Abstract
Chemical shifts contain important site-specific information on the structure and dynamics of proteins. Deviations from statistical average values, known as random coil chemical shifts (RCCSs), are extensively used to infer these relationships. Unfortunately, the use of imprecise reference RCCSs leads to biased inference and obstructs the detection of subtle structural features. Here we present a new method, POTENCI, for the prediction of RCCSs that outperforms the currently most authoritative methods. POTENCI is parametrized using a large curated database of chemical shifts for protein segments with validated disorder; It takes pH and temperature explicitly into account, and includes sequence-dependent nearest and next-nearest neighbor corrections as well as second-order corrections. RCCS predictions with POTENCI show root-mean-square values that are lower by 25–78%, with the largest improvements observed for 1Hα and 13C′. It is demonstrated how POTENCI can be applied to analyze subtle deviations from RCCSs to detect small populations of residual structure in intrinsically disorder proteins that were not discernible before. POTENCI source code is available for download, or can be deployed from the URL http://www.protein-nmr.org.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The chemical shift is the single most easy obtainable parameter from NMR experiments, can be measured with very high precision, and carries important information on molecular structure and dynamics. The chemical shift of a nucleus in a random coil polypeptide will depend on intrinsic factors, such as the identities of the nearest residues, as well as extrinsic factors such as pH, ionic strength and temperature. All these aspects affect the electronic and spatial structure of the peptide chain, and thereby alter the chemical shifts of the affiliated nuclei. As a consequence, a segment of a protein chain that is devoid of any structure imposed by long-range non-bonded interactions, such as hydrogen bonds, burial of hydrophobic side chains, and Coulombic interactions, can be considered to have a random coil structure, and the chemical shifts observed for this particular segment of amino acids across different proteins would be identical. The chemical shifts for the nuclei in the segment would then be considered random-coil chemical shifts (RCCSs), as these reflect the dynamically averaged chemical shifts experienced by rapid conformational dynamics on the free energy landscape governed by local interactions. Concurrently, deviations from RCCSs can be used to detect the presence of secondary structure formation (Williamson 1990; Spera and Bax 1991; Marsh et al. 2006; Camilloni et al. 2012; Kjaergaard and Poulsen 2012).
Intrinsically disordered proteins (IDPs) constitute a hitherto little-recognized, but important part of the protein universe (Ward et al. 2004; Dyson and Wright 2005; van der Lee et al. 2014; Wright and Dyson 2015). Due to the dynamic nature of IDPs, the single most powerful structure-determination technique, X-ray crystallography of crystals, is disqualified and NMR spectroscopy has become the prime tool for their investigation. Chemical shifts have become a prime source of information on IDP structure, as these characterize the protein at the residue level, and chemical shifts can be obtained by the simple and effective process of sequence-specific resonance assignment (Felli and Pierattelli 2012; Brutscher et al. 2015). To capture random coil chemical shifts, there are several sets of reference values in use, which have quite disparate origins: First, guest-host substitutions in small peptides, such as GGXGG were employed to obtain reference chemical shift values for the nuclei of the amino acid X (Richarz and Wüthrich 1978; Bundi and Wüthrich 1979; Braun et al. 1994; Wishart et al. 1995; Schwarzinger et al. 2000; Kjaergaard et al. 2011). However, these peptides do not sample conformational space in a representative way for all peptides (Kjaergaard and Poulsen 2011) and small differences between GGXGG (Schwarzinger et al. 2000) and GGXAG (Wishart et al. 1995) were responsible for divergent interpretations in the propensities of a fragment of the human protein tau, involved in human neurodegeneration (Eliezer et al. 2005; Mukrasch et al. 2005), clearly showing that nearest neighbor effects need critical evaluation (Tamiola et al. 2010). An alternative approach presented in the literature is the collection of a database of chemical shifts for proteins of known structure, and to classify those regions outside canonical secondary structure and loop regions as ‘coil’ (Wang and Jardetzky 2002; De Simone et al. 2009). Lamentably, this approach suffers from the heterogeneity of conditions used for protein structure determination and NMR data acquisition, as well as the lack of a clear definition of which regions would classify as representative of a total lack of structure, beyond that dictated by the sequence. As a potential solution, Tamiola et al. (2010) published a curated database of chemical shifts for IDPs, and used a statistical method to exclude chemical shifts that would mark local deviations from random coil behavior. Their method, called ncIDP, took into account the importance of neighboring residues in the sequence, as demonstrated by others (Braun et al. 1994; Wishart et al. 1995; Schwarzinger et al. 2001), and proved to be more appropriate for predicting the RCCSs of IDPs than existing methods at the time (Tamiola et al. 2010; Kjaergaard and Poulsen 2011; Kragelj et al. 2013). However, the small number of IDPs used to derive the ncIDP reference chemical shift database resulted in very little data for amino acids with low abundance, such as Trp and Cys, and ncIDP suffered from substantial variation in NMR sample conditions of the used entries. We demonstrated previously (Nielsen and Mulder 2016) that IDPs are a complex concatenation of regions with various magnitudes of order and disorder, and the ncIDP database might therefore still inadvertently suffer from heterogeneous composition by including fragments with residual order. To remedy this situation, we therefore devised a statistically robust procedure for assessing the degree of disorder for each residue (coined the CheZOD Z-score), and this metric is used herein for the compilation of a database containing exclusively disordered residues. In addition, others have previously shown that the effects of temperature and pH are highly significant (Merutka et al. 1995; Kjaergaard et al. 2011), and that these need to be properly accounted for, in order to arrive at an adequate reference dataset for RCCSs.
Herein we present POTENCI—Prediction Of TEmperature, Neighbor and pH Corrected shifts for Intrinsically disordered proteins—which predicts the RCCSs for the backbone nuclei as well as 13Cβ and 1Hβ for IDPs with a significantly higher accuracy than currently available methods. The algorithm takes pH and temperature explicitly into account, and includes sequence-dependent nearest and next-nearest neighbor corrections. A first such an empirical database, presented by Tamiola et al. (2010) in 2010, contained 6903 chemical shifts (reduced to 4439 after removing chemical shifts that were judged to be outliers) obtained for 14 proteins, and these were used to derive a model consisting of 20 random coil reference chemical shift values (with GXG as reference) and 40 (assumed independent) nearest neighbor corrections to predict the chemical shifts for backbone and 13Cβ nuclei from sequence for the central amino acid in any given tripeptide. The POTENCI database presented herein now contains 47,757 unique chemical shifts obtained from 137 proteins, comprising 9810 residues that are soundly classified as disordered under native conditions. The roughly ten-fold extension of the database size has allowed us to take a number of aspects into account, which was not possible previously. First, next-nearest neighbor corrections were included in the model, such that the prediction is based on pentapeptides, rather than tripeptides. This is especially relevant to account for ring current shifts due to aromatic side chains. Second, neighbor corrections no longer need to be assumed independent of the central amino acid, and center-type-specific corrections were extracted. Such correlated corrections were found to be important for Gly and Pro, in particular. This result was anticipated, as Gly and Pro sample backbone dihedral angle space very differently from the remaining amino acids (Ramachandran et al. 1963), and the resulting correction effects therefore vary significantly. Third, we used an electrostatic model (pepKalc; available from http://www.protein-nmr.org) (Tamiola et al. 2018) to compute the average protonation state of all titratable side chains along the sequence at a given pH and ionic strength, in order to apply appropriate corrections for Asp, Glu, His, Tyr, Cys, and Lys side chains (Arg can safely be considered to always be protonated in IDPs). Fourth, a correction for temperature is explicitly included, and this considerably and predominantly affects 15N chemical shift prediction. Using POTENCI, we are able to predict 1Hα, 1HN, 13Cα, 13Cβ, 13C′, and 15N RCCSs better than the current prominent approaches ncIDP by Tamiola, Acar and Mulder (TAM) (Tamiola et al. 2010) and RCCSs computed from the QQXQQ peptide database of Kjaergaard, Brander and Poulsen (KBP) (Kjaergaard et al. 2011; Kjaergaard and Poulsen 2011). Root-mean-square (rms) values are lower by 25–78% in comparison to the TAM and KBP approaches, with the largest improvement observed for 13C′ and 1Hα. More importantly, many deviations from RCCSs observed with ncIDP and the QQXQQ reference sets that might be considered signs of structuration are absent with POTENCI. The strong improvement afforded by POTENCI makes it a more reliable tool for detecting small, but relevant RCCS deviations in IDPs that may be correlated with functional outcomes.
Methods
Parameterization of predicted random coil chemical shift
The sequence-corrected RCCS for a nucleus in a pentapeptide, p = (i − 2, i − 1, i, i + 1, i + 2), with amino acid type, a i for position, i, at a given pH, with known pKas for the central triplet, t = (i − 1, i, i + 1) and temperature, T (in K), is calculated as:
where
and
where
here \({\delta _{RC}}\left( {{a_i},298~{\text{K}},~{\text{pH}}~7} \right)\) is the random coil chemical shift for residue type, a i , at position i, for the reference condition 298 K and pH 7, \(\Delta \left( p \right)\) is the sum of the linear correction factors for the nearest and next-nearest amino acid neighbors, \(\chi \left( p \right)\) is a correlated correction term (second-order effect) for the combination of amino acid types for the center residue and its nearest/next-nearest neighbor. The N- and C-terminal residues were not included in the dataset, but residues next to the termini were included. For these, next-terminal residues, the next-nearest neighbor past the N- and C-termini was treated as an extra type of amino acid in the calculation of Δk (for k = − 2 and 2). While, exhaustively, χk would need 1600 constants for parameterization, in practice most are negligible and some can be grouped (see below), meaning that only between 17 and 56 unique non-zero parameters were necessary here. The variation of the random coil chemical shift with temperature is accounted for by using linear temperature coefficients, β i , (for the residue type a i ) derived in Kjaergaard et al. (2011). \({\varepsilon _{{\text{pH}}}}\left( {t,{\text{pH}}} \right)\) corrects for the effect of non-neutral pH for titratable amino acid side chains in each triplet, t, where the correction for each residue, ε k , is derived using the difference between the chemical shift of the fully protonated and deprotonated states, \({\Delta}\delta _{{HA - A}}^{k}\), as determined in Platzer et al. (2014) (which is non-zero only for titratable amino acids and residue neighbors to titratable amino acids) and the fractional population of the protonated state, fHA, at the specified pH. fHA depends on the pKa and cooperativity constant, n H , which were both estimated using pepKalc (http://www.protein-nmr.org) (Tamiola et al. 2018) and the estimation of pKa and n H in pepKalc depend on the ionic strength.
Fitting of amino acid neighbor corrections
The chemical shift from a submitted sequence-specific assignment is first calibrated for temperature and pH as follows:
where \({\delta _{obs}}(i,I)\) is the observed chemical shift for residue i for the protein with id, I, and λ(I) is an offset correction for protein I, while the other terms are as defined above. The left-hand side of Eq. 7, containing only observed values and fixed parameters, is used to fit to the parameterization on the right-hand side containing the free variables. To prevent over-fitting of the experimental data, the smallest possible set of free parameters that provides an adequate fit of the experimental data is used. e.g. the reference offset correction is parameterized as:
e.g. only a subset of all proteins (the ones with id, \(I \in \Lambda\), where \(\Lambda\) is a subset of all protein ids) are reference corrected with offset ρ I . Hence, the values of \({\rho _I}~{\text{for}}~I \in \Lambda\) are to be determined in the fitting procedure. In addition, the exact nature of the subset, \(\Lambda\), is determined through optimization of repeated fits with different definitions of the subset (see below).
Rather than determining a weight for each possible amino acid neighbor and next-nearest neighbor, a principle component representation (Georgiev 2009) with possibly fewer than 20 parameters is used; the correction for the k’th amino acid neighbor of type ai+k is parameterized as:
where \({\alpha _j}({a_{i+k}})\) is the value of the j‘th principal component corresponding to the amino acid type, \(w_{j}^{k}\) (k = − 2, − 1, 1, 2 and j = 1, 2, …, q k ) are the adjustable weights, and γ k ≤ 20 is the number of principle components used (to be optimized).
The second-order amino acid neighbor correction term is parameterized using grouping of the amino acids and subset application:
where amino acids were grouped into 7 categories notated here with g(a) = “G”, “P”, “r”, “a”, “+”, “−”, “p” if the amino acid, a, is either G, P, F/Y/W (aromatic), L/I/V/M/C/A (aliphatic), K/R (positive), D/E (negative), or N/Q/S/T/H (polar), respectively, Π is the index set corresponding to the combined position and combination of groups that produces a significant chemical shift perturbation, and \(\omega _{{l,m}}^{k}\) are the adjustable weights (k = − 2, − 1, 1, 2) and l, m is one of the seven groups defined above. For example, the weight, \(\omega _{{G,r}}^{{ - 1}}\), corresponds to a correction to the chemical shifts for the central Gly residue Gly (l = g(G) = “G”) due to the presence of an aromatic residue (m = “r”), located at the position immediately before (k = − 1), alternatively denoted as the pentapeptide xrGxx, where “x” denotes any amino acid type.
To summarize, the chemical shifts are fitted for an assumed model of the significant parameters defined by the set of subsets:
where Γ and Π are the subsets defined above and Γ is the set defined by the limits, γ k
For such a given model, M, the number, N M of adjustable weights for fitting is:
where N RC is the number of fitted random coil chemical shifts for the center residue, \({\delta _{RC}}\left( {{a_i},298\,{\text{K}},~{\text{pH}}~7} \right)\), corresponding to the number of amino acids with assigned chemical shift for the particular nucleus, i.e. N A = 20 except for HN (n.a. for Pro) and Cβ/Hβ (n.a. for Gly) where in these cases N RC = 19. Note that for residues with two Hβ protons and Gly with two Hα protons, our method predicts the average of the chemical shifts. \({N_\Gamma } \le 4 \times 20\) is the number of parameters used for fitting the neighboring amino acids contribution, \({N_\Lambda } \le {N_P}\) is the number of proteins where the offset is corrected (N P is the total of number of proteins in the training set) and \({N_\Pi } \ll 4 \times 7 \times 7\) is the number of parameters used for parameterizing the contribution from combinations of center and neighbor amino acids (this number was significantly smaller than the maximum theoretical value in our fitting, see “results”).
For a given model, M, the derivation of the weights, \(\left( {{\delta _{RC}},w_{j}^{k},~\omega _{{l,m}}^{k},{\rho _I}} \right)\), (where \({\delta _{RC}}\) denotes the set of random coil chemical shifts for all the center residue types) that minimize the sum of squared differences between observed and predicted chemical shifts, reduces to a standard linear least squares fitting problem, which can be solved with procedures similar to those described in Tamiola et al. (2010). The optimal model must represent the best compromise between having the closest agreement between observed and predicted shifts and, at the same time, using the fewest possible number of free parameters. This is accomplished here by choosing the model with the lowest value of Akaike’s information criterion, AIC: (Akaike 1974, 1985)
where N tot is the number of chemical shift data points, rms is the resulting square root of the average of squared differences between observed and predicted shifts after performing the least squares fit, N M (Eq. 13) is the number of parameters used by the model, M, and r VIF ≥ 1 is a parameter that over-weights the number of model parameters relative to the classical AIC. r VIF can be interpreted as the variance inflation factor (Theil and Theil 1971) accounting for the (moderate) correlation between data points as discussed before in relation to chemical shifts (Nielsen et al. 2012) (values between 2.5 and 5.0 were used here). The optimal model having the lowest AIC was derived by varying the model definition systematically using a genetic algorithm (see Supplementary Methods for all details). The fitting procedure was coded in python using the numpy.linalg library for the least squares fitting routines and in-house developed procedures similar to ones described in Nielsen et al. (2016) for the genetic algorithm.
Briefly, the optimization algorithm consisted of five consecutive cycles of parameter fitting followed by outlier stripping using decreasing values of the variance inflation factor. In the first cycles, the aim was a robust fitting, whereas in the later cycles, the aim progressively changed towards selecting for the smallest residual error of fitting. At the end of each cycle, outliers were removed according to a principle of matching the observed data set quantiles to theoretical quantiles for a normal distribution. Following this principle, the absolute errors ε (difference between observed and predicted shifts) scaled by the standard deviation, σ, among all errors in the data set were identified:
The N data points were ranked according to their value of ε.
This means that for a rank, k, the fraction, F obs , of observed errors, ε < ε k is:
and εk is the k’th N-quantile, Q obs , for the data sample. For comparison, for the k’th ranked point with corresponding fraction, f k , the expected value of the error is the theoretical quantile, Q theo :
where F theo is the theoretical cumulative distribution function, which is the standard half-normal distribution here. To identify outliers in the data sample, we removed the points corresponding to the largest errors until the observed and theoretical quantiles were matched for ε = 3.0, i.e. until:
where the integer number for N × 0.0027 was used. At the end of each cycle, all data points were (re)-evaluated for possible outlier-stripping, including points removed in the preceding cycles. Each cycle consisted of 12,000 steps of subset redefinition followed by least squares fitting (for details, see Supplementary Methods). The parameters in the final cycle, leading to the lowest value of AIC, were retained. The parameters were optimized based on sets of experimentally assigned chemical shifts from the BMRB database. Residues classified as disordered are those with CheZOD Z-score < 3.0 (Nielsen and Mulder 2016), and these were used for the subsequent fitting. Since the definition of the Z-score itself depends on the sequence-corrected random coil chemical shift, the parameterization of the random coil shifts was performed in three iterations, each time revising the set of residues used for fitting, using progressively more data (see all details below).
Construction of the database of intrinsically disordered regions of proteins with chemical shifts
The random coil chemical shift prediction parameters were fitted in three iterations, each time using a new, and larger, set of chemical shifts from disordered residues. In the first two iterations, residues from the published CheZOD dataset, containing 119 proteins was used (Nielsen and Mulder 2016). In the third iteration, the dataset was expanded with another complementary set of disordered proteins. This complementary set was derived by considering all published chemical shift datasets in the BMRB database (retrieved on 27 Apr 2016), and applying procedures as described before (Nielsen and Mulder 2016) to ensure a sufficient number of disordered residues and native, non-denaturing conditions. To be more specific, we required at least 50 assigned chemical shifts, at least 40 residues, 4 ≤ pH ≤ 8 (eventually no entries had pH above 7.5) and 273 ≤ T ≤ 313 K, and calculated the Z-score for all residues and required at least 50% disordered residues (Z-score < 3.0). This procedure yielded 242 entries. These entries were manually curated, removing entries with biasing conditions such as denaturants or added co-factors, in order to focus on the correlation between sequence and chemical shift exclusively. Next, the remaining sequences were aligned using the EMBOSS implementation (http://www.ebi.ac.uk/Tools/psa/emboss_needle/) of the Needleman–Wunsch alignment algorithm (Needleman and Wunsch 1970) keeping entries only for sequences having < 50% mutual sequence identity and < 50% sequence identity to any sequence from entries used from the CheZOD dataset leading to a final “complementary dataset” of 84 entries and a total of 203 entries when combined with the original CheZOD database.
In each of the three iterations of data fitting, a residue from a candidate entry was included if either at least five consecutive residues were disordered (Z-score < 3) or just requiring Z-score < 3 for the particular residue if most residues in the full protein were disordered as quantified by f D < f min where f D is the fraction of disorder residues with Z-score < Zmin using Zmin = 3 and f D = 0.8 in the first iteration and Zmin = 4 and f D = 0.75 in the last two iterations. In the first iteration, neighbor-corrected RCCSs used as a basis to estimate the Z-score, were estimated using the method of Tamiola et al. (2010), which considers the center residue and the nearest-neighbor amino acid types using weights for the chemical shift atom types as described before (Nielsen and Mulder 2016). For the other iterations, the random coil chemical shifts were estimated using parameters from the previous iteration with corrections for nearest and next-nearest neighbors, and smaller chemical shift weights based on the RMSD of the refined fit from the first iteration. Specifically, we calculated the chemical shift chi square deviation, χ2, as the tripeptide sum of squared weighted differences between observed, \({\delta _{obs}}\left( {j,n} \right)\), and predicted, \({\delta _{pred}}\left( {j,n} \right)\), chemical shift based on the current prediction model, for residue j for nuclei, n, as:
Using the chemical shift standard deviations, σ Ζ (n), representative for the prediction RMSDs:
The chi square statistic was converted to a Z-score as described previously (Nielsen and Mulder 2016). The correlated effect of the pentapeptide amino acids on the chemical shifts were only included in the last iteration. In the first iteration, pH and temperature were not considered, but to avoid large effects on the chemical shifts, entries were only included when 6 ≤ pH ≤ 7.5 and 285 ≤ T ≤ 301 K. In contrast, in the final two iterations, entry inclusion was not restricted by pH or temperature, but the effect on the chemical shift was accounted for as described above. The progressively less stringent criteria for residue inclusion with each iteration was reflected in the number of included chemical shifts, using 2663, 4530 and 8846 15N chemical shifts for the first, second and third iteration, respectively. In the third iteration, the final residues and segments classified as disordered were selected based on the refined criteria. This database of residues and chemical shifts represents a reference set of protein sub-segments of validated disorder. 137 protein entries were included in this dataset containing 9810 residues and 47,757 chemical shifts spread across 743 residue segments in total. The complete validated disorder database is given in Table S1 in the Supplementary Material and the experimental conditions of pH and temperature pertinent to these entries are visualized for comparison to the Tamiola database in Fig. S1.
Construction of the database of structured proteins with chemical shifts
Another database of structured proteins was constructed to allow for comparison with the POTENCI database of disordered residues. This database was constructed by retrieving the entries from the RefDB database (Zhang et al. 2003) and calculating the CheZOD Z-scores along the sequences as described above. This database was culled by requiring (i) at least three assigned chemical shifts per residue (on average), (ii) no more than 30% sequence homology within the database as determined by the cullpdb procedure (Wang and Dunbrack 2003), (iii) no more than 40% sequence identity to any protein in the POTENCI database determined as described above using the EMBOSS implementation of the Needleman–Wunsch alignment (Needleman and Wunsch 1970), (iv) requiring the protein to be well structured as judged by having less than 10% disordered residues (fD < 0.1, with Z-score < 3.0). This procedure resulted in a final database having 630 entries and 80,517 residues. The database of structured proteins is compared to the POTENCI database in Results and an analysis of amino acid preferences in the two sets is visualized in Fig. 1.
Amino acid disorder promoting tendencies. The height of the bar for each amino acid, a, is equal to ln(fIDP(a)/fRefDB(a)) where fIDR(a) and fRefDB(a) are observed frequencies of amino acid, a, in our POTENCI database of validated disordered residues and a similar database of validated order build from amino acid sequences from proteins in the RefDB database (Zhang et al. 2003) having fIDR < 10% (see “methods”)
Results
A set of 137 protein entries with assigned chemical shifts were generated by extending the CheZOD database (Nielsen and Mulder 2016) as described in “methods”. The disordered residues were identified by calculating the Z-score as described before (Nielsen and Mulder 2016) (see “methods”) leading to a database of 9810 validated disordered residues. The experimental conditions pertinent to the entries used to construct the POTENCI database are visualized and compared with the Tamiola database in Fig. S1, correlating the fraction of disordered residues with the number of residues and the temperature vs. pH. The disordered residues were distributed across various parts of the protein sequences considered, and not only in one part or only in the ends, as also seen in our previous study. More specifically, 743 segments of disordered residues were identified equaling 5.4 segments per protein. The protein entries used contained between 15 and 100% disordered residues (see Table S1 for all details). In line with earlier observations (Romero et al. 2001), our database contains more frequently Gly and Pro as well as negatively charged and polar residues, and comparatively fewer apolar residues and Cys compared to structured proteins (see Fig. 1).
The database of disordered residues contained 47,757 chemical shifts, which were used to parameterize POTENCI as described in “methods”. The chemical shifts were corrected for pH and temperature and the possibility of misreferencing was considered (Eq. 7) following the procedures described in Methods. The fitting procedure converged after 60,000 steps, removing approximately 1% of the chemical shifts judged to be outliers in the process. The distribution of errors subsequent to parameter fitting were normally distributed, whereas, in contrast, the outliers removed according to the principle of quantile matching (see “methods” and Eq. 19) were clearly beyond the tails of the normal distribution (see Fig. 2). The resulting RMSDs for the training data were 0.4180, 0.1624, 0.1320, 0.1498, 0.05753, 0.02385, and 0.01873 ppm for 15N, 13Cα, 13Cβ, 13C′, 1HN, 1Hα and 1Hβ, respectively (see also Table 1).
Fitting performance and statistics. a Observed vs. predicted Hα shifts in the training set showing used data points in blue and points deemed to be outliers as green larger disks with black outline. b fraction of points, f = 1 − Fobs(nσ) (Eq. 17), with absolute scaled errors, (Eq. 15), ε > nσ (for Hα) as a function of nσ. showing data points subsequent to outlier-stripping as a solid blue curve, all data points—including outliers—with green dashes, and the curve corresponding to the standard cumulative normal distribution as a continuous black dashed line for reference. c Q–Q plot (Wilk and Gnanadesikan 1968) showing observed quantiles against theoretical quantiles (Eq. 18) for all nuclei using blue, red, black, green, cyan, magenta and yellow curves for C′, Cα, Cβ, Hα, HN, N and Hβ, respectively. The straight dashed line, y = x indicates that the error follows a standard half-normal distribution; points above the line in the right-hand side indicate heavy tail outliers
The POTENCI parameterization provided random coil chemical shifts, neighbor and next-nearest neighbor corrections for all residue types as well as additional corrections for correlations of central and neighboring residues. The random coil shifts are given in Table 2. The neighbor correction parameters were obtained by using fewer than the maximal number of parameters following the parsimonious robust fitting procedure described in “methods” [see Table 1 and Eqs. (8)–(10)]. In total between 61(Cβ) and 72(Hβ) parameters out of the possible 82 (including two to encode the termini) were used to account for the neighboring residues, whereas only between 17(Hβ) and 56(N) out the maximal 196 parameters for correlations between amino acids were required. Between 42(Hβ) and 94(HN) of the 137 proteins required offset corrections. As a result, a total sum of between 151(Hβ) and 222(HN) parameters were used to fit the chemical shifts. Consequently, the total number of parameters used per chemical shift data point was low, between 0.023 and 0.027 for the heavy atoms, 0.027 for HN, and 0.036/0.054 for Hα/Hβ, where fewer chemical shifts were available, suggesting a robust fit (all counts are available in Table 1).
The derived amino acid neighbor corrections show a few general and interesting trends (see Fig. 3): For example, aromatic neighboring residues produce upfield shifts for central residue proton chemical shifts, whereas beta-branched residues at position i − 1 lead to downfield shifts for central residue 15N and (to a lesser extent) HN shifts. Overall, Gly and Pro neighbors result in the largest absolute perturbations on the central residue chemical shifts, whereas Gln, Lys, and Arg have least influence. Furthermore, 13Cα and 13Cβ nuclei are the least affected, while 15N/1HN chemical shifts are mostly affected by neighbor i − 1, whereas 13C′ chemical shifts are most effected by neighbor i + 1. Finally, there is a clear trend that amino acid correction matrices are correlated in the following pairs: Cα/Cβ, 15N/HN, and Hα/Hβ. All neighbor corrections are provided in the Supplementary Material Table S2.
Visualization of amino acid neighbor corrections to random coil shifts. The scaled amino acid neighbor corrections, Δk/sn, (Eqs. 2 and 9) are shown for each amino acid and atom for residue i + k with k = − 2, − 1, 1, and 2, images visualized according to the color bar. The corrections were scaled by dividing with sn = 1.0, 4.0 and 10.0 for 1H, 13C and 15N, respectively. Values were truncated to an absolute maximum for 0.25 to enhance contrast in the visualization. This was necessary for: Pro i + 1 C′/Cα/Hα (full un-scaled corrections were − 1.91/− 2.02/0.283 ppm) and 15N Ile i-1 (2.75 ppm). See also Table S2 for all neighbor contributions
A somewhat similar picture is seen for the correlated neighbor contributions, \(\omega _{{l,m}}^{k}(n)\), Eq. (10), for residue groups, l and m, related to the central residue position, i, and neighbor i + k, respectively, for nucleus, n. The effects are largest when Gly or Pro are involved and largest for the nearest residue positions, (k = − 1 and 1). As expected, the contributions for k = − 2 and 2 are largest for 15N/1HN, and 13C′, respectively. The correlated contributions are visualized in Fig. 4. It is seen that a significant number of these are non-zero and are different for each individual nucleus. The three most important contributions are highlighted with circles and explained in detail in the figure legend. The 20 most significant correlation corrections are listed in Table 3. The full list of correlated neighbor contributions is provided in the Supplementary Material Table S3.
Visualization of correlated amino acid contributions to random coil chemical shifts. The matrix of images shows the contribution, \(\omega _{{l,m}}^{k}(n)\) (Eq. 10), for each nucleus, n, (rows, labels to the left) and sequence position, i + k, (columns, labels at the top). Each image shows the scaled corrections, \(\omega _{{l,m}}^{k}(n)/\sigma (n)\), (scales defined in Eq. 21) according to the color bar for all combinations of the central residue group type, l, (vertical axis for each image) and neighboring group, m, (horizontal axis for each image). The amino acid groups are described in “methods” below (Eq. 10) using labels: G(Gly) = ”G”, P(Pro) = ”P”, F/Y/W(aromatic) = ”r”, L/I/V/M/C/A(aliphatic) = ”a”, K/R(positive) = ”+”, D/E(negative) = ”−”, or N/Q/S/T/H(polar) = ”p”. Only a small subset of the possible correlated corrections was used (between 17 and 56 out of the possible 196) showing all non-used groups here as white pixels in the images. The scaled corrections were visualized using a truncation to an absolute value of 2.0. Colored circles highlight corrections with absolute values higher than 5.0, five further corrections were between 2.0 and 2.8 (see Table 3 below). The colored circles highlights contributions from (n, k, l, m) = (Cα, 1, Gly, Pro) (motif: xxGPx as in Table 3 below, “x” denotes any amino acid, green circle), (n, k, l, m) = (Cβ, 1, Pro, Pro) (xxPPx, purple) and (n, k, l, m) = (HN, − 2, Gly, aromatic) (rxGxx, black)
Predicted chemical shift can now be derived, using the values from the tables indicated above. A few examples for representative pentapeptides are provided in Fig. S4 in the Supplementary Material for reference and the predictions are compared to those using ncIDP (Tamiola et al. 2010) and the method of Kjaergaard et al. (2011), Kjaergaard and Poulsen (2011).
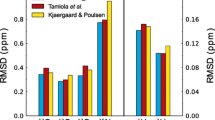
To evaluate the performance of POTENCI, 12 representative proteins were selected from the training set for cross-validation, i.e. POTENCI was parameterized with all proteins except one, I, and this parameterization was applied to derive the predicted shift for protein I. This procedure was repeated leaving each of the 12 proteins out from the cross-validation set one-by-one. The only exception from this procedure was BMRB ID, 6968, which was not used in the full training or the leave-one-out sets. The 12 proteins (listed in Table 4) were selected for their high degree of disorder and large number of available chemical shifts, and to also represent cases with low temperature and low pH. Following this principle, the set contained chemical shifts for all 6 different nuclei, between 1026 (1Hα) and 1681 (13C′) chemical shifts in total, with temperatures between 273.0 and 300.1 K and pH between 4.5 and 7.0. The performance of POTENCI on the cross-validation set was compared to the performance of the two currently most accurate predictors, ncIDP (Tamiola et al. 2010) and the method of Kjaergaard et al. 2011), Kjaergaard and Poulsen (2011) derived from the QQXQQ peptide library (henceforth referred to as the QQXQQ or KBP method). A few chemical shifts were identified having very large errors, corresponding to assignment errors or oxidized Cys, for all three methods (see Table S5). These chemical shifts (10 cases, Table S5) with absolute errors > 1.5 and 4.5 ppm for 13C and 15N, respectively, and 0.6 and 0.25 ppm for 1HN and 1Hα, respectively, were excluded from the comparison. Furthermore, the protein with BMRB ID 15563, was obviously assigned using a non-standard reference, and therefore LACS (Wang et al. 2005) was used to re-reference the 13C chemical shifts using offset corrections of 2.74 ppm for 13Cα and 13Cβ and 3.07 ppm for 13C′. The result of the cross-validation and comparison between method performance is visualized in Fig. 5. The results on the cross-validation is slightly higher than the training set statistics, yielding RMSDs between observed and predicted shifts of the cross-validation set (Table 4) of 0.1861, 0.1677, 0.1862, 0.5341, 0.0735, and 0.0319 ppm for 13C′, 13Cβ, 13Cα, 15N, 1HN and 1Hα, respectively. However, the RMSDs for the other methods are significantly higher (see Fig. 5) obtaining RMSDs which are between 22.4% (13Cα) and 83.7% (1Hα) higher than POTENCI. Throughout this paper, we equate a higher accuracy to a lower RMSD between observed and predicted shifts.
Performance of POTENCI and other methods on the 12 protein cross-validation set (see Table 4) showing the RMSD between observed and predicted shifts for each nucleus and method. For the predictions by the method of Kjaergaard et al., GGXGG-derived neighbor corrections were used for glycines
Statistical analysis of the errors in the cross-validation set reveals that POTENCI performs better than the two other methods across the full error range (see Fig. 6). Furthermore, it is observed that the errors are not precisely normal-distributed with a scale parameter corresponding to the RMSDs for the final evaluation in the training set (Table 1), but, rather, the smallest errors appear to follow a normal distribution, whereas the largest errors are much larger than expected from the normal distribution (see Fig. 6). These over-dispersed points could either be due to method-specific bias on the predicted shifts or due to experimental bias in the assigned chemical shifts. Here we consider two types of experimental biases, namely (i) constant offset from misset chemical shift reference and (ii) over-dispersion caused by residual structure. To analyze (i) we included one additional parameter, λ, for comparing the observed and predicted shift minimizing (Eq. 22 below).
Distribution of errors. Plots for each nucleus, showing the fraction of points (y-axis) above an error threshold as a function of the error threshold as in Fig. 2b (this time the absolute unscaled error in ppm is shown). The different methods have color coding as in Fig. 5 and highlighting the theoretical curve (broken magenta outline) for a normal distributed data set with a standard deviation equal to the RMSDs for the final evaluation in the training set (Table 1)
Performance of POTENCI and other methods RMSD with and without experimental data bias remedies. RMSDs for POTENCI and other methods showed with colored bars as in Fig. 5 with or without method adapted offset correction (Eq. 22) and/or stripping off residues with residual structure (see text and definition in Table S1)
where \({\delta _{obs}}\left( {i,I} \right)\) and \({\delta _{pred}}\left( {i,I,m} \right)\) is the observed and predicted shift, respectively, for residue, i, and protein, I, and method, m, and λ is a phenomenological offset correction for protein, I, adapted for the method, m. To test for (ii) we excluded all residues in the cross-validation data set corresponding to residues with supposed residual order based on the criterion of the CheZOD Z-score > 3, hence, only retaining the same residues as used in the training set (see Table S1) (note that the particular entries were not used for deriving the cross-validation parameters leaving each protein entry out one-by-one). Between 5.4 and 7.6% of the data points were removed by this criterion (see Table S6). Firstly, inclusion of the adaptive-method-specific chemical-shift offset revealed optimized RMSDs (Eq. 22) for POTENCI that were about 11% lower than the RMSD without offset correction (see Fig. 7 and Table S6). The other methods also showed improved performance by including an offset correction, in particular 1Hα RMSDs were much lower (Fig. 7 and Table S6) suggesting problems for the predictions with constant bias, which is significant compared to the prediction RMSD for 1Hα for these methods. Secondly, stripping off the supposedly residually-structured residues leads again to improved RMSDs of ca. 10% for POTENCI. This improvement was also found for the other methods (Fig. 7 and Table S6). Finally, the inclusion of both experimental bias remedies at the same time yielded significantly improved RMSDs for all methods. In particular, POTENCI shows RMSDs of 0.1457, 01248, 0.1494, 0.4131, 0.0563, and 0.0254 ppm for 13C′, 13Cβ, 13Cα, 15N, 1HN and 1Hα, respectively. Still, these RMSDs are significantly higher for the other methods by between 25 and 78% relative to POTENCI. We note also that the remedied RMSDs for POTENCI are on par with the RMSDs in the training set (Table 1). Analyzing the distribution of errors (Fig. S2) reveals that remedying the data using either the offset correction or the residue stripping, still feature a remaining part of the errors being over-dispersed. However, including both remedies leads to errors that are very close to being completely normal distributed (see Fig. S2).
Discussion
We have presented here a method, POTENCI, for predicting random coil chemical shifts from protein sequence. Our analysis revealed that POTENCI outperforms the currently most accurate methods as outlined above. Below we compare POTENCI with several other methods in greater detail, discuss the validation of POTENCI, the origin of the high accuracy of POTENCI, potential applications of POTENCI, and the mechanism of neighbor effects on chemical shifts.
Comparison of POTENCI with other methods
To compare more specifically the performance of POTENCI with other methods and analyze how the improved accuracy impacts on the interpretation of dynamics along to sequence, we analyze the specific errors in the prediction for all methods sequence-specifically for one protein in the cross-validation set, Hsp12, the heat shock protein from Saccharomyces cerevisiae (Singarapu et al. 2011) (BMRB ID 17483, Fig. 8). It is clear again that POTENCI produces much lower digressions compared to the other methods and uniform deviations along the sequence except a slightly larger variation for segment 75–83 and for the C-terminal residues 105–108. On the other hand, ncIDP (Tamiola et al. 2010) clearly produces upfield-biased Hα RCCSs (seen as biased positive errors shown with green dots in Fig. 8c). The QQXQQ method appears to have overall larger errors for certain residues in the sequence. It should be noted that larger differences between observed and predicted RCCSs can both be interpreted as limitations of the model but also as amplified fluctuations in secondary chemical shifts indicating residual ordered structure. An interpretation of increased order can be assisted by calculation of the CheZOD Z-score (Eq. 20) (black curve, Fig. 8). For comparison, the two other methods have higher background levels of error (RCCSs difference) fluctuations (i.e. higher minimum Z-score) and more regions with larger errors (and higher Z-score), which would make it more difficult to identify true segments of increased ordered structure. Interestingly, segment 75–83 is part of the only ordered structured segment (helix IV) that forms both in SDS micelles and in the presence of DPC (Singarapu et al. 2011), and, as shown here for the first time, also seems to form partially in aqueous solution.
Weighted errors for RCCS prediction for protein Hsp12 (BMRB ID 17483). The signed scaled error between observed and predicted shifts, ε = (δobs − δpred)/σ, is shown as a function of the position in the sequence (using scaling by Eq. 21) as dots colored as described in the legend to Fig. 2d for POTENCI (a), the method of Kjaergaard et al. (2011), Kjaergaard and Poulsen (2011) (b) and the method of Tamiola et al. (2010) (c). The CheZOD Z-score calculated based on the sum of the squared scaled errors (Eq. 20) as described in “methods” and (Nielsen and Mulder 2016) is shown as a black curve. Lines for ε = 0 and Z = 3 are shown for reference. d Predicted dynamical and structural properties: order parameter, S2, by RCI [green curve (Berjanskii and Wishart 2005)], neighbor-corrected structural propensities [ncSPC, black curve (Tamiola and Mulder 2012)], and secondary structure probabilities as predicted by δ2Δ (Camilloni et al. 2012) shown as a red curve for α-helix and a blue curve for β-sheet (displaying the negative of the probability here)
We compared the POTENCI-derived CheZOD Z-score with other methods for inferring structure and dynamics from chemical shifts (see Fig. 8d). The predicted order parameter, S2, by RCI (Berjanskii and Wishart 2005) does not agree well with the Z-score and appears to have larger background noise. The reason for this might be that RCI applies a truncation of the RCCS difference. The neighbor-corrected structural propensities [ncSPC, black curve (Tamiola and Mulder 2012)], which is based on ncIDP RCCS predictions, show positive values (indicative of helix population) for residues 74–85 matching well with the region for helix IV discussed above. On the other hand, ncSPC reveals longer regions with negative signs, suggesting a propensity for beta-sheet formation, which is not reflected in the Z-score. We argue that the derived increased propensity for beta-sheets is due to the upfield bias in ncIDP predicted 1Hα RCCS. With some similarities, δ2Δ (Camilloni et al. 2012) predicts a small region near helix IV with higher propensity for helix formation and a region in the middle and near the C-terminal with increased population of beta-sheet (Fig. 8d).
The origin of the high accuracy of POTENCI
As outlined above, POTENCI predicts the chemical shifts significantly better, with an uncertainty approaching the measurement error for 1Hα and 1Hβ. The predictions have almost an order of magnitude lower error compared to the chemical shift prediction error for structured proteins based on their sequence and structure (Meiler 2003; Neal et al. 2003; Shen and Bax 2007; Kohlhoff et al. 2009; Han et al. 2011). This reflects that chemical shifts for IDPs correspond to a statistical distribution of conformations that are highly specific to the local sequence of amino acids. By analyzing the sources of the accuracy of POTENCI a lot can be learnt about what limits the accuracy of chemical shift prediction and its interpretation in terms of structure and dynamics for IDPs.
Whereas earlier library methods based on guest-host substitutions of amino acids into short peptides (Richarz and Wüthrich 1978; Bundi and Wüthrich 1979; Braun et al. 1994; Wishart et al. 1995; Schwarzinger et al. 2000; Kjaergaard et al. 2011; Kjaergaard and Poulsen 2011) only sampled a small biased region in sequence and condition space, the POTENCI parameters were derived from a diverse database of full length protein sequences studied under native conditions that reflect representative sampling of conformational space. Notably, with POTENCI we observe that correlated contributions were important for the prediction (Fig. 4), i.e. certain amino acid neighbors, such as Pro, influence the chemical shifts and local conformational sampling idiosyncratically, dependent on the nature of the central amino acid. Taking this contribution into account leads to improvement of chemical shift prediction relative to library methods, where it would require as many as 8000 tripeptides or 3,200,000 pentapeptides to be synthesized and measured to compile the analogue of the data presented here.
Tamiola et al. (2010) adapted a statistical approach, conceptually very similar to the one presented here, addressing the inherent limitation of library methods by analyzing a small database of IDPs, the ncIDP database. The burgeoning assignment of ever more IDPs allowed us to compile a much larger database of proteins, comprising 137 entries compared to a mere 14 in the ncIDP database (see Fig. S1). More specifically, after removing outliers in both databases, the POTENCI database retained 47,159 chemical shifts, whereas the ncIDP database consisted of 4439 chemical shifts, i.e. a factor of over ten times more data points. This indicates that POTENCI could apply more than ten times as many parameters, maintaining the same parameter-to-data ratio, and thereby study more subtle correlations between local sequence and chemical shifts. While this is of course a simplistic interpretation, since not all parameters are equally important, our application of AIC [“methods”, Eq. (14)] allowed us to increase the number of adjustable parameters as long as it decreased the fitting rms significantly. Indeed, POTENCI included a significant number of correlated contributions as described above, but also correction terms for next-nearest neighbors. This effect is very important, since the correction terms for the next-nearest neighbors, in some cases, were almost as large as those for the direct neighbors.
POTENCI also incorporates the effect of pH, which leads to partial or full protonation of titratable side chains, on the chemical shifts of the central and neighboring residues [“methods”, Eqs. (5) and (6)]. We analyzed the errors in the chemical shift predictions for human SRC residues 1–85 (Perez et al. 2009) (BMRB ID 15563), studied at pH 4.5 both with and without using the pH corrections. Neglecting these corrections lead to large errors in chemical shifts for all histidines (see Fig. 9), seen as spikes in the derived Z-scores that could potentially be wrongly interpreted as residual order. For comparison, the KBP method is capable of accounting for pH, whereas ncIDP is not.
Weighted errors for RCCS prediction for the protein human SRC (1-85) with BMRB ID 15563 at pH 4.5. The signed scaled error between observed and predicted shifts and the CheZOD Z-score is shown along the sequence as in Fig. 8 in the main text. Predictions without using pH correction are shown in panel b with the amino acid sequence shown for reference. Histidine residues H25, H47, and H90 are highlighted with yellow bars
POTENCI also applies residue-specific corrections for the temperature dependence of the chemical shifts (“methods”, Eq. 4). