Abstract
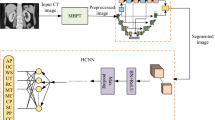
Renal segmentation is one of the most fundamental and challenging task in computer aided diagnosis systems. In order to overcome the shortcomings of automatic kidney segmentation based on deep network for abdominal CT images, a two-stage semantic segmentation of kidney and space-occupying lesion area based on SCNN and ResNet models combined with SIFT-flow transformation is proposed in paper, which is divided into two stages: image retrieval and semantic segmentation. To facilitate the image retrieval, a metric learning-based approach is firstly adopted to construct a deep convolutional neural network structure using SCNN and ResNet network to extract image features and minimize the impact of interference factors on features, so as to obtain the ability to represent the abdominal CT scan image with the same angle under different imaging conditions. And then, SIFT Flow transformation is introduced, which adopts MRF to fuse label information, priori spatial information and smoothing information to establish the dense matching relationship of pixels so that the semantics can be transferred from the known image to the target image so as to obtain the semantic segmentation result of kidney and space-occupying lesion area. In order to validate effectiveness and efficiency of our proposed method, we conduct experiments on self-establish CT dataset, focus on kidney organ and most of which have tumors inside of the kidney, and abnormal deformed shape of kidney. The experimental results qualitatively and quantitatively show that the accuracy of kidney segmentation is greatly improved, and the key information of the proportioned tumor occupying a small area of the image are exhibited a good segmentation results. In addition, our algorithm has also achieved ideal results in the clinical verification, which is suitable for intelligent medicine equipment applications.
Similar content being viewed by others

Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
With the continuous advancement of medical imaging technology, the intelligent medical imaging processing should be conducive to accurate determination of the conditions including spatial position and size of focus, etc. as well as the corresponding relationship between focus and its surrounding tissues and helping the medical staffs to make the accurate qualitative and quantitative analysis on pathological tissues and organs so as to have a more accurate diagnosis on the health condition and therapeutic schedule of tissues and organs [1]. Hence, there will certainly be of great clinical value if a set of fast and accurate image segmentation algorithm is designed to free the clinical doctors from the tedious and boring task [2].
The kidney and space-occupying lesion area segmentation in medicine aided diagnosis system has achieved good results in ideal conditions [3]. However, the semantic segmentation effect of organ in abdominal CT scan image is not very good in the case of the low contrast, irregular shape, uneven gray, interference factors adjacent tissue. How to realize semantic segmentation of CT images under complex conditions is an problem to be solved in the development of intelligent diagnosis system and is the focus of this paper.
Kidney segmentation is a specific direction in the field of abdominal medical imaging segmentation and meanwhile has its particularity. Currently, certain research results have been achieved in the full-automatic segmentation algorithm specific to kidney. But some problems demanding prompt solutions are still faced [4]. For example, (1) The tissues and organs adjacent to kidney in the medical imaging have the similar tissue density, which leads to obscure boundary, as shown in Fig. 1; (2) The size and shape of the kidney image of the same individual in different tomography images may change; (3) There exist differences of size and shape of kidney among different individuals; (4) The gray value of kidney in the sequence image may fluctuate due to the influences of noise interference and other factors;(5) In the CT sequence image, kidney motion or deformation may occur due to breathing or abdominal movement; (6) The differences of renal carcinoma in size, position and gray, etc. may also influence the accuracy of segmentation of kidney. In other words, the same tissue has the problem of inconsistent intensity between different patients, different modalities, and even different frames in the same modality. In consequence, kidney image segmentation is one of the most fundamental and challenging task in computer aided diagnosis systems.
The semantic comprehension of image cannot be separated from the segmentation technology. The image is essentially the two-dimensional matrix formed by a series of pixel while semantic segmentation actually focuses on grouping these pixels in image according to the different expression meanings in the image [5]. It is usually called as segmentation. Aiming at the problem of kidney segmentation, extensive and in-depth research has been carried out, including various methods based on level set, graph cut, feature abstraction, and deep learning [6]. The kidney features mainly include artificial features such as texture features, local feature, and word-bags feature. [7] However, the distinguishability of artificial features is limited, the generalization is relatively poor, and it is difficult to select effective features. Deep learning, specially convolutional neural networks, has outperformed the state of the art in many image recognition and target detection tasks in the field of computer vision. Also, CNN has excellent performance in the semantic segmentation of natural images. This provides a novel way to automatically and accurately segment the kidney and space-occupying lesion area. [8]
Before deep learning isn’t applied in the field of semantic segmentation yet, most of researchers of semantic segmentation conduct modelling and calculation by adopting the graph method in accordance with the image pixel’s own low-level visual cues. Training isn’t required, thus the computation complexity is lower. However, these methods cannot be applied in the complicated abdominal CT scan image. Especially in the case that the artificial aided information cannot be provided, the segmentation effect is unsatisfactory. After deep learning enters the field of computer vision, the semantic segmentation technology also steps into a new era. The training method based on convolutional neural network (CNN) can greatly improve the accuracy of semantic segmentation [9]. The excellent algorithms including the fully convolutional neural network, Dilated Convolution and the post-processing operation represented by conditional random fields [10] etc. are proposed. SegNet based on Caffe framework, modifies VGG-16 to generate the network structure model of open-source semantic segmentation on the basis of FCN [11]. DeepLab conduct processing on the basis of FCN. Its semantic segmentation process can be divided into two steps: a rough category fraction is obtained through FCN and the size of original image is obtained through linear/nonlinear interpolation; the detail optimization is conducted on the segmentation results of step 1 by utilizing full-connection CRF. On the basis of FCN, PSPNet [12] introduced Spatial Pyramid Pooling to expand the feature pixel-level into the special Spatial Pyramid Pooling designed in the paper, and meanwhile combines local feature with global information so as to provide the relatively accurate prediction results specific to the semantic scene segmentation. The practice also draws lessons from the method adopted for acquiring global scene features during extraction of complicated scene features. In addition, a type of optimization method based on deep supervision Loss is also proposed in the paper.
The tissues and organs adjacent to kidney in the medical imaging have similar tissue density, which causes obscure boundary and greater differences of individuals and furthers a very difficult for semantic segmentation. Accordingly, in order to solve the problem that it is not accurate to conduct direct semantic segmentation, it is necessary to propose a semantic transfer model through seeking the matched images of known segmentation results and establishment of close connection. If it is needed to retrieve the corresponding known image through the unknown image, features extraction of image is required. In this paper, feature extraction is conducted by using deep convolutional neural network SCNN and ResNet, and meanwhile metric learning is added to make sure that features can better describe image, where the images at different imaging angles are divided into the same category, and the images at the identical angle are partitioned into different categories. After image retrieval is completed, the pixel matching method based on SIFTflow transformation is proposed. Under the premise of registration parameters between two known images, how to make the abdominal CT image pixels of different individuals correspond to known image pixels is the research contents in our paper. Hence, we uses MRF to integrate pixel information, spatial prior information and smoothing information to obtain the relationship between target image and known image so as to transfer semantic meanings of known image into the target image to gain an exact segmentation of kidney and space-occupying lesion area under different conditions.
Related works
Essentially our proposed model is based on a metric learning approach to construct a deep convolutional neural network structure using SCNN and ResNet network so as to extract image features and minimize the impact of interference factors on features. Thus, we will only discuss the most related SCNN and ResNet model [13] .
Siamese convolutional neural network
Siamese convolutional neural network (SCNN) is a type of similar measurement methods in essence, and is relatively suitable for being used for recognition and classification under the circumstance that there are more data categories but less sample data of each category. SCNN focuses on learning a similarity measurement from datasets and then using the measurement to make comparisons and alignment on the samples of unknown category. The method aims at mapping input to a target feature space through a function and using the relatively simple distance function to make the similarity comparison in the space. During training, the loss function values of a pair of samples coming from the same category (label = 1) is minimized (making samples of the same category closer) while the loss function values of a pair of samples coming from different categories (label = 0) is maximized (making samples of different categories farther).
Similarity measure function for Siamese convolutional neural network can be written as
where GW(X1) is a differentiable mapping function, and its parameter is W. The goal of the optimization process is to find a group of W, which makes the function value is smaller when X1 and X2 belong to the same category; the function value is bigger when X1 and X2 do not belong to the same category; that is to say, we can get min(EW(X1, X2)) for the paired training data when X1 and X2 belong to the same category; we can get max(EW(X1, X2)) if X1 and X2 are from different class.
To sum up, the main difference between SCNN and prior traditional CNN lies in that the paired samples will be input instead of the single sample. Meanwhile, each sample isn’t the label with the category mark any longer but one label will be given to each sample [14] . The label will indicate whether this pair of samples belongs to the same category or not. Two images in the samples input into network respectively enter the identical network. Two networks share weight W, and meanwhile the similarity measurement is conducted on output so as to get Loss function to direct network learning through back-propagation [15] .
In comparison to other algorithms, SCNN fades the label and the category never trained may also be classified through the network structure [16] . Thus it has a very good expansibility. Moreover, in terms of the datasets with a smaller data volume, it can also show a very good effect while many other algorithms cannot achieve.
Siamese network structure is as shown in Fig. 2. For two different input X1 and X2, the corresponding low-dimensional space feature vector GW(X1) and GW(X2) are obtained through CNN. After it is substituted into equation, a comparison will be made through energy function EW(X1, X2). The samples are input in pair, so the network structure is symmetrical if the same two mapping functionsGare used and the equal weight W is shared. Therefore, it is called Siamese architecture.
Essentially, SCNN is a type of dimensionality reduction method. There is an assumption that Loss function is only related to input data and weightW. Then, loss function can be defined as:
where p is the training sample number. X1 and X2 indicate a pair of images. Y is denoted as the corresponding label. (Y, X1, X2)i formed by a pair of pictures and the corresponding label is denoted as the sample i. When Y is 0, the right side of the equation is LG(EW(X1, X2)i). In addition, the loss value is the loss function of image sample with the same category LG, otherwise it is represented as LI. The goal focuses on reducing the loss function value and the energy EW of image sample of the same category as much as possible and meanwhile increasing the energy of image sample of different categories, thus it is necessary to design LG into monotone increasing function and LI into monotone decreasing function so as to achieve the performance.
