Abstract
Currently available treatments for Parkinson’s disease offer limited symptomatic relief to patients and do not halt the progress of the disease. Several studies have reported that the use of a multi-targeting approach for treating neurodegenerative diseases may prove more beneficial for patients. In this study, two successful drug targets for treating Parkinson’s disease known for their potential to slow down neuronal loss were selected. Natural products have long been known to be effective and safe in treating various diseases. Therefore, we used computational approaches to screen the North, East, and South African Natural Products databases for novel dual-targeting drug candidates against MAO-B and AA2AR. A hybrid virtual screening was performed through pharmacophore modelling and molecular docking followed by ADME/Toxicity evaluation. Our results revealed two furanoisoflavones with equally favourable binding affinities and interaction profiles for MAO-B and AA2AR as well as desirable pharmacokinetic properties for drugs acting on the brain. Molecular dynamics simulations were conducted to assess the stability of the lead compounds and the reference drugs over time. The findings emphasized the notable stability of the suggested drugs in comparison to safinamide. Notably, 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone established a crucial hydrogen bond with Gln-206, a characteristic interaction observed in most MAO-B inhibitors. Regarding AA2AR, while the interaction strength with Asn-253 may not match that of the reference drug, simulation results indicated a parallel trend in protein interaction, suggesting its potential as an antagonist. The findings from this study could potentially act as starting points for refining and developing natural products into disease-modifying remedies for Parkinson’s disease patients. Nevertheless, it is imperative to conduct experimental assays to substantiate these discoveries.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Parkinson’s disease (PD) is second only to Alzheimer’s disease in prevalence [1]. The progressive loss of dopamine-producing neurons in the brain is thought to have a complex nature involving multiple pathways such as dopamine metabolism, oxidative phosphorylation, the ubiquitin-proteasome system, and mitochondrial dysfunction [2]. The development of dual-targeting ligands for monoamine oxidase B (MAO-B) and adenosine A2A receptor (AA2AR) has garnered significant attention in recent years, supported by a growing body of computational and experimental studies [3,4,5]. Computational approaches, including molecular docking, structure-activity relationship (SAR) analyses, and pharmacokinetic property predictions, have been instrumental in elucidating the structural determinants for dual MAO-B/AA2AR inhibition and identifying promising lead compounds. Molecular docking has revealed key binding interactions and insights into selectivity, while SAR studies have identified critical structural modifications, such as substitutions at the C8 position of caffeine, that enhance dual activity [3]. Virtual screening campaigns, leveraging the structural information from computational studies, have facilitated the discovery of novel scaffolds with potent dual MAO-B/AA2AR inhibitory activity. Additionally, molecular dynamics simulations have assessed the stability of ligand-receptor complexes, refining our understanding of the molecular interactions involved [4]. Experimental validation of the dual-targeting concept has yielded promising results. Several studies have reported the development of potent dual MAO-B/AA2AR inhibitors based on various scaffolds, including caffeine analogues, indanone derivatives, and 2-aminopyridine-3-carbonitriles [3, 6]. Notably, C8-substituted caffeine analogues containing a 4-phenylbutadiene moiety have demonstrated remarkable potency and selectivity for both targets. These findings highlight the potential of rational drug design approaches in the development of novel therapeutic agents for Parkinson’s disease and other neurological disorders. Furthermore, existing drugs like safinamide (primarily a MAO-B inhibitor) has been shown to exhibit off-target activity against the other receptor, suggesting the potential for repurposing this compound for dual-targeting therapy. The most recent medication approved for PD treatment is istradefylline, which is often used as an adjunctive treatment in combination with levodopa/carbidopa for people with PD who experience motor fluctuations despite standard therapy [7]. Istradefylline acts as a potent AA2AR antagonist with a Ki value of 2.2 nM [8]. Studies have shown that it has MAO-B inhibitory activity, although in the micromolar range (IC50 = 28 µM) which prompts the search for novel potent compounds [9]. MAO-B is another relevant target in PD, as its blockade increases dopamine in the brain while also contributing to a reduced production of reactive oxygen species (ROS), such as hydrogen peroxide (H2O2) which results in mitochondrial dysfunction [10].
MAO is a mitochondrial flavin adenine dinucleotide (FAD)-dependent enzyme that catalyzes the degradation of some neurotransmitters such as dopamine [11]. It is expressed in two isoforms: MAO-A and MAO-B, sharing 70% of sequence similarity [12]. The development of MAO-A inhibitors has been abandoned due to their tyramine-related side effects such as cardiovascular and hypertensive crisis [13, 14]. Therefore, a new generation of selective MAO-B inhibitors has been of interest for a while, and they have proven to be relevant, especially when considering the age-related increase in MAO-B activity in parkinsonian brains [14].
AA2AR is considered a relevant target in PD, given the fact that its blockade increases dopamine signaling, which otherwise would be inhibited by adenosine [15]. Moreover, an experimental study has shown that AA2AR antagonists may prevent the loss of dopaminergic neurons, suggesting their neuroprotective activities [16].
Natural products have long been used in traditional medicine for their therapeutic potential and structural diversity [17]. Numerous studies have demonstrated the potent MAO inhibitory activity of various herbal compounds, including flavonoids, xanthones, coumarins [18]. These natural compounds have emerged as promising sources for the development of novel synthetic MAO inhibitors [19].
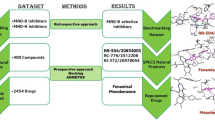
The present study aims to search for novel potent natural compounds as dual MAO-B inhibitors/AA2AR antagonists that could exert combined symptomatic relief and neuroprotective activities for PD patients. The selection of African natural products is driven by their diverse chemical compositions and historical use in traditional medicine, presenting compelling prospects for uncovering novel therapeutic agents. This approach not only aims to advance drug discovery initiatives but also highlights the significance of biodiversity conservation in exploring new treatments for complex neurological conditions such as Parkinson’s disease. A multi-stage virtual screening approach, combining pharmacophore, docking-based, and ADME/Toxicity screening, was conducted to study at the molecular level the interactions of natural products from North, East, and South Africa with the structures of MAO-B and AA2AR (Fig. 1). The stability of the lead compounds was further assessed through 100 ns molecular dynamics simulations and compared to the reference drugs.
2 Materials and Methods
2.1 Protein Preparation and Grid Generation
The crystallographic structures of MAO-B (Chain A in complex with safinamide, SAG) and AA2AR (in complex with ZM-241385, ZMA) were fetched and retrieved from the RCSB Protein Data Bank (https://www.rcsb.org/) at a resolution of 1.7 Å, with PDB IDs: 2V5Z and 5IU4 respectively [20, 21]. Co-crystallized ligands and water molecules were removed from the proteins as they were not involved in the ligand binding. The target proteins were optimized using protein preparation wizard in Maestro 12.5 [22]. Missing loops and side-chain atoms were filled using Prime [23,24,25]. Explicit hydrogens were added to the structures. Protonation states were optimized for all residues using PROPKA at pH = 7.0 [26]. Receptor grid generation was used to generate the active site grid box with a size of 15 Å at the centroid of the active site residues formed by: Tyr-60, Pro-102, Pro-104, Leu-164, Phe-168, Leu-171, Cys-172, Ile-198, Ile-199, Gln-206, Ile-316, Tyr-326, Phe-343, Tyr-398 and Tyr-43 for MAO-B [27], and Asn-253, Ser-277, His-278, Thr-88, Phe-168, Glu-169, Met-177, Leu-249, Ile-274 for AA2AR [28].
2.2 Ligands’ Database Preparation
The chemical structures of 6,511 ligands were retrieved and downloaded from the Northern and Eastern African Natural Products Databases (NANPDB and EANPDB) in SDF-3D format [29, 30]. A total of 1,017 natural compounds were retrieved from the South African Natural Compounds Database (SANCDB) which is a free database containing compounds isolated from the plant and marine life in and around South Africa [31]. Possible ionization states were generated for all ligands at physiological pH of 7.0 ± 2.0 using LigPrep module of Maestro 12.5 and OPLS3e force field [32]. QikProp module was employed to calculate physicochemical properties and assess the drug-likeness of the selected ligands [33]. Primary filtration was conducted based on Lipinski’s rule of five to eliminate all compounds that present any violation of the five rules of orally active drugs [34, 35].
2.3 Pharmacophore Modelling
Pharmacophore modelling was conducted using the Phase module of Maestro 12.5 [36]. Twenty-five known inhibitors of MAO-B were sourced from ChEMBL database (Table S1 in Supporting Information) [37]. Selected inhibitors were prepared with LigPrep at the default settings and added to the Phase module to develop pharmacophore models, with hypothesis settings set to match 25% and a preferred minimum number of features of five. As for AA2AR, a five-point pharmacophore model previously published within our group was used as a query for virtual screening [28].
2.4 Structure-based Virtual Screening
Structure-based virtual screening of the filtered ligands was conducted against MAO-B and AA2AR by employing Glide module in Maestro 12.5 using Glide SP (standard precision) docking protocol with default parameters [38]. To further reduce the number of obtained hits, the top-ranking compounds which displayed a binding score of − 10.0 kcal/mol or less were then subject to a molecular docking study using Extra precision (XP) in Glide module [39].
2.5 ADME/Toxicity Properties Prediction
In silico ADME/Toxicity prediction is a rapid tool that can help to identify drug-like compounds by calculating their pharmacokinetic parameters, physicochemical properties, and toxicity profiles. This can significantly reduce the time and resources required for the overall drug development process. In the present study, the QikProp tool was used to predict pharmacokinetic parameters such as water solubility, human oral absorption, central nervous system (CNS) activity, brain/blood partition coefficient (QPlogBB), human serum albumin binding, and Madin-Darby Canine Kidney (MDCK) cell permeability [33, 40] Organ toxicity and toxicity endpoints were predicted using ProTox-3.0 web server for the prediction of toxicity of chemicals [41].
2.6 Molecular Dynamics Simulations
Two promising compounds with high affinity for MAO-B) and AA2AR, along with favourable pharmacokinetic properties, were selected for molecular dynamics (MD) simulations to investigate and compare the stability of their protein-ligand complexes over time relative to reference drugs. The Desmond module of Schrödinger’s suite (2020-3) was employed to perform 100 ns MD simulations [42, 43]. Utilizing the System Builder panel within Desmond, a water-soaked solvated system was constructed for each protein. The OPLS3e force field was employed, and for both proteins, a 10 Å orthorhombic box was employed in conjunction with the Single Point Charge (SPC) solvent model. Randomly adding enough counterions (Na+ and Cl−) neutralized the system, and the addition of 0.15 M NaCl maintained an isosmotic state. The OPLS3e force field parameters were utilized as the default Desmond protocol to subject the solvated model system to energy minimization [44, 45]. Subsequently, the system was equilibrated throughout the simulation duration via the NPT ensemble at a constant temperature of 300 K and a pressure of 1 atm, employing the Nose-Hoover thermostat algorithm and Martyna-Tobias-Klein Barostat algorithm, respectively [46]. A total of 100 ns simulations were conducted, during which 1,000 frames were recorded. Finally, the Simulation Interaction Diagram (SID) tool was used to analyse the MD simulation trajectory [47]. The Prime module was also utilized to perform MM-GBSA free energy calculations [48] to ascertain the relative binding free energies and energy characteristics of individual ligand, receptor, and complex structures that influence overall binding energies following the formula below:
Where, ΔGsolv represents the difference in GBSA solvation energy between the protein-ligand complex and the combined solvation energies of the apo protein and the ligand.
ΔEMM represents the difference in minimized energies between the protein-ligand complex and the combined energies of the apo protein and the ligand.
ΔGSA represents the difference in surface area energies between the complex and the combined surface area energies of the apo protein and the ligand.
3 Results
3.1 Pharmacophore Model Validation
Pharmacophore hypotheses were generated using Phase module for MAO-B (Table S2 in Supporting Information). The most accurate pharmacophore model consisted of one hydrophobic group, two aromatic rings and two hydrogen bond acceptors, as for AA2AR a previously pharmacophore model within our group consisting of three aromatic rings, one hydrogen bond donor and one hydrogen bond acceptor was selected as shown in Fig. 2. Pharmacophore hypotheses for MAO-B were validated using 68 active ligands and 6,931 decoys downloaded from the DUD-E database (https://dude.docking.org/) [49]. The performance of the best pharmacophore models is depicted in Table 1. The receiving operating characteristic (ROC) plots and percentage screening results are shown in Figure S1 in Supporting Information. The prepared compounds were initially screened using the generated MAO-B pharmacophore model to remove the compounds that did not match the pharmacophoric sites. This reduced the dataset to 942 compounds, which were then screened using the AA2AR pharmacophore. At this stage, 426 out of the 4,201 compounds were retained.
Proposed 3D-pharmacophore models to screen for MAO-B inhibitors (A) and AA2AR antagonists (B). Hypothesis consisted of 5 pharmacophoric features: one hydrophobic group, two aromatic rings and two hydrogen bond acceptors for MAO-B; three aromatic rings, one hydrogen bond donor and one hydrogen bond acceptor for AA2AR
3.2 Glide Molecular Docking Results
The accuracy of the Glide docking protocol was assessed by redocking the native ligands and comparing the resulting poses to the experimental conformations as illustrated in Fig. 3. The RMSD values between the native and redocked structures were calculated, resulting in values of 1.26 Å for safinamide and 1.67 Å for ZM-241,385. These low RMSD values indicate a good agreement between the predicted and experimental binding poses, thereby validating the reliability of the Glide docking protocol. Structure-based virtual screening was conducted on the selected compounds against MAO-B and AA2AR active sites using SP mode in Glide. Twenty compounds displayed a docking score of − 10.0 kcal/mol or lower against the selected protein targets. Glide Extra Precision (XP) mode was employed to further predict and score the orientations of each drug with respect to the binding sites of MAO-B and AA2AR. The compounds were ranked by their docking scores for both targets, then a consensus rank was calculated for each compound as shown in Table 2. The chemical structures of the retained compounds are displayed in Fig. 4. Five compounds which displayed binding scores of − 7.0 kcal/mol or higher were omitted from the results.
3.3 ADME/Toxicity Evaluation Results
Table 3 summarizes the ADME properties of the selected natural products as predicted by QikProp. All compounds exhibit aqueous solubility values within the recommended range (− 6.5 to 0.5), aligning with 95% of known drugs. Additionally, the natural compounds demonstrate superior predicted human oral absorption compared to reference inhibitors, suggesting enhanced bioavailability. Predicted CNS permeability displayed the highest values for eight compounds belonging mainly to isoflavones which is an important parameter to consider in the development of drugs acting on the CNS. Alternatively, brain/blood partition coefficient (logBB) was also predicted, the highest values were observed for 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone, and maximaisoflavone H since these parameters are correlated. Moreover, MDCK cells are a good mimic for the blood-brain barrier, predicted apparent MDCK cell permeability is considered great if > 500 and poor if < 25. The predicted MDCK values show that most compounds have medium to high cell permeability, particularly for 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone, and maximaisoflavone H which yielded high values of 2546.71 and 2393.63, respectively. Prediction of binding to human serum albumin (QPlogKhsa) yielded values ranging between − 0.93 and 0.68 which are within the recommended range (− 1.5 and 1.5) for 95% of known drugs.
The heatmap shown in Fig. 5 illustrates the toxicity profiles of the studied compounds across 11 toxicity endpoints. The toxicity endpoints include hepatotoxicity, neurotoxicity, nephrotoxicity, respiratory toxicity, cardiotoxicity, carcinogenicity, immunotoxicity, mutagenicity, cytotoxicity, ecotoxicity, and clinical toxicity. The legend indicates the class of toxicity and the prediction toxicity associated with each colour. Dark blue indicates high probability, while light yellow signifies low probability.
The toxicity heatmap analysis indicates that most of the studied natural products have a low likelihood of causing neurotoxicity, in contrast to the reference drugs. Cardiotoxicity appears inactive for the majority of compounds, though the probability varies. While carcinogenicity is also assessed, the probability is very low across all compounds. The most concerning endpoint is immunotoxicity, which shows a high probability of activity for the natural compounds. To confirm these findings, further in vitro cytotoxicity assays are necessary to assess the toxic effects on immune cells. Overall, certain compounds, like compound 11, exhibit fewer “active” toxiccity predictions, potentially making them safer alternatives.
3.4 Lead Candidates’ Analysis
To discover potential dual MAO-B/AA2AR inhibitors from natural products, visual inspection was conducted on the highest-ranking compounds, the latter revealed that most compounds belong to flavonoids, isoflavones, alkaloids and anthraquinones chemical classes.
Two compounds that exhibited good docking scores and favourable interactions for MAO-B and AA2AR binding sites as well as suitable ADME profiles for CNS-drugs were selected for further analysis. Binding conformations and protein-ligand interactions analysis were conducted on 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone and maximaisoflavone H using PyMOL and Ligplot + v2.2 as shown in Figs. 6 and 7 respectively [50, 51].
Literature investigation revealed the presence of 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone and maximaisoflavone H in Milletia dura and Milletia oblata respectively which both belongs to the Leguminosae family and can be found in East tropical Africa as ornamental trees [52,53,54].
Analysis of the redocked safinamide revealed that it interacts through hydrogen bonding with Gln-206 as mentioned in the literature [3]. Various hydrophobic interactions were observed consisting of the following residues: Pro-102, Trp-119, Leu-164, Leu-167, Phe-168, Leu-171, Tyr-326, Pro-103, Cys-172, Pro-104, Ile-198, Ile-199, Tyr-435, Phe-343, Tyr-398, Tyr-188. These amino acids form the entrance and substrate cavity of MAO-B binding site [55].
ZM-241,385 was also docked to MAO-B cavity to gain an insight into its mode of interaction, results show a docking score of − 9.9 kcal/mol with a similar hydrophobic interactions profile as seen in other ligands.
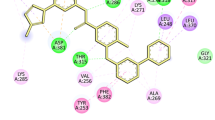
7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone is a furanoisoflavone which exhibited a docking score of − 10.5 and − 10.3 kcal/mol for MAO-B and AA2AR, respectively. The furan ring linked to the isoflavone skeleton was found to be directed towards the entrance cavity and involved in numerous hydrophobic interactions.
Maximaisoflavone H is another furanoisoflavone that is characterized by its furan ring which is linked to the bicyclic scaffold of the isoflavone skeleton. This compound interacts in a similar fashion as seen in other isoflavones but with the furan ring occupying the substrate cavity and is sandwiched by the aromatic cage forming three 𝜋-𝜋 stacking interactions with the residues Ty-398 and Tyr-435. Another 𝜋-𝜋 stacking interaction was observed through the phenyl ring and the gating residue Tyr-326 [56, 57].
Analysis of the redocking of the co-crystallized adenosine A2A antagonist, ZM-241,385, revealed that the latter is surrounded by a network of amino acids: Leu-85, Phe-168, Glu-169, Met-177, Trp-246, Leu-249, His-250, Asn-253, His-264, Leu-267, Met-270. These residues constitute the inhibitory binding cavity [58].
Upon further investigation, ZM-241,385 was found to interact with two polar residues Glu-169 and Asn-253 via the formation of non-covalent hydrogen bonds. Hydrophobic interactions also played into anchoring the antagonist, whereby an adjacent aromatic amino acid, Phe-168, was reported to form 𝜋-𝜋 stacking interactions with the heterocyclic core and the furan ring of the antagonist. These two components are deemed crucial to a good ligand-binding pocket interaction, seeing that they are responsible for mediating the receptor’s biological activity [59].
Safinamide was also docked into AA2AR binding pocket. Although results showed a docking score of − 9.1 kcal/mol, an indicating sign of a good binding affinity, safinamide showed little to no resemblance to ZM-241,385 binding profile. The inhibitor formed hydrophobic interactions with several residues from the orthosteric region of the receptor [60], this includes but not limited to: Ile-66, Val-84, Leu-85, Leu-167, Met-270, Tyr-271. While Glu-169 contributed to hydrophilic interactions via the formation of one hydrogen bond with the inhibitor’s oxygen group.
Docking results of the selected ligands revealed a score of − 10.3 and − 10.4 kcal/mol for 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone and maximaisoflavone H respectively with a similar binding mode to the native antagonist, where hydrogen bonding with Asn-253 was maintained.
The binding pattern of 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone showed that it interacted with the polar residue Asn-253 via its endocyclic oxygen atom. Besides, two additional 𝜋-𝜋 stacking interactions involving Phe-168 and Tyr-271 took place.
Although structurally different, maximaisoflavone H shared some resemblance in the binding mode to that of 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone. The ligand’s exocyclic oxygen atom was involved in the hydrogen bonding interaction with Asn-253. Regarding 𝜋-𝜋 interactions, the ligand gained an interaction with Hid-250, while maintaining two interactions with Phe-168.
3.5 Molecular Dynamics Simulations Analysis
The lead compounds obtained from the virtual screening study could be further validated using molecular dynamics simulations to assess the stability of the selected natural products with MAO-B and AA2AR under dynamical conditions. The predicted binding poses as well as the stability of 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone, and maximaisoflavone H in complex with MAO-B and AA2AR were evaluated and compared to reference inhibitors, safinamide and ZM-241,385 using 100 ns molecular dynamics simulations regarding various parameters such as root mean square deviation (RMSD), root-mean square fluctuation (RMSF), protein-ligand interactions analysis, and cross-correlation matrices.
3.5.1 Root-mean Square Deviation
The protein and ligand RMSD were evaluated to check their structural stability over time and measure the conformational changes of the given complexes as well as describe whether the simulation is in equilibrium. The RMSD plots of C-alpha atoms and the ligands with respect to the proteins are displayed in Fig. 8.
The analysis of MAO-B protein in complex with 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone and maximaisoflavone H remained significantly stable with slight structural deviations equilibrating around 2.4 Å in the last 20 ns in Complex B whereas it equilibrated much faster in Complex C after the first 40 ns. Meanwhile, the reference complex displayed higher deviations reaching an RMSD value of 3.5 Å throughout the simulation time.
However, C-alpha atoms of the reference complex A showed notable structural deviations which are stabilized at around 3.5 Å after the first 20 ns. Moreover, visual inspection of MD trajectory showed that the selected compounds are stable in the binding site demonstrating negligeable deviations with RMSD value averaging around 0.4 Å in Complex B and C in contrast to the reference inhibitor which is stabilized at around 1.5 Å.
On the other hand, the RMSD analysis of C-alpha atoms of AA2AR revealed a similar pattern in deviation which converges at 2.8 Å in the three complexes. Furthermore, an inspection of the selected ligands’ RMSD revealed that the reference inhibitor with respect to protein is averaging at around 1.2 Å. However, these atomic deviations are less noticeable in the selected compounds with an average RMSD value of 0.8 Å which can be attributed to their limited flexibility.
RMSD analysis of C-alpha atoms and ligand RMSD with respect to protein during MD simulation. (A, B, C) MAO-B in complex with safinamide, 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone, and maximaisoflavone H respectively; (D, E, F) AA2AR in complex with ZM-241,385, 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone, and maximaisoflavone H, respectively
3.5.2 Root-mean Square Fluctuation
The RMSF analysis of C-alpha atoms of the studied proteins was used to evaluate the average atomic fluctuations for a given amino acid during the simulation (Fig. 9). RMSF is an indicator that reflects the level of flexibility and the stability of any given residue during a simulation [61, 62]. Figure 9 shows that residues of the three studied ligands with MAO-B enzyme remained stable throughout the simulation time especially at the level of residues involved in ligand binding. The results show that the highest fluctuations were observed around 4.5 Å for the three complexes. However, these highly fluctuating protein residues are not involved in ligand interactions as they are in the C-terminal region which indicates that the conformational change was slight. For AA2AR, the RMSF analysis indicates a consistent pattern of fluctuations across all three complexes. Most fluctuating residues peak at 2.4 Å. However, the amino acids interacting with the ligands exhibit fluctuations around 1.2 Å, suggesting a stable binding pocket. The regions with the highest mobility are dispersed around the sequence regions, specifically residues 280–300 at the N-terminal position and residues 208–219. This heightened mobility is due to the undetermined nature of these sequence segments [63].
Protein root mean square fluctuation (RMSF) plot of MAO-B and AA2AR in complex with the selected compounds. (A, B, C) MAO-B in complex with safinamide, 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone, and maximaisoflavone H respectively; (D, E, F) AA2AR in complex with ZM-241,385, 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone, and maximaisoflavone H, respectively
3.5.3 Protein-Ligand Interactions
Monitoring the protein-ligand contacts throughout the simulation highlighted the contribution of each amino acid during the protein-ligand interactions. The protein-ligand diagrams of MAO-B complexes (Fig. 10) showed that most interactions with MAO-B active site consists of hydrophobic forces, the hydrogen bond involving Gln-206 present in the reference inhibitor, safinamide, is preserved and stronger in MAO-B complex with 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone, however this interaction is not present in maximaisoflavone H in complex C. The most important hydrophobic interactions were mainly involving Leu-171, Ile-199, Tyr-326, Tyr-398, and Tyr-435.
Figure 11 plots the different type of interactions between the binding pocket residues of AA2AR and the docked ligands. The plot provides valuable insights into the preservation of key contacts (H-bonding, hydrophobic interactions and water mediated contacts) throughout the simulation.
Complex D displays a strong H-bonding interaction involving Asn-253, an equally important π-π stacking interaction with Phe-168 and a water mediated interaction with Glu-169. The last two interactions were conserved through the simulation for maximaisoflavone H and 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone, although the latter displayed a more dampened interaction with Asn-253, maximaisoflavone H completely lost its H-bonding interaction with Asn-253. Intriguingly, another H-bonding interaction can be seen with Asn-181 in complex F, which was not depicted earlier in the molecular docking study.
Protein-ligand interactions histograms of safinamide (A), 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone (B) and maximaisoflavone H (C) in complex with MAO-B. Residues involved in the interactions are presented in the x axis, the y axis presents the normalized value of the temporal length of the interactions during the simulation
Protein-ligand interactions histograms of ZM-241,385 (D), 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone (E) and maximaisoflavone H (F) in complex with AA2AR. Residues involved in the interactions are presented in the x axis, the y axis presents the normalized value of the temporal length of the interactions during the simulation
3.5.4 MM-GBSA Free Energy Calculations
Table 4 shows the mean values and standard deviations of all free energy components including the binding free energy taken from MM-GBSA analysis for the six complexes, A-F. The binding free energy is a measure of the overall strength of the interaction between the protein and ligand. The largest contributors to the binding energy for all complexes are the electrostatic energy, polar solvation energy, and van der Waals energy. The electrostatic energy is the energy of interaction between charged atoms and is typically negative for protein-ligand complexes due to the formation of favourable electrostatic interactions. The polar solvation energy is the energy of interaction between the polar atoms of the ligand and the surrounding solvent molecules. It is typically positive, as it represents the energy required to remove the ligand from the solvent. The van der Waals energy is the energy of attraction between non-polar atoms and is typically negative for protein-ligand complexes. The other energy components, such as the covalent interaction energy and lipophilic interaction energy, are generally smaller in magnitude. The covalent interaction energy is the energy of interaction between covalently bonded atoms and is typically negative for protein-ligand complexes due to the formation of new covalent bonds. The lipophilic interaction energy is the energy of interaction between non-polar atoms and the surrounding solvent molecules. It is typically negative, as it represents the energy released when non-polar atoms are removed from the solvent. Complex B exhibited the lowest binding free energy for both MAO-B and AA2AR complexes. 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone emerged as the most stable complex energetically, with binding free energies of -139.40 ± 6.66 and − 74.23 ± 2.95 kcal/mol for MAO-B and AA2AR, respectively.
ΔGbinding: Binding energy; ΔGcoulomb: Coulombic interaction energy; ΔGcovalent: Covalent interaction energy; ΔGhbond: Hydrogen bonding energy; ΔGlipo: Lipophilic interaction energy; ΔGsolv: Solvation free energy; ΔGvdW: van der Waals energy.
3.5.5 Dynamic Cross-correlation Matrices
A contour plot of the dynamic cross-correlation matrices (DCCMs) constitutes a heatmap where atomic motions with strong correlations are represented by high correlation values [64, 65]. The DCCMs provide insights into the correlated motions between different regions of the protein-ligand complex during the MD simulations. By analyzing these matrices, we can identify regions that exhibit concerted motions, which are critical for understanding the dynamic behavior of the system and the stability of the interactions between the protein and the ligand. This information helps us pinpoint areas that are crucial for maintaining the binding affinity and overall structural integrity of the complex. Strong positive correlations between specific pairs of atoms in the DCCM indicate coordinated motions, suggesting that these atoms move together over time. Conversely, strong negative correlations indicate anti-correlated motions, suggesting that changes in one atom’s position are associated with opposite changes in another atom’s position at a later time.
The DCCMs plots in Fig. 12 show the cross-correlations between the residues of the MAO-B protein (complexes A, B, and C) and the AA2AR (complexes D, E, and F). Cross-correlations are a measure of how correlated the motions of two residues are. A high cross-correlation indicates that the two residues move together, while a low cross-correlation indicates that the two residues move independently. The plots for complexes A, B, and C show that there are strong cross-correlations between many of the residues in the MAO-B protein. This suggests that the MAO-B protein is a rigid structure, with the residues moving together as a unit. The plots for complexes D, E, and F show that there are also strong cross-correlations between many of the residues in the adenosine A2A receptor. However, the cross-correlations are not as strong as in the MAO-B protein, suggesting that the adenosine A2A receptor is a more flexible structure.
3.5.6 Principal Component Analysis
Principal Component Analysis (PCA) is a technique used to analyze the movement of biomolecular systems in MD simulations [66]. It is a dimensionality reduction technique that can be used to identify the most important features of a dataset. In the context of protein-ligand complexes, PCA can be used to identify the most important motions of the complex. The PCA plots shown in Fig. 13 show the projection of the studied complexes (A-F) onto the first two principal components (PCs). The PCs are ordered by their eigenvalues, which represent the amount of variance they explain. The first PC typically explains the most variance, followed by the second PC, and so on. In MAO-B, the three complexes exhibit distinct collective motions along the first PC, as evidenced by their separation along this axis. This separation suggests that the complexes undergo different global movements. The second PC, with its tighter range of values between − 20 and 20, likely corresponds to more localized motions, such as the movement of the ligand binding pocket. The AA2AR plot shows that while the complexes exhibit some separation along the first PC, it is less pronounced compared to MAO-B. Visual analysis reveals a similar pattern of motion between complexes D and F, forming a U shape. In contrast, complex E displays a reversed pattern, suggesting a distinct motion compared to the other two complexes.
3.5.7 Post-MD Complexes Analysis
We performed superimposition of all the investigated complexes and the binding poses of each ligand throughout the 100 ns MD simulation time. Figure 14 illustrates the superimposition of the studied MAO-B and AA2AR complexes with reference and proposed lead compounds at a 10-ns interval, respectively. Visual analysis of the proteins revealed compactness and a high level of stability throughout the simulation. Moreover, the ligands remained stable and bound within their respective binding sites. The most notable movement was noticed in maximaisoflavone H, which deviated from its initial docked conformation. A similar occurrence was found in AA2AR in complex with 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone.
Table 5 summarizes the RMSD values of the ligand poses throughout the 100 ns MD simulation. Notably, all ligands exhibited RMSD values below 2 Å, with the most significant deviations observed during the initial nanoseconds. This initial fluctuation is expected as the system adjusts to the simulation environment. However, for the remainder of the simulation, RMSD fluctuations remained consistently below 1 Å, indicating a high degree of stability for the ligand-protein complexes.
4 Discussion
The results of this work emphasize the dual-targeting potential of furanoisoflavones on MAO-B and AA2AR and may lead to a better understanding of the reported neuroprotective properties of flavonoids against Parkinson’s disease [67]. In summary, this study’s aim was the bioprospecting for small molecules found in natural sources present in North, East, and South Africa using the state-of-the-art virtual screening approaches [68]. The key contribution of this study is the computational exploration of the entire African natural products which revealed two natural compounds belonging to the isoflavone chemical class. The two drugs displayed favourable affinities for the selected targets: MAO-B and AA2AR as well as desirable pharmacokinetic properties for CNS-acting drugs. The selected candidates in complex with MAO-B and AA2AR were subject to molecular dynamics simulations and binding free energy calculations. MMGBSA binding free energy confirmed the role of van der Waals binding free energy (ΔGvdW) as a considerable contributor to the stability of MAO-B inhibitors and AA2AR antagonists as mentioned in the literature [69, 70]. The analysis of the RMSD of the selected complexes throughout the simulation time revealed that 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone with the dioxolane ring linked to the monocycle of the flavonoid scaffold is more stable when bound to MAO-B in contrast to maximaisoflavone H which displayed better stability when bound to AA2AR. This suggestion is further emphasized in the RMSF plots which displayed notable fluctuations in AA2AR when bound to 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone. The results of the protein-ligand diagrams displayed a strong presence of a hydrogen bond with Gln-206 in 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone in complex with MAO-B. Gln-206 is recognized as a hydrogen bond acceptor for most MAO-B inhibitors and contributes significantly to their stability in the substrate cavity [71]. As for AA2AR, the key hydrogen bond with Asn-253 was maintained during the whole simulation for the reference antagonist [3]. This interaction was slightly present in 7,3’-dimethoxy-4’,5’-methylenedioxyisoflavone which may explain its weak stability to AA2AR. However, for maximaisoflavone H, a new hydrogen bond with Asn-181 was observed for 20% of the simulation time. Furthermore, hydrophobic interactions were observed which conferred to the stability of maximaisoflavone H. By analyzing the DCCMs, we observed strong positive correlations between specific residues in MAO-B, indicating coordinated motions and a rigid protein structure. In contrast, the DCCMs of the AA2AR displayed weaker correlations, suggesting a more flexible structure. The PCA analysis suggests different global and local dynamics between the MAO-B and AA2AR complexes. In the MAO-B system, the distinct separation of the three complexes along the first principal component indicates different overall motions. The tighter range of values on PC2 suggests more localized movements, potentially related to the ligand binding pocket. The AA2AR system displays less pronounced separation along the first PC, indicating similarities in global motions. However, the U-shaped pattern formed by complexes D and F and the reversed pattern of complex E reveal distinct local motions within the AA2AR complexes. Finally, the visual inspection revealed strong compactness and minimal structural changes in the proteins, demonstrating their stability. While some ligands exhibited initial fluctuations during the first few nanoseconds, they quickly settled into stable binding poses with RMSD values consistently below 1 Å for most of the simulation. These findings provide strong evidence for the stability and tight interactions between the proposed lead compounds and their respective target proteins, MAO-B and AA2AR.
Recent studies have similarly investigated dual-target approaches and novel compounds for Parkinson’s disease, emphasizing the significance of our findings. For instance, one study examined the dual-targeting inhibitory potential of Phenylxanthine derivatives on MAO-B and AA2AR, demonstrating high affinities and promising pharmacokinetic properties, further supporting the dual-targeting strategy in PD therapy [72]. Another study highlighted the MAO-B inhibitory potential of rutin, a natural flavonoid, suggesting its strong binding interactions and potential as a lead compound for PD treatment [73]. Additionally, the FDA’s approval of istradefylline marks a significant advancement in PD treatment, reinforcing the importance of targeting the AA2AR [74]. Furthermore, the exploration of indanone derivatives as MAO-B inhibitors also showed promising results, supporting the development of new compounds to inhibit dopamine degradation in PD [75]. Furthermore, previous studies have explored the inhibitory effects of other isoflavones on MAO-B. For example, genistein and daidzein have been tested on MAO-B. Genistein inhibits MAO-B with an IC50 of 6.81 µM and MAO-A with an IC50 of 4.31 µM, acting as a time-independent, reversible, and competitive inhibitor of both enzymes. Genistein’ inhibition of MAO-B is more potent than its inhibition of MAO-A, and it also inhibits MAO-B tyramine oxidation and hydrogen peroxide production more effectively than MAO-A. Conversely, daidzein exhibits weak inhibitory effects on both MAO-A and MAO-B, with IC50 values of 304.05 and 356.87 µM, respectively [76]. These findings highlight that while genistein shows significant reversible inhibition of MAO-B, daidzein has minimal inhibitory effects on MAO isozymes. Although genistein is known to bind to the adenosine A1 receptor subtype, its effects on AA2AR have not been extensively studied [77]. Biochanin A is another isoflavone compound found in various plants such as red clover, chickpeas, and soybeans. Several studies have investigated the effects of Biochanin A on MAO-B. Biochanin A has been shown to be a potent, reversible, and selective inhibitor of MAO-B. In one study, Biochanin A inhibited MAO-B with an IC50 of 0.003 µg/mL, demonstrating high potency and 38-fold selectivity for MAO-B over MAO-A. The inhibition of MAO-B by Biochanin A was found to be competitive and reversible, with a low Ki of 3.8 nM [78].
To the best of our knowledge, this is the first study to investigate all the natural products isolated from African medicinal flora and marine life as drug candidates for Parkinson’s disease. However, some study limitations should be acknowledged such as lack of experimental data. This poses an area for future work that should focus on bioactivity validation of the selected compounds, in vitro blood-brain barrier permeability, and cell viability assays. Finally, future research should also focus on the further optimization of the proposed natural products into a potent and selective dual-target-directed drug for PD patients.
5 Conclusion
The present study aimed to identify novel potent compounds from natural sources capable of simultaneously inhibiting MAO-B and AA2AR. This approach holds promise for providing more effective and beneficial treatments for patients with Parkinson’s disease (PD). Among the screened natural products, two compounds with the highest binding affinities, as determined by molecular docking, and favourable ADME profiles were selected. Structural analysis revealed promising interactions with both MAO-B and AA2AR binding sites. To further assess the stability of these compounds, molecular dynamics simulations were conducted, confirming their ability to effectively bind and inhibit the action of both MAO-B and AA2AR.
The findings of this study suggest a potential shift of interest towards the development of novel antiparkinsonian drugs with neuroprotective properties derived from natural compounds. However, to validate these results, additional experimental studies such as bioactivity assays and in vitro ADME profiling are warranted.
References
Draoui A, El Hiba O, Aimrane A, Khiat AE, Gamrani H (2020) Parkinson’s disease: From bench to bedside, Rev Neurol (Paris), vol. 176, no. 7–8, pp. 543–559, Sep. https://doi.org/10.1016/J.NEUROL.2019.11.002
Tan SH et al (2019) Emerging pathways to neurodegeneration: dissecting the critical molecular mechanisms in Alzheimer’s disease, Parkinson’s disease. Biomed Pharmacotherapy 111. https://doi.org/10.1016/j.biopha.2018.12.101
Azam F, Madi AM, Ali HI (2012) Molecular docking and prediction of pharmacokinetic properties of dual mechanism drugs that block MAO-B and adenosine a 2A receptors for the treatment of Parkinson′s disease. J Young Pharmacists 4(3):184–192. https://doi.org/10.4103/0975-1483.100027
Boulaamane Y, Ibrahim MA, Britel MR, Maurady A (2022) In silico studies of natural product-like caffeine derivatives as potential MAO-B inhibitors/AA2AR antagonists for the treatment of Parkinson’s disease. J Integr Bioinform 19:20210027
Jaiteh M et al (Jun. 2018) Docking screens for dual inhibitors of Disparate Drug targets for Parkinson’s Disease. J Med Chem 61(12):5269–5278. https://doi.org/10.1021/acs.jmedchem.8b00204
Koch P et al (2018) Probing substituents in the 1- and 3-position: Tetrahydropyrazino-annelated water-soluble xanthine derivatives as multi-target drugs with potent adenosine receptor antagonistic activity, Front Chem, vol. 6, no. JUN, https://doi.org/10.3389/fchem.2018.00206
Berger AA et al (2020) Istradefylline to treat patients with parkinson’s disease experiencing ‘off’ episodes: a comprehensive review. Neurol Int 12(3). https://doi.org/10.3390/neurolint12030017
Azam F, Ibn-Rajab IA, Alruiad AA (2009) Adenosine A2A receptor antagonists as novel anti-parkinsonian agents: a review of structure-activity relationships. Pharmazie 64(12). https://doi.org/10.1691/ph.2009.9632
Kumar M, Chatterjee J, Rani D, Kumar R (2022) FDA approved five-membered ring fused pyrimidine-based derivatives and their biological properties. Fused Pyrimidine-Based Drug Discovery. https://doi.org/10.1016/B978-0-443-18616-5.00006-5
Dias V, Junn E, Mouradian MM (2013) The role of oxidative stress in parkinson’s disease. J Parkinson’s Disease 3(4). https://doi.org/10.3233/JPD-130230
Tipton KF (2018) 90 Years of Monoamine Oxidase: Some Progress and Some Confusion, J Neural Transm, vol. 125, no. 11, pp. 1519–1551, Nov. https://doi.org/10.1007/s00702-018-1881-5
Naoi M, Riederer P, Maruyama W (2016) Modulation of monoamine oxidase (MAO) expression in neuropsychiatric disorders: genetic and environmental factors involved in type A MAO expression. J Neural Transm 123. no. 210.1007/s00702-014-1362-4
Flockhart DA (2012) Dietary restrictions and drug interactions with monoamine oxidase inhibitors: An update, Journal of Clinical Psychiatry, vol. 73, no. SUPPL. 1. https://doi.org/10.4088/JCP.11096su1c.03
Foley P, Gerlach M, Youdim MBH, Riederer P (2000) MAO-B inhibitors: multiple roles in the therapy of neurodegenerative disorders? Parkinsonism Relat Disord 6(1). https://doi.org/10.1016/S1353-8020(99)00043-7
Al-Attraqchi OHA, Attimarad M, Venugopala KN, Nair A, Al-Attraqchi NHA (2019) Adenosine A2A receptor as a potential drug target - current status and future perspectives. Curr Pharm Des 25(25). https://doi.org/10.2174/1381612825666190716113444
Ikram M, Park TJ, Ali T, Kim MO (2020) Antioxidant and neuroprotective effects of caffeine against Alzheimer’s and Parkinson’s disease: insight into the role of Nrf-2 and A2AR signaling. Antioxidants 9(9). https://doi.org/10.3390/antiox9090902
Yuan H, Ma Q, Ye L, Piao G (2016) The traditional medicine and modern medicine from natural products. Molecules 21(5). https://doi.org/10.3390/molecules21050559
Vina D, Serra S, Lamela M, Delogu G (2013) Herbal Natural products as a source of Monoamine Oxidase inhibitors: a review. Curr Top Med Chem 12(20). https://doi.org/10.2174/156802612805219996
Carradori S, Secci D, Petzer JP (2018) MAO inhibitors and their wider applications: a patent review, Expert Opin Ther Pat, vol. 28, no. 3, pp. 211–226, Mar. https://doi.org/10.1080/13543776.2018.1427735
Binda C et al (2007) Nov., Structures of human monoamine oxidase B complexes with selective noncovalent inhibitors: Safinamide and coumarin analogs, J Med Chem, vol. 50, no. 23, pp. 5848–5852, https://doi.org/10.1021/JM070677Y/SUPPL_FILE/JM070677Y-FILE002.PDF
Segala E et al (Jul. 2016) Controlling the dissociation of ligands from the Adenosine A2A receptor through Modulation of Salt Bridge strength. J Med Chem 59(13):6470–6479. https://doi.org/10.1021/ACS.JMEDCHEM.6B00653/SUPPL_FILE/JM6B00653_SI_001.PDF
Schrödinger (2018) Maestro| Schrödinger, Schrödinger Release 2018-1
Jacobson MP et al (2004) A Hierarchical Approach to All-Atom protein Loop Prediction. Proteins: Struct Function Genet 55(2). https://doi.org/10.1002/prot.10613
Jacobson MP, Friesner RA, Xiang Z, Honig B (2002) On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol 320(3). https://doi.org/10.1016/S0022-2836(02)00470-9
Greenwood JR, Calkins D, Sullivan AP, Shelley JC (2010) Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des 24:6–7. https://doi.org/10.1007/s10822-010-9349-1
Olsson MHM, SØndergaard CR, Rostkowski M, Jensen JH (2011) PROPKA3: consistent treatment of internal and surface residues in empirical p K a predictions. J Chem Theory Comput 7(2). https://doi.org/10.1021/ct100578z
Boulaamane Y, Ahmad I, Patel H, Das N, Britel MR, Maurady A (2023) Structural exploration of selected C6 and C7-substituted coumarin isomers as selective MAO-B inhibitors. J Biomol Struct Dyn 41(6):2326–2340. https://doi.org/10.1080/07391102.2022.2033643
Touati I et al (2023) Identification of novel dual acting ligands targeting the adenosine A2A and serotonin 5-HT1A receptors. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2023.2270753
Ntie-Kang F et al (2017) NANPDB: a resource for Natural products from northern African sources. J Nat Prod 80(7). https://doi.org/10.1021/acs.jnatprod.7b00283
Ntie-Kang F et al (2013) AfroDb: a select highly potent and diverse natural product library from African medicinal plants. PLoS ONE 8(10). https://doi.org/10.1371/journal.pone.0078085
Hatherley R, SANCDB et al (2015) A South African natural compound database. J Cheminform 7(1). https://doi.org/10.1186/s13321-015-0080-8
Roos K et al (2019) OPLS3e: extending Force Field Coverage for Drug-Like Small molecules. J Chem Theory Comput 15(3). https://doi.org/10.1021/acs.jctc.8b01026
Schrödinger (2015) QikProp
Lipinski CA (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technol 1. no. 410.1016/j.ddtec.2004.11.007
Pollastri MP (2010) Overview on the rule of five. Curr Protoc Pharmacol 49(1):9–12
Dixon SL, Smondyrev AM, Rao SN (2006) PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem Biology Drug Des 67. no. 510.1111/j.1747-0285.2006.00384.x
Gaulton A (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40:1100–1107
Halgren TA et al (2004) Mar., Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening, J Med Chem, vol. 47, no. 7, pp. 1750–1759, https://doi.org/10.1021/JM030644S/SUPPL_FILE/JM030644S_S.PDF
Friesner RA et al (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49(21). https://doi.org/10.1021/jm051256o
Ioakimidis L, Thoukydidis L, Mirza A, Naeem S, Reynisson J (2008) Benchmarking the reliability of QikProp. Correlation between experimental and predicted values. QSAR Comb Sci 27(4). https://doi.org/10.1002/qsar.200730051
Banerjee P, Kemmler E, Dunkel M, Preissner R (2024) ProTox 3.0: a webserver for the prediction of toxicity of chemicals, Nucleic Acids Res, vol. pp. 1–8, Apr. 2024, https://doi.org/10.1093/NAR/GKAE303
Release S, Research DES (2019) Desmond Molecular Dynamics System, DE Shaw Research, New York, NY.
Phillips JC et al (2005) Dec., Scalable molecular dynamics with NAMD, J Comput Chem, vol. 26, no. 16, pp. 1781–1802, https://doi.org/10.1002/JCC.20289
Boulaamane Y, Kandpal P, Chandra A, Britel MR, Maurady A (2023) Chemical library design, QSAR modeling and molecular dynamics simulations of naturally occurring coumarins as dual inhibitors of MAO-B and AChE. J Biomol Struct Dyn 18:1–18
Md S, Rahman et al (2021) Exploring the role of Monoamine Oxidase Activity in Aging and Alzheimer’s Disease. Curr Pharm Des 27(38). https://doi.org/10.2174/1381612827666210612051713
Boulaamane Y et al (2024) Antibiotic discovery with artificial intelligence for the treatment of Acinetobacter baumannii infections. mSystems May. https://doi.org/10.1128/MSYSTEMS.00325-24
Boulaamane Y, Jangid K, Britel MR, Maurady A (2023) Probing the molecular mechanisms of α-synuclein inhibitors unveils promising natural candidates through machine-learning QSAR, pharmacophore modeling, and molecular dynamics simulations. Mol Div, vol. 1–17
Mulakala C, Viswanadhan VN (2013) Could MM-GBSA be accurate enough for calculation of absolute protein/ligand binding free energies? J Mol Graph Model, vol. 46, pp. 41–51, Nov. https://doi.org/10.1016/J.JMGM.2013.09.005
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking, J Med Chem, vol. 55, no. 14, pp. 6582–6594, Jul. https://doi.org/10.1021/jm300687e
Delano LW (2002) Pymol: An open-source molecular graphics tool., CCP4 Newsletter on protein crystallography, vol. 40, no. 1
Laskowski RA, Swindells MB (Oct. 2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51(10):2778–2786. https://doi.org/10.1021/CI200227U/ASSET. http://IMAGES/MEDIUM/CI-2011-00227U_0006.GIF"
Fern K (2024) Tropical Plants Database, tropical.theferns.info.http://-05-25. > Add, 2024
Derese S, Yenesew A, Midiwo JO, Heydenreich M, Peter MG (2003) A new isoflavone from stem bark of Millettia dura. Bull Chem Soc Ethiop 17(1). https://doi.org/10.4314/bcse.v17i1.20933
Derese S et al (2014) 4′-Prenyloxyderrone from the stem bark of Millettia Oblata ssp. teitensis and the antiplasmodial activities of isoflavones from some Millettia species. Phytochem Lett 8(1). https://doi.org/10.1016/j.phytol.2014.02.001
De Colibus L, Li M, Binda C, Lustig A, Edmondson DE, Mattevi A (2005) Three-dimensional structure of human monoamine oxidase A (MAO A): Relation to the structures of rat MAO A and human MAO B, [Online]. Available: www.pnas.orgcgidoi10.1073pnas.0505975102
Milczek EM, Binda C, Rovida S, Mattevi A, Edmondson DE (2011) The ‘gating’ residues Ile199 and Tyr326 in human monoamine oxidase B function in substrate and inhibitor recognition, FEBS J, vol. 278, no. 24, pp. 4860–4869, Dec. https://doi.org/10.1111/J.1742-4658.2011.08386.X
Rauhamäki S et al (2018) Structure-activity relationship analysis of 3-phenylcoumarin-based monoamine oxidase B inhibitors. Front Chem 6(no MAR). https://doi.org/10.3389/fchem.2018.00041
Jaakola VP et al (2008) The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist, Science (1979), vol. 322, no. 5905, https://doi.org/10.1126/science.1164772
Azam F, Prasad MVV, Thangavel N, Ali HI (2011) Molecular docking studies of 1-(substituted phenyl)-3-(naphtha [1, 2-d] thiazol-2-yl) urea/thiourea derivatives with human adenosine A2A receptor. Bioinformation 6(9). https://doi.org/10.6026/97320630006330
Caliman AD, Miao Y, McCammon JA (2018) Mapping the allosteric sites of the A 2A adenosine receptor. Chem Biology Drug Des 91(1). https://doi.org/10.1111/cbdd.13053
Zeng L, Guan M, Jin H, Liu Z, Zhang L (2015) Integrating pharmacophore into membrane molecular dynamics simulations to improve homology modeling of G protein-coupled receptors with ligand selectivity: A2A adenosine receptor as an example. Chem Biol Drug Des 86(6). https://doi.org/10.1111/cbdd.12607
Rodríguez D, Piñeiro Á, Gutiérrez-De-Terán H (2011) Molecular dynamics simulations reveal insights into key structural elements of adenosine receptors. Biochemistry 50(19). https://doi.org/10.1021/bi200100t
Yu H, Dalby PA (2020) A beginner’s guide to molecular dynamics simulations and the identification of cross-correlation networks for enzyme engineering, in Methods in Enzymology, vol. 643, https://doi.org/10.1016/bs.mie.2020.04.020
Kasahara K, Fukuda I, Nakamura H (2014) A novel approach of dynamic cross correlation analysis on molecular dynamics simulations and its application to Ets1 dimer-DNA complex. PLoS ONE 9(11). https://doi.org/10.1371/journal.pone.0112419
Sittel F, Jain A, Stock G (2014) Principal component analysis of molecular dynamics: on the use of cartesian vs. internal coordinates. J Chem Phys 141(1). https://doi.org/10.1063/1.4885338
David CC, Jacobs DJ (2014) Principal component analysis: a method for determining the essential dynamics of proteins. Methods Mol Biol 1084. https://doi.org/10.1007/978-1-62703-658-0_11
Jung UJ, Kim SR (2018) Beneficial effects of flavonoids against Parkinson’s Disease. J Med Food 21. no. 510.1089/jmf.2017.4078
Santana K et al (2021) Applications of virtual screening in Bioprospecting: facts, shifts, and perspectives to explore the chemo-structural diversity of Natural products. Front Chem 9. https://doi.org/10.3389/fchem.2021.662688
Kumar S et al (2021) Navigating into the Chemical Space of Monoamine Oxidase inhibitors by Artificial Intelligence and Cheminformatics Approach. ACS Omega 6(36). https://doi.org/10.1021/acsomega.1c03250
Kim SK et al (2003) Modeling the Adenosine receptors: comparison of the binding domains of A2A agonists and antagonists. J Med Chem 46(23). https://doi.org/10.1021/jm0300431
Binda C, Li M, Hubá lek F, Restelli N, Edmondson DE, Mattevi A (2003) Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures, [Online]. Available: www.pnas.orgcgidoi10.1073pnas.1633804100
Mettai M et al (2023) Molecular docking/dynamics simulations, MEP analysis, bioisosteric replacement and ADME/T prediction for identification of dual targets inhibitors of Parkinson’s disease with novel scaffold. Silico Pharmacol 11(1). https://doi.org/10.1007/s40203-023-00139-3
Azam F et al (2019) Dec., Rutin as promising drug for the treatment of Parkinson’s disease: an assessment of MAO-B inhibitory potential by docking, molecular dynamics and DFT studies, Mol Simul, vol. 45, no. 18, pp. 1563–1571, https://doi.org/10.1080/08927022.2019.1662003
Chen JF, Cunha RA (2020) The belated US FDA approval of the adenosine A2A receptor antagonist istradefylline for treatment of Parkinson’s disease, Purinergic Signal, vol. 16, no. 2, pp. 167–174, Jun. https://doi.org/10.1007/s11302-020-09694-2
Mettai M, Daoud I, Melkemi N (2023) In Silico Approaches for the Study of New Anti-Parkinson’s Agents, https://doi.org/10.3390/ecsoc-27-16067
Jacobson KA, Moro S, Manthey JA, West PL, Ji XD (2002) Interactions of flavones and other phytochemicals with adenosine receptors. Adv Exp Med Biol 505. https://doi.org/10.1007/978-1-4757-5235-9_15
van Rensburg HDJ, Terre’Blanche G, Legoabe LJ (Aug. 2023) Synthesis and Adenosine RA1/A2A receptor Affinity of selected aurones and 2-Benzylidene-1-Indanones. Pharm Chem J 57(5):729–734. https://doi.org/10.1007/S11094-023-02944-8/METRICS
Zarmouh NO, Eyunni SK, Soliman KFA (2017) The Benzopyrone Biochanin-A as a reversible, competitive, and selective monoamine oxidase B inhibitor. BMC Complement Altern Med 17(1). https://doi.org/10.1186/s12906-016-1525-y
Acknowledgements
We would like to thank Francisco Javier Luque Garriga, Professor in the Department of Chemical Physics, University of Barcelona, Spain, for his assistance. His contribution is appreciated and gratefully acknowledged.
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
No potential conflict of interest was reported by the authors.
Human Ethics and Consent to Participate Declarations
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Boulaamane, Y., Touati, I., Qamar, I. et al. In silico Discovery of Dual Ligands Targeting MAO-B and AA2AR from African Natural Products Using Pharmacophore Modelling, Molecular Docking, and Molecular Dynamics Simulations. Chemistry Africa (2024). https://doi.org/10.1007/s42250-024-01074-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42250-024-01074-2