, McGlinchey S, Nowotka M, Papadatos G, Santos R, Overington JP (2014) The ChEMBL bioactivity database: an update. Nucleic Acids Res 42:D1083–D1090. https://doi.org/10.1093/nar/gkt1031
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 55(14):6582–6594. https://doi.org/10.1021/jm300687e
Lagarde N, Zagury J-F, Montes M (2015) Benchmarking data sets for the evaluation of virtual ligand screening methods: review and perspectives. J Chem Inf Model 55:1297–1307. https://doi.org/10.1021/acs.jcim.5b00090
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23(1):3–25. https://doi.org/10.1016/S0169-409X(96)00423-1
Bemis GW, Murcko MA (1996) The properties of known drugs. 1. Molecular frameworks. J Med Chem 39(15):2887–2893. https://doi.org/10.1021/jm9602928
Good AC, Oprea TI (2008) Optimization of CAMD techniques 3. Virtual screening enrichment studies: a help or hindrance in tool selection. J Comput Aided Mol Des 22(3–4):168–178. https://doi.org/10.1007/s10822-007-9167-2
Réau M, Langenfeld F, Zagury J-F, Lagarde N, Montes M (2018) Decoys selection in benchmarking datasets: overview and perspectives. Front Pharmacol 9:11. https://doi.org/10.3389/fphar.2018.00011
Hawkins PCD, Skillman AG, Warren GL, Ellingson BA, Stahl MT (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the protein databank and Cambridge structural database. J Chem Inf Model 50(4):572–584. https://doi.org/10.1021/ci100031x
Halgren TA (1999) MMFF VI. MMFF94s option for energy minimization studies. J Comput Chem 20(7):720–729. https://doi.org/10.1002/(SICI)1096-987X(199905)20:7%3c720:AID-JCC7%3e3.0.CO;2-X
Hawkins PCD, Skillman AG, Nicholls A (2007) Comparison of shape-matching and docking as virtual screening tools. J Med Chem 50(1):74–82. https://doi.org/10.1021/jm0603365
Nel MS, Petzer A, Petzer JP, Legoabe LJ (2016) 2-Heteroarylidene-1-indanone derivatives as inhibitors of monoamine oxidase. Bioorg Chem 69:20–38. https://doi.org/10.1016/j.bioorg.2016.09.004
Binda C, Wang J, Pisani L, Caccia C, Carotti A, Salvati P, Edmondson DE, Mattevi A (2007) Structures of human monoamine oxidase B complexes with selective noncovalent inhibitors: safinamide and coumarin analogs. J Med Chem 50:5848–5852. https://doi.org/10.1021/jm070677y
McGann M (2011) FRED pose prediction and virtual screening accuracy. J Chem Inf Model 51(3):578–596. https://doi.org/10.1021/ci100436p
McGann M (2012) FRED and HYBRID docking performance on standardized datasets. J Comput Aided Mol Des 26(8):897–906. https://doi.org/10.1007/s10822-012-9584-8
Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143(1):29–36. https://doi.org/10.1148/radiology.143.1.7063747
Xia J, Jin H, Liu Z, Zhang L, Wang XS (2014) An unbiased method to build benchmarking sets for ligand-based virtual screening and its application to GPCRs. J Chem Inf Model 54(5):1433–1450. https://doi.org/10.1021/ci500062f
Avram SI, Crisan L, Bora A, Pacureanu LM, Avram S, Kurunczi L (2013) Retrospective group fusion similarity search based on eROCE evaluation metric. Bioorg Med Chem 21(5):268–1278. https://doi.org/10.1016/j.bmc.2012.12.041
Cabrera-Pérez MÁ, Pham-The H (2018) Computational modelling of human oral bioavailability: What will be next? Expert Opin Drug Discov 13(6):509–521. https://doi.org/10.1080/17460441.2018.1463988
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7(1):42717. https://doi.org/10.1038/srep42717
Talevi A (2015) Multi-target pharmacology: possibilities and limitations of the “skeleton key approach” from a medicinal chemist perspective. Front Pharmacol 6:205. https://doi.org/10.3389/fphar.2015.00205
Rohrer SG, Baumann K (2009) Maximum unbiased validation (MUV) data sets for virtual screening based on PubChem bioactivity data. J Chem Inf Model 49(2):169–184. https://doi.org/10.1021/ci8002649
Kumar A, Zhang KYJ (2018) Advances in the development of shape similarity methods and their application in drug discovery. Front Chem 6:315. https://doi.org/10.3389/fchem.2018.00315
Lee Y-H, Lin Y-C, Feng C-H, Tseng W-L, Lua C-Y (2017) A derivatization-enhanced detection strategy in mass spectrometry: analysis of 4-hydroxybenzoates and their metabolites after keratinocytes are exposed to UV radiation. Sci Rep 7:39907. https://doi.org/10.1038/srep39907
Son S, Ma J, Kondou Y, Yoshimura M, Yamashita E, Tsukihara T (2008) Structure of human monoamine oxidase A at 2.2-Å resolution: the control of opening the entry for substrates/inhibitors. Proc Natl Acad Sci USA 105(15):5739–5744. https://doi.org/10.1073/pnas.0710626105
Hubálek F, Binda C, Khalil A, Li M, Mattevi A, Castagnoli N, Edmondson DE (2005) Demonstration of isoleucine 199 as a structural determinant for the selective inhibition of human monoamine oxidase B by specific reversible inhibitors. J Biol Chem 280(16):15761–15766. https://doi.org/10.1074/jbc.M500949200
Geha RM, Chen K, Wouters J, Ooms F, Shih JC (2002) Analysis of conserved active site residues in monoamine oxidase A and B and their three-dimensional molecular modeling. J Biol Chem 27(19):17209–17216. https://doi.org/10.1074/jbc.M110920200
Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M (2008) DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 36:D901–D906. https://doi.org/10.1093/nar/gkm958
Acknowledgements
The authors thank ChemAxon Ltd. (Marvin Sketch and Instant JChem), OpenEye Ltd., and BIOVIA software Inc. (Discovery Studio Visualizer) for providing academic license, and Dr. Simona Funar-Timofei, “Coriolan Dragulescu” Institute of Chemistry for providing access to STATISTICA software. The authors wish to thank Schrödinger Inc. for providing an academic trial license to complete the calculations for this paper. Project No. 1.2 of the “Coriolan Dragulescu” Institute of Chemistry, Romanian Academy, Timisoara, financially supported the current work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors indicate no potential conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Crisan, L., Istrate, D., Bora, A. et al. Virtual screening and drug repurposing experiments to identify potential novel selective MAO-B inhibitors for Parkinson’s disease treatment. Mol Divers 25, 1775–1794 (2021). https://doi.org/10.1007/s11030-020-10155-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-020-10155-6