Abstract
Computer-aided detection (CAD) can help colonoscopists reduce their polyp miss-rate, but existing CAD systems are handicapped by using either shape, texture, or temporal information for detecting polyps, achieving limited sensitivity and specificity. To overcome this limitation, our key contribution of this paper is to fuse all possible polyp features by exploiting the strengths of each feature while minimizing its weaknesses. Our new CAD system has two stages, where the first stage builds on the robustness of shape features to reliably generate a set of candidates with a high sensitivity, while the second stage utilizes the high discriminative power of the computationally expensive features to effectively reduce false positives. Specifically, we employ a unique edge classifier and an original voting scheme to capture geometric features of polyps in context and then harness the power of convolutional neural networks in a novel score fusion approach to extract and combine shape, color, texture, and temporal information of the candidates. Our experimental results based on FROC curves and a new analysis of polyp detection latency demonstrate a superiority over the state-of-the-art where our system yields a lower polyp detection latency and achieves a significantly higher sensitivity while generating dramatically fewer false positives. This performance improvement is attributed to our reliable candidate generation and effective false positive reduction methods.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Discrete Cosine Transform
- Candidate Location
- Convolutional Neural Network
- Vote Scheme
- Discrete Cosine Transform Coefficient
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Colon cancer most often develop from colonic polyps. However, polyp grow slowly and it typically take years for polyps to develop into cancer, making colon cancer amenable to prevention. Colonoscopy is the preferred procedure for preventing colon cancer. The goal of colonoscopy is to find and remove polyps before turning into cancer. Despite its demonstrated utility, colonoscopy is not a perfect procedure. A recent clinical study [5] reports that a quarter of polyps are missed during colonoscopy. Computer-aided polyp detection can help colonoscopists reduce their polyp miss-rate, in particular, during long and back-to-back procedures where fatigue and inattentiveness may result in miss detection of polyps.
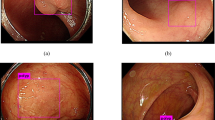
Significant variation in visual characteristics of polyps. (a) Color and appearance variation of the same polyp due to varying lighting conditions. (b) Texture and shape variation among polyps. Note how the distance between the polyps and colonoscopy camera determines the availability of polyp texture. (c) Other polyp-like structures in the colonoscopic view (Color figure online).
However, designing a high-performance system for computer-aided polyp detection is challenging: (1) Polyps appear differently in color, and even the same polyp, as shown in Fig. 1(a), may look differently due to varying lighting conditions. (2) Polyps have large inter- and intra-morphological variations. As shown in Fig. 1(b), the shapes of polyps vary considerably from one to another. The intra-shape variation of polyps is caused by various factors, including the viewing angle of the camera and the spontaneous spasms of the colon. (3) Visibility of the texture on the surface of polyps is also varying due to biological factors and distance between the polyps and the colonoscopy camera. This can be seen in Fig. 1(b) where texture visibility decrease as the polyps distance from the capturing camera. The significant variations among polyps suggest that there is no single feature that performs the best for detecting all the polyps.
As a result, to achieve a reliable polyp detection system, it is critical to fuse all possible features of polyps, including shapes, color, and texture. Each of these features has strengths and weaknesses. Among these features, geometric shapes are most robust because polyps, irrespective of their morphology and varying levels of protrusion, have at least one curvilinear head at their boundaries. However, this property is not highly specific to polyps. This is shown in Fig. 1(c) where non-polyp structures exhibit similar geometric characteristics to polyps. Texture features have the weakness of limited availability; however, when visible, they can distinguish polyps from some non-polyp structures such as specular spots, dirt, and fecal matter. In addition, temporal information is available in colonoscopy and may be utilized to distinguish polyps from bubbles or other artifacts that only briefly appear in colonoscopy videos.
Our key contribution of this paper is an idea to exploit the strengths of each feature and minimize its weaknesses. To realize this idea, we have developed a new system for polyp detection with two stages. The first stage builds on the robustness of shape features of polyps to reliably generate a set of candidate detections with a high sensitivity, while the second stage utilizes the high discriminative power of the computationally expensive features to effectively reduce false positive detections. More specifically, we employ a unique edge classifier coupled with a voting scheme to capture geometric features of polyps in context and then harness the power of convolutional deep networks in a novel score fusion approach to capture shape, color, texture, and temporal information of the candidates. Our experimental results based on the largest annotated polyp database demonstrate that our system achieves high sensitivity to polyps and generates significantly less number of false positives compared to state-of-the-art. This performance improvement is attributed to our reliable candidate generation and effective false positive reduction methods.
2 Related Works
Automatic polyp detection in colonoscopy videos has been the subject of research for over a decade. Early methods, e.g., [1, 3] for detecting colonic polyps utilized hand-crafted texture and color descriptors such as LBP and wavelet transform. However, given large color variation among polyps and limited texture availability on the surface of polyps (See Fig. 1), such methods could offer only a partial solution. To avoid such limitations, more recent techniques have considered temporal information [6] and shape features [2, 7, 9–11], reporting superior performance over the early polyp detection systems. Despite significant advancements, state-of-the-art polyp detection methods fail to achieve a clinically acceptable performance. For instance, to achieve the polyp sensitivity of 50 %, the system suggested by Wang et al. [11] generates 0.15 false positives per frame or approximately 4 false positive per second. Similarly, the system proposed in [10], which is evaluated on a significantly larger dataset, generates 0.10 false positives per frame. Clearly, such systems that rely on a subset of polyp characteristics are not clinically viable—a limitation that this paper aims to overcome.
3 Proposed Method
Our computer-aided polyp detection system is designed based on our algorithms [7, 8, 10], consisting of 2 stages where the first stage utilizes geometric features to reliably generate polyp candidates and the second stage employs a comprehensive set of deep features to effectively remove false positives. Figure 2 shows a schematic overview of the suggested method.
Our system consists of 2 stages: candidate generation and classification. Given a colonoscopy frame (A), we first obtain a crude set of edge pixels (B). We then refine this edge map using a classification scheme where the goal is to remove as many non-polyp boundary pixels as possible (C). The geometric features of the retained edges are then captured through a voting scheme, generating a voting map whose maximum indicates the location of a polyp candidate (D). In the second stage, a bounding box is estimated for each generated candidate (E) and then a set of convolution neural networks—each specialized in one type of features—are applied in the vicinity of the candidate (F). Finally, the CNNs are aggregated to generate a confidence value (G) for the given polyp candidate.
3.1 Stage 1: Candidate Generation
Our unique polyp candidate generation method exploits the following two properties: (1) polyps have distinct appearance across their boundaries, (2) polyps, irrespective of their morphology and varying levels of protrusion, feature at least one curvilinear head at their boundaries. We capture the first property with our image characterization and edge classification schemes, and capture the second property with our voting scheme.
Constructing Edge Maps. Given a colonoscopy image, we use Canny’s method to extract edges from each input channel. The extracted edges are then put together in one edge map. Next, for each edge in the constructed edge map, we determine edge orientation. The estimated orientations are later used for extracting oriented patches around the edge pixels.
Image Characterization. Our patch descriptor begins with extracting an oriented patch around each edge pixel. The patch is extracted so that the containing boundary is placed vertically in the middle of the patch. This representation allows us to capture desired information across the edges independent of their orientations. Our method then proceeds with forming \(8\times 16\) sub-patches all over the extracted patch. Each sub-patch has 50 % overlap with the neighboring sub-patches. For a compact representation, we compress each sub-patch into a 1D signal S by averaging intensity values along each column. We then apply a 1D discrete cosine transform (DCT) to the resulting signal:
where
With the DCT, the essential information of the intensity signal can be summarized in a few coefficients. We discard the DC component \(C_0\) because the average patch intensity is not a robust feature—it is affected by a constant change in patch intensities. However, the next 3 DCT coefficients \(C_1-C_3\) are more reliable and provide interesting insight about the intensity signal. \(C_1\) measures whether the average patch intensity along the horizontal axis is monotonically decreasing (increasing) or not, \(C_2\) measures the similarity of the intensity signal against a valley (ridge), and finally \(C_3\) checks for the existence of both a valley and a ridge in the signal. The higher order coefficients \(C_4-C_{15}\) may not be reliable for feature extraction because of their susceptibility to noise and other degradation factors in the images.
The selected DCT coefficients \(C_1-C_3\) are still undesirably proportional to linear illumination scaling. We therefore apply a normalization treatment. Mathematically,
Note that we use the norm of the selected coefficients for normalization rather than the norm of entire DCT coefficients. By doing so, we can avoid the expensive computation of all the DCT components. The final descriptor for a given patch is obtained by concatenating the normalized coefficients selected from each sub-patch.
The suggested patch descriptor has 4 advantages. First, our descriptor is fast because compressing each sub-patch into a 1D signal eliminates the need for expensive 2D DCT and that only a few DCT coefficients are computed from each intensity signal. Second, due to the normalization treatment applied to the DCT coefficients, our descriptor achieves invariance to linear illumination changes, which is essential to deal with varying lighting conditions (see Fig. 1). Third, our descriptor is rotation invariant because the patches are extracted along the dominant orientation of the containing boundary. Fourth, our descriptor handles small positional changes by selecting and averaging overlapping sub-patches in both horizontal and vertical directions.
Edge Classification. Our classification scheme has 2 layers. In the first layer, we learn a discriminative model to distinguish between the boundaries of the structures of interest and the boundaries of other structures in colonoscopy images. The structures of interest consists of polyps, vessels, lumen areas, and specular reflections. Specifically, we collect a stratified set of \(N_1=100,000\) oriented patches around the boundaries of structures of interest and r andom structures in the training images, \(S^1=\{(p_i,y_i)|y_i\in \{p,v,l,s,r\}, i=1,2,...,N_1\}\). Once patches are extracted, we train a five-class random forest classifier with 100 fully grown trees. The resulting probabilistic outputs can be viewed as the similarities between the input patches and the predefined structures of interest. Basically, the first layer receives low-level image features from our patch descriptor and then produces mid-level semantic features.
In the second layer, we train a 3-class random forest classifier with 100 fully grown trees. Specifically, we collect \(N_2=100,000\) pairs of oriented patches, of which half are randomly selected from the polyp boundaries and the rest are selected from random non-polyp edge segments. For an edge pixel at angle \(\theta \), one can obtain two oriented image patches \(\{p_i^1, p_i^2 \}\) by interpolating the image along the two possible normal directions \(\{n_i^1, n_i^2 \}\). As shown in Fig. 3(a), for an edge pixel on the boundary of a polyp, only one of the normal directions points to the polyp region. Our classification scheme operates on each pair of patches with two objectives: (1) to classify the underlying edge into polyp and non-polyp categories, and (2) to determine the desired normal direction among \(n_i^1\) and \(n_i^2\) such that it points towards the polyp location. Henceforth, we refer to the desired normal direction as “voting direction”.
(a) A pair of image patches \(\{ p_i^1, p_i^2 \}\) extracted from an edge pixel. The green and red arrows show the two possible normal directions \(\{ n_i^1, n_i^2 \}\) for a number of selected edges on the displayed boundary. The normal directions are used for patch alignment. (b) The suggested edge classification scheme given a test image. The edges that have passed the classification stage are shown in green. The inferred normal directions are visualized with the blue arrows for a subset of the retained edges (Color figure online).
Once image pairs are collected, we order the patches \(\{ p_i^1, p_i^2 \}\) within each pair according to the angles of their corresponding normal vectors, \( \angle n_i^1\) \(<\) \(\angle n_i^2\). In this way, the patches are represented in a consistent order. Each pair of ordered patches is then assigned a label \(y_i\in \{0,1,2\}\), where “0” indicates that the underlying edge does not lie on a polyp boundary, “1” indicates that the edge lies on a polyp boundary and that \(n_i^1\) is the voting direction, and “2” indicates that the edge lies on a polyp boundary but \(n_i^2\) shows the voting direction. Mathematically, \(S^2=\{(p_i^1, p_i^2,y_i)|y_i\in \{0,1,2\}, i=1,2,...,N_2\}\). To generate semantic features, each pair of ordered patches undergoes the image characterization followed by the first classification layer. The resulting mid-level features are then concatenated to form a feature vector \(f_i\). This process is repeated for \(N_2\) pairs of ordered patches, resulting in a labeled feature set, \(\{(f_i,y_i)|y_i\in \{0,1,2\}, i=1,2,...,N_2\}\), which is needed to train the second classifier. We train a 3-class classifier to learn both edge labels and the voting directions (embedded in \(y_i\)). Figure 3(b) illustrates how the suggested edge classification scheme operates given a test image.
Candidate Localization. Our voting scheme is designed to generate polyp candidates in regions surrounded by curvy boundaries. The rationale is such boundaries can represent the heads of polyps. In our voting scheme, each edge that has passed the classification stage, casts a vote along its voting direction (inferred by the edge classifier). The vote cast by the voter v at a receiver pixel \(r=[x,y]\) is computed as

where the exponential and cosinusoidal functions enable smooth vote propagation, which we will later use to estimate a bounding box around each generated candidate. In Eq. 2, \(C_{v_i}\) is the classification confidence, \(\vec {vr}\) is the vector connecting the voter and receiver, \(\sigma _F\) controls the size of the voting field, and \(\angle {\vec {n^*} \vec {vr}}\) is the angle between the voting direction \(\vec {n^*}\) and \(\vec {vr}\). Figure 4(a) shows the voting field for an edge pixel lying at 135 degree. As seen, due to the condition set on \(\angle {\vec {n^*} \vec {vr}}\), the votes are cast only in the region pointed by the voting direction.
It is essential for our voting scheme to prevent vote accumulation in the regions that are surrounded by low curvature boundaries. For this purpose, our voting scheme first groups the voters into 4 categories according to their voting directions, \(V^k\text {=}\{ v_i | \frac{k\pi }{4}\) \(<\) \( mod(\angle n_i^*,\pi )\) \(<\) \(\frac{(k+1)\pi }{4} \},\) \(k=0...3\). Our voting scheme then proceeds by accumulating votes of each category in a separate voting map. To produce the final voting map, we multiply the accumulated votes generated in each category. A polyp candidate is then generated where the final voting map has the maximum vote accumulation (MVA). Mathematically,
Comparing Fig. 4(b) and (c) clarifies how the suggested edge grouping mitigates vote accumulation between parallel lines, assigning higher temperature to only regions surrounded by curvy boundaries. Another important characteristic of our voting scheme is the utilization of voting directions. As shown in Fig. 4(d), casting votes along both possible normal directions can result in mislocalized candidates; however, incorporating voting directions allows for more accurate candidate localization (Fig. 4(e)).
(a) The generated voting map for an edge pixel lying at 135 degree. (b) Without edge grouping, all the votes are accumulated in one voting map, which results in undesirable vote accumulation between the parallel lines. (c) With the suggested edge grouping, higher temperature is assigned to only within the curvy boundaries. (d) Casting votes along both possible normal directions can result in a candidate placed outside the polyp region. (e) Casting votes only along the inferred voting directions results in a successful candidate localization. (f) A narrow band is used for estimating a bounding box around candidates. (g) A synthetic shape and its corresponding voting map. The isocontours and the corresponding representative isocontour are shown in blue and white, respectively (Color figure online).
3.2 Stage 2: Candidate Classification
Our candidate classification method begins with estimating a bounding box around each polyp candidate followed by a novel score fusion framework based on convolutional neural networks (CNNs) [4] to assign a confidence value to each generated candidate.