ResNet model
The key idea of Resnet network is to introduce the residual block, where it superimposes the constant mapping layer on the basis of a shallow network to carry out residual learning, improves the precision of deep feature extraction, and solves the problem of vanishing gradient. Assume that the original input samples of the Resnet network are obtained after multi-layer network mapping [16] . Therefore, the residual function is shown in Fig. 3. It can be seen that after an identity mapping, the input is superimposed on the convolution output to form a jump connection that can skip one or more layers, eliminating the vanishing gradient problem, and the network deep can be made into hundreds of layers.
The identity mapping is superimposed on the network, and even increasing the number of layers of the network does not degrade the performance of the network [17] . The structure of Fig. 3 can simply cause the weights of multiple nonlinear layers to be zero to approximate an identity mapping, whose output can be expressed as
The x and y are denoted as the input and output result of the sub-block,respectively; H(x, Wi) is the residual mapping. The introduced path xin the above equation neither introduces additional parameters nor increases computational complexity. The simulation results show that the Resnet network is easier to converge than a simple network of the same scale, and can obtain better output results without being affected by the network deep.
Improved SCNN network combined with ResNet-18
In this paper, a SCNN structure integrated with ResNet-18 CNN is proposed. By combining two ResNets to form a network with shared weights W [18], the data is input into the network in a pairwise manner, and finally the network training is guided by a relative loss function [19] .
The partial structure of the adopted ResNet-18 is shown in Fig. 4(a), and then the network structure integrated with SCNN is shown in Fig. 4(b). Compared to the network structure of ResNet-18, the following improvements are made in this paper:
-
1.
The data input of ResNet-18 data is that one sample corresponds to one label. In this paper, a pair of sample of data image and data image_p are respectively input, and also labeled with “Whether this pair of samples belong to the same category or not”. As inputting is divided into two parts, a contact layer (The layer’s name is data all) is needed to integrate them.
-
2.
ResNet is connected with a fully-connected layer fc1000 for feature extraction after the fifth convergence layer. In the network structure of this paper, the layer is removed, and meanwhile is replaced with a slice layer (Named feat each). Data needs to be segmented once on this layer into two corresponding features through network output due to use of single network. Finally, the label of this pair of feature and input data are substituted into the contrastive loss function to generate Loss so as to direct or guide network training [27].
Analysis of SCNN loss function
SCNN uses a contrastive loss function to supervise the network training:
where fi and fj are the normalized feature, and ‖fi − fj‖ is a measurement of similarity between fi and fj.If fij = 1, it means fi and fj belong to the same class, the loss function value was their Euclidean distance. The larger the distance, the larger the Loss value, so that the network can be controlled by back propagation so as to adjust the network parameters, making the features more similar [20] .
When fij = − 1, it means fi and fj don’t belong to the same category, there is a hyper-parameter [23] m that can be set by itself, indicating the minimum distance among features of different categories. When the distance between two features is greater than m, this shows that network already can differentiate these two features. Thus the value of max is 0 and Loss function value is 0; when the distance between two features is less thanm, Loss value is \( m-{\left\Vert {f}_i-{f}_j\right\Vert}_2^2 \) and Loss function value isn’t 0. Thus back-propagation is needed to constrain network parameters so as to make the distance between two features greater.
Establishment of metric learning and loss function
The tissues and organs adjacent to kidney have the similar density, which leads to burred boundary. Moreover, there exist huge differences in imaging individual. Accordingly, the robustness must be considered during feature representation of abdominal CT image. In this paper, SCNN contrastive loss function is improved, metric learning is conducted on data features of abdominal image under various imaging condition, and then alignment is made in the metric learning space [28].
The classical metric learning focuses on dimensionality reduction of training data through PCA [29]. However, PCA cannot retain the information of data structure well. PLS regression technology [24] can not only realize dimensionality reduction but also map the training data to a relatively compact space.
There is an assumption that the data space where kidney image is is A under a normal condition and the data space where kidney image is is B under the fuzzy noise condition. Suppose that A containsnfeature vectors withddimensions \( {X}^{(a)}=\left[{x}_1^{(a)},\cdots, {x}_n^{(a)}\right] \),
and the corresponding training data labels \( {Y}^{(a)}=\left[{y}_1^{(a)},\cdots, {y}_n^{(a)}\right] \); B contains m feature vectors with the d dimensions \( {X}^{(b)}=\left[{x}_1^{(b)},\cdots, {x}_n^{(b)}\right] \), and the corresponding training data labels \( {Y}^{(b)}=\left[{y}_1^{(b)},\cdots, {y}_n^{(b)}\right] \). When \( {y}_i^{(a)}={y}_j^{(b)} \), \( {x}_i^{(a)} \) and \( {x}_j^{(b)} \) belongs to the same class, which is called the positive sample pair; When \( {y}_i^{(a)}\ne {y}_j^{(b)} \), \( {x}_i^{(a)} \) and \( {x}_j^{(b)} \) are not in the same class,which is called a negative sample pair.
Next, PLS can be applied into two data space mappings. Firstly, define a matrix P with d × p(d < p) whered is the dimension of the original data, p is the data dimension in transformation space. Then the trained data from PLS can be mapped to the subspace, where we assume that the data is \( {\overline{X}}^{(a)} \)和\( {\overline{X}}^{(b)} \) in the new space, and \( {\overline{X}}^{(a)}={P}^{\mathrm{T}}{X}^{(a)} \), \( {\overline{X}}^{(b)}={P}^{\mathrm{T}}{X}^{(b)} \). In the new subspace, we define a positive semidefinite matrix W with dimension p × p, and W = VVT where V ∈ Rp × q and q < p. In this space, the distance between feature samples of training data can be defined as:
In order to facilitate comparison and analysis, the Loss function is defined as follows:
where θij=1 if \( {y}_i^{(a)}={y}_j^{(b)} \); θij= − 1 if \( {y}_i^{(a)}\ne {y}_j^{(b)} \); c is a constant.
It is assumed that the number of positive sample pairs in training data is N+, the number of negative sample pairs in training data is N−. Thus, the objective function based on metric learning can be described as:
where αij = 1/N+ if θij = 1; αij = 1/N−, if θij = − 1. Finally, It can be shown that Accelerated Proximal Gradient algorithm [21] can be used to solve the above objection function. When the objective function converges to a minimum value, the corresponding optimal value V∗ is applied to the test set.
There are two matrices in above metric learning results where the mapping matrix P is used to preserve the Latent Structure, and the optimal mapping matrix V∗ is for making the metric learning distance more adaptable. If there are s test samples in space A, the test sample can be represented as \( {Z}^{(a)}=\left[{z}_1^{(a)},\cdots, {z}_s^{(a)}\right] \); if there are t test samples in space B, the test sample can be represented as\( {Z}^{(b)}=\left[{z}_1^{(b)},\cdots, {z}_t^{(b)}\right] \). These two mapping matrices V∗ and P are applied into the test dataset.
where the dimensions of test samples in space A and B are all d.
In order to further reduce the difference between the test data features in spaces A and B, it is necessary to register the test subspace \( {\tilde{Z}}^{(a)} \) in A and the test subspace \( {\tilde{Z}}^{(b)} \) in B. Thus, the bregman matrix divergence is minimized to obtain the registration matrix R:
where Q(a) ∈ Rq × k and Q(b) ∈ Rq × k respectively represent the left singular matrix of \( {\tilde{Z}}^{(a)} \) and \( {\tilde{Z}}^{(b)} \), ‖‖F denotes Frobenius norm [22]. So the closed solution \( {R}_{\ast }={Q}_{(a)}^T{Q}_{(b)} \) of R can be obtained by calculation.
Similarly, R∗ can be adopted to map \( {\tilde{Z}}^{(a)} \) and \( {\tilde{Z}}^{(b)} \) into a sub-space.
where C(a) ∈ Rk × s and C(b) ∈ Rk × t represent the test data features in spaces A and B, respectively.
Finally, k dimension feature descriptions of the testing image are obtained after the mapping on a testing abdominal CT image in the given space B through above features. According to the descriptions, the most matched kidney and space-occupying lesion area can be found in the space A so as to conduct subsequent semantic segmentation and label transfer.
Semantic segmentation model
Despite the recent success of deep-learning based semantic segmentation, deploying a pre-trained abdominal CT segmentation model to a renal whose images are not presented in the training set would not achieve satisfactory performance due to dataset biases [23]. Instead of collecting a large number of annotated images of each renal of interest to train or refine the segmentation model, we propose a two-stage semantic segmentation of kidney and space-occupying lesion area based on SCNN and ResNet models combined with SIFT-Flow transformation. Next, we will analyze the details of the model.
The image in known database assumed to have been labeled with semantic meanings. Meanwhile SCNN is adopted to have realized image retrieval and correspondence of known semantic meanings and the pixel region on unknown image. Next, it is needed to conduct semantic segmentation on new unknown abdominal image, establish dense matching of pixel between two CT images [24], and transfer the known semantic meanings of kidney to the image to be segmented so as to complete semantic segmentation of kidney or its space-occupying lesion area.
SIFT Flow is adopted to realize dense matching in this paper. As to the method, histogram intersection kernel is firstly used to find the most similar image to the input image from database, and then dense feature sampling is conducted in two images to construct dense matching [25].
SIFT-flow fusion model
Due to the known space information of organs and viscera in abdominal image, kidney and space-occupying area studied in this paper have stronger spatial prior information; furthermore, there shall be a smooth transition among data label information of image. Hence, Markov Random Field (MRF) model is integrated with spatial prior information, smooth transition and dense matching information to form transfer model.
SIFT-Flow describes the matching degree according to estimation of objective function. Intuitively, SIFT descriptions need to be aligned on both ends of flow. In addition, flow shall be a set of smooth vectors except for the part of object edge (Disorderly and unsystematic condition as well as serious intersection each other are not allowed). Essentially, it focuses on finding the differential displacement w(p) = (u(p), v(p)) of all pixels to form dense matching pair. Given original pixelsp = (x, y), target pixels p1 = (x1, y1), p1 = (x1, y1), then we can get ∣u(p) ∣ = ∣ x − x1∣ and ∣v(p) ∣ = ∣ y − y1∣.
Based on above situation, the target transition energy function of SIFTflow is defined as:
where ε includes the spatial neighborhood of all pixels, and \( q=\left(\tilde{x},\tilde{y}\right) \) indicates the points within neighborhood of p.
Above energy functions totally include 3 items, respectively representing data item, displacement item and smooth item. The data item in formula includes transfer vector. Whether the label on both ends of w(p) is matched or not; the significance of displacement item lies in that displacement value should be ensured smaller as far as possible in the case that no other information can be compared; Smooth item requires that the vector w(p) of adjacent pixels shall be similar to the greatest extent. In terms of the objective function, the paradigm L1 is used in data item and smooth item, and t and d are used as the threshold.
Finally, the algorithm based on dual-layer loopy belief propagation is used to solve the objective function in terms of SIFT-flow. Accordingly, the energy transfer function can be defined as:
where ε includes the spatial neighborhood of all pixels, \( q=\left(\tilde{x},\tilde{y}\right) \) indicates the points within neighborhood of p. λ is the regularization parameter. Belief propagation method is utilized to minimize energy function E(w) so as to obtain the optimal solutionw.
Multi-feature semantic integration
In this paper, the structure and principle of Convolutional Neural Networks and ResNet are investigated deeply, the SCNN training network structure model integrated with ResNet is proposed, and the metric learning method is adopted to learn features. Through the improved SIFT-Flow semantic transfer model, the penalty items of label matching, spatial prior information and smoothing information, etc. are integrated with MRF to finally get the objective function.
Label matching
In terms of target SIFT figure I1, each point (Pixel) has its SIFT value. These values constitute SIFT field s1 of I1; vice versa. Currently, SIFT field s1 of I1, label field L1 and SIFI field S2 of I2 have been known. The target of semantic segmentation is to speculate the label field L2 of I2 based on above formula.
In order to speculate the label field L2 of I2, the dense matching relationship of I1 and I2 is utilized and the spatial structure prior information of current kidney image and spatial smoothing information of I2 are combined so as to obtain the label field L2of I2. According to the dense correspondence, the punishment formula is defined as:
where Γ(I2, p) indicates the labeling result of pixel p on CT image I2.
Spatial prior information
In order to utilize the spatial prior information, the penalty function of spatial prior information shall be firstly set up and then Log is added for smoothing so as to obtain:
where H(p) is the prior probability that the pixel pbelongs to a certain kind. The prior probability is obtained through training of centralized pictures. The author makes a statistics of position information of spatial histogram on each semantic category by utilizing labeling image of training set so as to obtain the spatial histogram distribution of each semantic category. Each figure indicates the spatial position distribution of one semantic category in all training sets. The deeper the color, the higher the probability that the semantic category occurs at the position.
Smoothing information
In order to integrate smoothing information, the smoothing information penalty function is established and a penalty item of smoothing information Ψ(Γ(I2, p), Γ(I2, q)) is defined. Among it, Γ(I2, p) and Γ(I2, q) are the corresponding label of the pixels in two adjacent fields.
where r is a constant not related to image. It is only used for regulating to ensure that the index item in the function can adapt to different contrasted conditions.
Semantic integration
In order to realize accurate segmentation of kidney and space-occupying lesion, the above dense label’s corresponding information, spatial prior information and smoothing information are integrated by utilizing Probabilistic Markov Random Field Model to finally constitute the objective function of semantic label transfer:
Experimental analysis
In this section, we report the characteristics and the segmentation results of the proposed semantic deep model qualitatively and quantitatively. For the evaluation metrics, we employ the dice ratio (DR) score, and the Kappa index (KI). Large DR and KI indicate high segmentation accuracy. In addition, we also use Centroid Distance indicates the distance between the central pixels of the new method segmentation result and the manual result, and we adopt and compute the precision-recall (PR) curves for additional comparisons, which have been widely used for object detection and segmentation problems on general image.
Data and experimental setup
Medical CT scan images from the French IRCAD International Medical Center database and self-built database are adopted as training images, where 15,500 CT images of 363 subjects with kidney tumors are used to implement and test our proposed model. In addition, the data of 128 patients with a single unilateral renal tumour between January 2011 and December 2017 were retrospectively analyzed and collected form Changzhou No.1 People’s Hospital. This study satisfied the requirements of the institutional review board for a retrospective study. 1130 kidney images and corresponding kidney labels in the space-occupying lesion area are got by adopting manually-labeling method. Except for the target area, the rest is marked as the background, that is to say that the datasets labeled can be used in training and testing process for semantic segmentation.
To illustrate the proposed segmentation method more clearly and fully demonstrate the performance gains from the combination of SCNN and ResNet, the proposed method is compared with five state-of-the-art methods which include BK-CNN [24], K-ResNet [25], and ConvNet [26]. Except K-ResNet algorithm, all the results are based on the source codes or executables released by the original authors. The default parameters are employed in the comparison algorithms. We try to realize K-ResNet. Unfortunately, our results are inconsistent with the original literature. To make a fair comparison, all the evaluation indexes of K-ResNet are from the literature [25].
The Abdominal CT image studies used in our study were axially acquired by a Siemens CT Scanner. Each image has isotropic in-plane resolutions. The slice thickness varies from 0.8 mm to 2.0 mm. In our study, we applied a dual-plateaus histogram equalization to each image to standardize the intensity scale. The training platform is the Keras framework under Ubuntu 14.04.5with an Intel Core i7 8100 at 3.06 GHz, 1080 Ti GPU and 256 GB memory. Training takes approximately 60 h on a 1080 Ti GPU.
Implementation details
In this work, all the abdominal CT scanning images are produced by utilizing the open source Keras framework, and the codes will be released upon acceptance. In order to eliminate the interference of the difference in imaging angle, the image-based registration was computed using the Advanced Normalization Tools (ANTS) software [26]. The registration sequentially optimized an affine transform and a deformable transform between the pair of images, using the Mattes mutual information metric [27].
In the following deep-structure experiments, we use mini-batch size 16 and the Adam optimizer with learning rate of 5 × 10−6, β1 = 0.9, β2 = 0.98 to optimize the network. Moreover, the rest training parameters are the initial teaming rate is 0.05. At intervals of a certain iterative times, learning rate is decreased. Due to the limited server memory capacity, batch size is set to be 32. In consideration of characteristic of SCNN, a pair of image needs to be input once. Thus the batch size inputted into network every time is actually 2×32 = 64. After about 300,000 times of loop iteration, the obtained network model is regarded as the experimental network model.
Comparison of quantitative evaluation
We apply our proposed model to segment these images in our self-build Dataset. In Table 1, we list average values of dice ratio (DR) score, and the Kappa index (KI) of all test images using different methods. As shown in Table 1, there is a severe performance drop in the four images compared to its original performance on images. Interestingly, we observe a trend that the farther the distance between the unknown image and the known image, the severer the performance degradation. This implies that different visual appearances from different angle due to semantic differences would dramatically impact the accuracy of the segmentation algorithm. It also justifies the necessity of an effective image retrieve method for the renal segmentation which can alleviate the discrimination.
The experiment result shows that our algorithm had higher Dice scores than other algorithms, and lower mean boundary distance than K-ResNet and ConvNet. Although partially attributable to higher variability due to similar tissue density, the observed differences in median accuracy metrics are generally smaller than for other organs. Finally, the experimental data show that the result of automatic segmentation through our proposed network is more accurate than comparison segmentation algorithms. Therefore, the model also has more robustness.
It is not surprising that our method shows outstanding performances for the test image containing some tumor. Tables 1 and 2 show a comparison of different segmentation deep model for renal and lesion segmentation in abdomen CT scans. The experiment results show the normal or abnormal kidney segementation performance for the multi-organ methods have huge different. Importantly, compared to most of these existing method, our proposed framework doesn’t rely on any atlas nor detection stage for the segmentation. We note also that the ConvNet needs to adopt the lots of semantic remark so as to compute a segmentation,and these model cannot better adapt and segment the space-occupying lesion area in complex CT background. Next, we will briefly analyze the evaluation indicators.
As for Dice index, our proposed method outperforms the K-ResNet, BK-CNN and ConvNet, and on average it is superior to BK-CNN by 0.1~0.2, to K-ResNetby 0.03~0.13 and to ConvNet by 0.1~0.23 in Table 1. For Kappa index, the proposed method outperforms the K-ResNet and ConvNet, and it is superior to K-ResNet by 0.04~0.3 in Table 2. It is obvious that our proposed semantic deep model outperforms other methods in terms of the higher dice ratio score, the Kappa index, and the smallest value of distance measurements which reflect high quality segmentation.
Figures 5, 6 and 7 show the PR curves of our method and its comparison algorithms for further comparison. It shows that the deep semantic learning can improve the segmentation performance for each specific class: the normal abdominal CT image and abnormal renal CT images with space-occupying lesion where Fig. 5 is quantitative value of segmentation results for all test CT images, while Figs. 6 and 7 are normal renal CT images or abnormal renal CT images with space-occupying lesion. Our proposed algorithm performed better for renal segmentation studies and that the deep semantic based step-wise integration approach improved upon the results produced by any of the deep models. We observe that our proposed deep semantic renal segmentation is surprisingly accurate when imaging differences lead to inconsistent gray levels. And the Dice scores and recall for our segmentation models are in fact higher than the fine existing comparison algorithms; however, the precision is slightly lower. We believe this effect arises from the fact that kidneys are relatively smooth organs, which our semantic technique is able to yield very high-quality segmentation performance.
Therefore, the experimental results that our proposed deep semantic segmentation method achieved the best overall performance across all the measurement and improved upon the existing methods with a large margin for both normal renal and abnormal renal.
Visual comparisons
The kidney and space-occupying lesion area segmentation are the least accurate for all existing algorithms and all metrics. In addition, since the structure, shape, size of different kidneys are quite different, and the available slices of deep network training are less, the learned knowledge is not enough to cope with the variability of the kidney and space-occupying lesion area, and the obtained model is trained on some data, and then tested on other data, resulting in poor recognition ability when segmenting renal area. Therefore, we propose a deep semantic model to improve the segmentation performance. Sample slices are illustrating the results of the framework for kidney segmentation in Fig. 8.
For ease of analysis, the segmentation results are mapped directly into the original CT image, where the red represents space-occupying lesion area, and blue is for the kidney area. The qualitative results of different methods on three abdomen images are shown in the Fig. 5, where Fig. 5(a) is the original kidney CT image; Fig. 5(b) is the ground-truth image approved by experts; Fig. 5(c) is the segmentation result of the our proposed network structure in this paper; Fig. 5(d) is the segmentation results from BK-CNN deep model; Fig. 5(e) is the segmentation result of ConvNet model; Obviously, our proposed segmentation is similar to the ground truth in most cases. For tumors with simple texture, such as the first row in Fig. 5, ConvNet works well. However, in other cases, ConvNet cannot achieve appealing performance. BK-CNN does not achieve competitive results especially in the small tumor cases. In addition, it can also be seen that the segmentation effect of the deep semantic model proposed in this paper from the perspective of complete tumor analysis is better, and the tumor area and the normal tissue area can be clearly distinguished, which are closer to the ground-truth label value, while the segmentation result of BK-CNN is smoother at the boundary, and the segmentation within the tumor is relatively unsatisfactory compared with the ConvNet network. This may also be why BK-CNN is not sensitive enough to the details.
Despite these promising results, there are still limitations in our algorithm. First, deep semantic segmentation-based algorithms are inferior for detecting dim -small structures, which may lead to inaccurate segmentation of some thin and low-contrast objects (the third row in Fig. 8. Second, tumors with heterogeneous intensities or small sizes residing at the kidney’s edge might be under-segmented. This is mainly caused by the high boundary term effect generated by the improved SIFT-flow semantic transfer mode. Third, false segmentation of the initial slice would increase the overall segmentation error. In the future, we will be committed to solving these problems and evaluate our algorithm with more clinical datasets.
Conclusion
In this paper, we propose a unified framework utilizing a two-stage semantic segmentation of kidney and space-occupying lesion area based on SCNN and ResNet models combined with SIFT-flow algorithm, which performs joint global and class-wise alignment by leveraging soft labels from source and target-domain data. In addition, our method uniquely identifies and introduce static-object priors, which are retrieved from known images. The metric learning method is adopted to learn features. Through the improved SIFT-flow semantic transfer model, the penalty items of label matching, spatial prior information and smoothing information, etc. are integrated with MRF to finally get the objective function. The experimental results qualitatively and quantitatively show that the accuracy of kidney segmentation is greatly improved, and the key information of the proportioned tumor occupying a small area of the image are exhibited a good segmentation results.
References
Hu, P., Wu, F., Peng, J., Bao, Y., Chen, F., and Kong, D., Automatic abdominal multiorgan segmentation using deep convolutional neural network and time-implicit level sets. Int. J. Comput. Assist. Radiol. Surg., 2016a. https://doi.org/10.1007/s11548-016-1501-5.
Li, W., Jia, F., and Hu, Q., Automatic segmentation of liver tumor in CT images with deep convolutional neural networks. J. Comput. Commun. 3(11):146–151, 2015.
Vivanti, R., Ephrat, A., Joskowicz, L., Karaaslan, O., Lev-Cohain, N., and Sosna, J., Automatic liver tumor segmentation in follow-up CT studies using convolutional neural networks. Proc. Patch-Based Methods Med. Image Process. Workshop, MICCAI’2015, 54–61, 2015.
Ben-Cohen, A., Diamant, I., Klang, E., Amitai, M., and Greenspan, H., Deep learning and data labeling for medical applications. In: Proceedings of the International Workshop on Large-Scale Annotation of Biomedical Data and Expert Label Synthesis. Lect. Notes Comput. Sci. 10008:77–85, 2016. https://doi.org/10.1007/978-3-319-46976-8_9.
Christ, P. F., Elshaer, M. E. A., Ettlinger, F., Tatavarty, S., Bickel, M., Bilic, P., Rempfler, M., Armbruster, M., Hofmann, F., D’Anastasi, M. et al., Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: Proceedings of the medical image computing and computer-assisted intervention. Lect. Notes Comput. Sci. 9901:415–423, 2016. https://doi.org/10.1007/978-3-319-46723-8_48.
Dou, Q., Chen, H., Jin, Y., Yu, L., Qin, J., and Heng, P.-A., 3D deeply supervised network for automatic liver segmentation from CT volumes. IEEE Trans. Biomed. Eng. 64(7):1558–1567, 2016a.
Hu, P., Wu, F., Peng, J., Liang, P., and Kong, D., Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys. Med. Biol. 61:8676–8698, 2016b.
Lu, F., Wu, F., Hu, P., Peng, Z., and Kong, D., Automatic 3D liver location and segmentation via convolutional neural network and graph cut. Int. J. Comput. Assist. Radiol. Surg. 12:171–182, 2017.
Lu, X., Xu, D., and Liu, D., Robust 3d organ localization with dual learning architectures and fusion. In: Proceedings of the deep learning in medical image analysis (DLMIA). Lect. Notes Comput. Sci. 10008:12–20, 2016.
Ravishankar, H., Sudhakar, P., Venkataramani, R., Thiruvenkadam, S., Annangi, P., Babu, N., and Vaidya, V., Understanding the mechanisms of deep transfer learning for medical images. In: Proceedings of the Deep Learning in Medical Image Analysis (DLMIA). Lect. Notes Comput. Sci. 10008:188–196, 2016b.
Thong, W., Kadoury, S., Piché, N., and Pal, C.J., Convolutional networks for kidney segmentation in contrast-enhanced CT scans. Comput. Methods Biomech. Biomed. Eng. Imag. Vis. 1–6, 2016..
Farag, A., Lu, L., Roth, H.R., Liu, J., Turkbey, E., and Summers, R.M., A bottom-up approach for pancreas segmentation using cascaded superpixels and (deep) image patch labeling. arxiv:1505.06236, 2015.
Roth, H. R., Lu, L., Farag, A., Shin, H.-C., Liu, J., Turkbey, E. B., and Summers, R. M., DeepOrgan: Multi-level deep convolutional networks for automated pancreas segmentation. In: Proceedings of the medical image computing and ComputerAssisted intervention. Lect. Notes Comput. Sci. 9349:556–564, 2015b.
Cai, J., Lu, L., Zhang, Z., Xing, F., Yang, L., and Yin, Q., Pancreas segmentation in MRI using graph-based decision fusion on convolutional neural networks. Proc. Med. Image Comput. Computer-Assist. Interven. Lect. Notes Comput. Sci. 9901:442–450, 2016a.
Roth, H. R., Lu, L., Farag, A., Sohn, A., and Summers, R. M., S holistically-nested networks for automated pancreas segmeings of the medical image computing and computer-Assi. Lect. Notes Comput. Sci. 9901:451–459, 2016a.
Tajbakhsh, N., Gurudu, S. R., and Liang, J., A comprehensive computer-aided polyp detection system for colonoscopy videos. Proc. Inform. Process. Med. Imag. Lect. Notes Comput. Sci. 9123:327–338, 2015b. https://doi.org/10.1007/978-3-319-19992-4_25.
Liu, J., Wang, D., Wei, Z., Lu, L., Kim, L., Turkbey, E., and Summers, R.M., .Colitis detection on computed tomography using regional convolutional neural networks. Proc. IEEE Int. Symp. Biomed. Imag. 863–866, 2016a.
Nappi, J.J., Hironaka, T., Regge, D., and Yoshida, H., .Deep transfer learning of virtual endoluminal views for the detection of polyps in CT colonography. Proc. Med. Imag. 97852B, 2016.
Tachibana, R., Näppi, J. J., Hironaka, T., Kim, S. H., and Yoshida, H., Deep learning for electronic cleansing in dual-energy ct colonography. Proc. SPIE Med. Imag. 9785:97851M, 2016.
Zhang, R., Zheng, Y., Mak, T. W. C., Yu, R., Wong, S. H., Lau, J. Y. W., and Poon, C. C. Y., Automatic detection and classification of colorectal polyps by transferring lowlevel CNN features from nonmedical domain. IEEE J. Biomed. Health Inf. 21:41–47, 2017.
Liao, S., Gao, Y., Oto, A., and Shen, D., Representation learning: A unified deep learning framework for automatic prostate mr segmentation. In: Proceedings of the medical image computing and computer-assisted intervention. Lect. Notes Comput. Sci. 8150:254–261, 2013.
Cheng, J.-Z., Ni, D., Chou, Y.-H., Qin, J., Tiu, C.-M., Chang, Y.-C., Huang, C.-S., Shen, D., and Chen, C.-M., Computer-aided diagnosis with deep learning architecture: Applications to breast lesions in US images and pulmonary nodules in CT scans. Nat. Sci. Rep. 6:24454, 2016a.
Guo, Y., Gao, Y., and Shen, D., Deformable MR prostate segmentation via deep feature learning and sparse patch matching. IEEE Trans. Med. Imag. 35(4):1077–1089, 2016.
Milletari, F., Navab, N., and Ahmadi, S.-A., V-Net: fully convolutional neural networks for volumetric medical image segmentation. arxiv:1606.04797, 2016b.
Yu, L., Yang, X., Chen, H., Qin, J., and Heng, P.A., Volumetric convnets with mixed residual connections for automated prostate segmentation from 3D MR images. Proc. Thirty-First AAAI Conf. Artif. Intell., 2017c.
Azizi, S., Imani, F., Ghavidel, S., Tahmasebi, A., Kwak, J. T., Xu, S., Turkbey, B., Choyke, P., Pinto, P., Wood, B., Mousavi, P., and Abolmaesumi, P., Detection of prostate cancer using temporal sequences of ultrasound data: A large clinical feasibility study. Int. J. Comput. Assist. Radiol. Surg. 11(6):947–956, 2016.
Shah, A., Conjeti, S., Navab, N., and Katouzian, A., Deeply learnt hashing forests for content based image retrieval in prostate MR images. Proc. SPIE Med. Imag. 9784:978414, 2016.
Zhu, Y., Wang, L., Liu, M., Qian, C., Yousuf, A., Oto, A., and Shen, D., MRI based prostate cancer detection with high-level representation and hierarchical classification. Med. Phys. 44(3):1028–1039, 2017.
Cha, K. H., Hadjiiski, L. M., Samala, R. K., Chan, H.-P., Cohan, R. H., Caoili, E. M., Paramagul, C., Alva, A., and Weizer, A. Z., Bladder cancer segmentation in CT for treatment response assessment: Application of deep-learning convolution neural network-a pilot study. Tomography 2:421–429, 2016.
Acknowledgements
This paper is supported by the Jiangsu Committee of Health on the subject (No. H2018071).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
We declare that we have no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors. Informed consent was obtained from all individual participants included in the study.
Additional information
This article is part of the Topical Collection on Image & Signal Processing
Rights and permissions
About this article
Cite this article
Xia, Kj., Yin, Hs. & Zhang, Yd. Deep Semantic Segmentation of Kidney and Space-Occupying Lesion Area Based on SCNN and ResNet Models Combined with SIFT-Flow Algorithm. J Med Syst 43, 2 (2019). https://doi.org/10.1007/s10916-018-1116-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-018-1116-1