Bounding Box Estimation. To measure the extent of the polyp region, we estimate a narrow band around each candidate, so that it contains the voters that have contributed to vote accumulation at the candidate location. In other words, the desired narrow band will enclose the polyp boundary and thus can be used to estimate a bounding box around the candidate location. As shown in Fig. 4(f), the narrow band B consists of a set of radial lines \(\ell _\theta \) parameterized as \( \ell _\theta : MVA+t[\cos (\theta ), \sin (\theta )]^T, t\in [t_\theta -\frac{\delta }{2},t_\theta +\frac{\delta }{2}] \), where \(\delta \) is the bandwidth, and \(t_\theta \) is the distance between the candidate location and the corresponding point on the band skeleton at angle \(\theta \). Once the band is formed, the bounding box is localized so that it fully contains the narrow band around the candidate location (see Fig. 4(f)). The bounding box will be later used for data augmentation where we extract patches in multiple scales around the polyp candidates.
To estimate the unknown \(\delta \) and \(t_\theta \) for a given candidate, we use the isocontours of the corresponding voting map. The isocontour \(\varPhi _c\) of the voting map V is defined as \(\varPhi _c=\{(x,y)|V(x,y)=c\times M\}\) where M denotes the maximum of the voting map and c is a constant between 0 and 1. As shown in Fig. 4(g), the isocontours of the voting map, particularly those located farther away from the candidate, have the desirable feature of following the shape of the actual boundary from which the votes have been cast at the candidate location. Therefore, one can estimate the narrow band’s parameters from the isocontours such that the band encloses the object’s boundary. However, in practice, the shape of far isocontours are undesirably influenced by other nearby voters in the scene. We therefore obtain the representative isocontour \(\bar{\varPhi }\) by computing the median shape of the isocontours of the voting map (see Fig. 4(g)). We have experimented with different sets of isocontours and found out that as long as their parameter c is uniformly selected between 0 and 1, the resulting representative isocontour serves the desired purpose.
Let \(d_{iso}^i\) denotes the distance between the \(i^{th}\) point on the representative isocountour \(\bar{\varPhi }\) and the candidate location. We use \(d_{iso}^i\) to predict \( d_{obj}^i\), the distance between the corresponding point on the object boundary and the candidate location. For this purpose, we employ a second order polynomial regression model
where \(b_0\), \(b_1\), and \(b_2\) are the regression coefficients that are estimated using a least square approach. Once the model is constructed, we take the output of the model \(d_{obj}\) at angle \(\theta \) with respect to MVA as \(t_{\theta }\) and the corresponding prediction interval as the bandwidth \(\delta \).
Probability Assignment. We propose a score fusion framework based on convolutional neural networks (CNNs) that can learn and integrate color, texture, shape, and temporal information of polyps in multiple scales for more accurate candidate classification. We choose to use CNNs because of their superior performance in major object detection challenges. The attractive feature of CNNs is that they jointly learn a multi-scale set of image features and a discriminative classifier during a supervised training process. While CNNs are known to learn discriminate patterns from raw pixel values, it turns out that preprocessing and careful selection of the input patches can have a significant impact on the performance of the subsequent CNNs. Specifically, we have found out that partial illumination invariance achieved by histogram equalizing the input patches significantly improves the performance of the subsequent CNNs and that curse of dimensionality caused by patches with more than 3 channels results in CNNs with inferior performance.
Considering these observations, we propose a 3-way image presentation that is motivated by the three major types of polyp features suggested in the literature: (1) for color and texture features, we collect histogram-equalized color patches \(P_{C}\) around each polyp candidate; (2) for temporal features, we form 3-channel patches \(P_{T}\) by stacking histogram-equalized gray channel of the current frame and that of the previous 2 frames; (3) for shape in context, we form 3-channel patches \(P_{S}\) by stacking the gray channel of the current frame and the corresponding refined edge channel and voting channel produced in the candidate generation stage (see Fig. 2).
We collect the three sets of patches \(P_{C}\), \(P_{T}\), and \(P_{S}\) from candidate locations in the training videos, label each individual patch depending on whether the underlying candidate is a true or false positive, and then train a CNN for each set of the patches. Figure 5(a) shows the test stage of the suggested score fusion framework. Given a new polyp candidate, we collect the three sets of patches in multiple scales and orientations around the candidate location, apply each of the trained CNNs on the corresponding patches, and take the maximum response for each CNN, resulting in three probabilistic scores. The final classification confidence is computed by averaging the resulting three scores.
4 Experiments
For evaluation, we have used 40 short colonoscopy videos. We have randomly halved the database at video level into the training set containing 3800 frames with polyps and 15100 frames without polyps, and the test set containing 5700 frames with polyps and 13200 frames without polyps. Each colonoscopy frame in our database comes with a binary ground truth image. For performance evaluation, we consider a detection as a true (false) positive if it falls inside (outside) the white region of the ground truth image.
Our candidate generation stage yielded a sensitivity of 73.6 % and 0.8 false positives/frame. For candidate classification, we used Krizhevsky’s GPU implementation [4] of CNNs. With data augmentation, we collected 400,000 32\(\,\times \,\)32 patches for \(P_{C}\), \(P_{T}\), and \(P_{S}\) where half of the patches were extracted around false positive candidates and the rest around true positive candidates. Specifically, for a candidate with an \(N\times N\) bounding box, we extracted patches at three scales \(sN\times sN\) with \(s\in \{1,1.2,1.4\}\) and then resized them to 32\(\,\times \,\)32 patches. Furthermore, we performed data augmentation [4] by extracting patches at multiple orientations and translation in each given scale. We have used the layout shown in Fig. 5(b) for all the CNNs used in this paper.
Figure 6(a) shows FROC analysis of the suggested system. As seen, our system based on the suggested score fusion approach shows a relatively stable performance over a wide range of voting fields. For comparison, we have also reported the performance of our system based on individual CNNs trained using color patches (\(P_C\)), temporal patches (\(P_T\)), and shape in context patches (\(P_S\)). We have also experimented with the channel fusion approach where color, shape, and temporal patches are stacked for each polyp candidate followed by training one CNN for the resulting 9-channel training patches. To avoid clutter in the figure, only their best performance curves obtained by \(\sigma _F=70\) are shown. As seen in Fig. 6(a), the proposed score fusion framework yields the highest performance, achieving 50 % sensitivity at 0.002 FPs/frame, outperforming [10] with 0.10 FPs/frame at the same sensitivity.
FROC analysis is widely used for evaluating computer-aided detection systems designed for static datasets such as CT scans and mammograms. However, for temporal or sequence-based datasets such as colonoscopy videos, it has the drawback of excluding the factor of time. While it is desirable for a polyp detection system to detect as many polyp instances as possible, it is also important to measure how quickly a polyp is detected after it appears in the video. We therefore employ a new performance curve [8] that measures the polyp detection latency with respect to the number of false positives. Briefly, if \(t_1\) denotes the arrival frame of the polyp, \(t_2\) denotes the frame in which the polyp is detected, and fps is the frame rate of the video, the detection latency is then computed as \(\varDelta T=(t_2-t_1)/fps\). As with FROC, we change a threshold on the detection confidences and then at each operating point measure the median polyp detection latency of the test positive shots and the number of false positives in the entire test set. As seen in Fig. 6(b), different variations of our system yield significantly less number of false positives than our previous work [10] at nearly all operating points.
On a desktop computer with a 2.4 GHz quad core Intel and an Nvidia GeForce GTX 760 video card, our system processes each image at 2.65 s, which is significantly faster than [11] with run-time of 7.1 s and [2] with run-time of 19 s. We should note that a very large fraction of the computation time (2.6 s) is caused by the candidate generation stage and that the candidate classification based on CNNs is extremely fast because CNNs are only applied to the candidate location in each frame. We expect a significant speedup of our system using parallel computing optimization.
5 Conclusion
We proposed a new computer-aided polyp detection system for colonoscopy videos. Our system was based on context-aware shape features to generate a set of candidates and convoluational neural networks to reduce the generated false positives. We evaluated our system using the widely-used FROC analysis, achieving 50 % sensitivity at 0.002 FPs/frame, outperforming state-of-the-art systems [10, 11], which generate 0.15 FPs/frame and 0.10 FPs/frame at 50 % sensitivity, respectively. We also evaluated our system using a latency analysis, demonstrating a significantly lower polyp detection latency than [10] particularly in low false positive rates.
References
Alexandre, L.A., Nobre, N., Casteleiro, J.: Color and position versus texture features for endoscopic polyp detection. In: International Conference on BioMedical Engineering and Informatics, BMEI 2008, vol. 2, pp. 38–42. IEEE (2008)
Bernal, J., Snchez, J., Vilario, F.: Towards automatic polyp detection with a polyp appearance model. Pattern Recogn. 45(9), 3166–3182 (2012)
Karkanis, S.A., Iakovidis, D.K., Maroulis, D.E., Karras, D.A., Tzivras, M.: Computer-aided tumor detection in endoscopic video using color wavelet features. IEEE Trans. Inform. Technol. Biomed. 7(3), 141–152 (2003)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012), https://code.google.com/p/cuda-convnet/
Leufkens, A., van Oijen, M., Vleggaar, F., Siersema, P.: Factors influencing the miss rate of polyps in a back-to-back colonoscopy study. Endoscopy 44(05), 470–475 (2012)
Park, S.Y., Sargent, D., Spofford, I., Vosburgh, K.: A-Rahim, Y.: A colon video analysis framework for polyp detection. IEEE Trans. Biomed. Eng. 59(5), 1408–1418 (2012)
Tajbakhsh, N., Chi, C., Gurudu, S.R., Liang, J.: Automatic polyp detection from learned boundaries. In: 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI), pp. 97–100. IEEE (2014)
Tajbakhsh, N., Gurudu, S.R., Liang, J.: Automatic polyp detection in colonoscopy videos using an ensemble of convolutional neural networks. In: Biomedical Imaging (ISBI), 2015 IEEE 12th International Symposium on. pp. 79–83. IEEE (2015)
Tajbakhsh, N., Gurudu, S.R., Liang, J.: A classification-enhanced vote accumulation scheme for detecting colonic polyps. In: Yoshida, H., Warfield, S., Vannier, M.W. (eds.) Abdominal Imaging 2013. LNCS, vol. 8198, pp. 53–62. Springer, Heidelberg (2013)
Tajbakhsh, N., Gurudu, S.R., Liang, J.: Automatic polyp detection using global geometric constraints and local intensity variation patterns. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014, Part II. LNCS, vol. 8674, pp. 179–187. Springer, Heidelberg (2014). http://dx.doi.org/10.1007/978-3-319-10470-6_23
Wang, Y., Tavanapong, W., Wong, J., Oh, J., de Groen, P.: Part-based multi-derivative edge cross-section profiles for polyp detection in colonoscopy. IEEE J. Biomed. Health Inform. PP(99), 1–1 (2013)
Acknowledgment
This research has been supported by an ASU-Mayo Clinic research grant.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Tajbakhsh, N., Gurudu, S.R., Liang, J. (2015). A Comprehensive Computer-Aided Polyp Detection System for Colonoscopy Videos. In: Ourselin, S., Alexander, D., Westin, CF., Cardoso, M. (eds) Information Processing in Medical Imaging. IPMI 2015. Lecture Notes in Computer Science(), vol 9123. Springer, Cham. https://doi.org/10.1007/978-3-319-19992-4_25
Download citation
DOI: https://doi.org/10.1007/978-3-319-19992-4_25
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-19991-7
Online ISBN: 978-3-319-19992-4
eBook Packages: Computer ScienceComputer Science (R0)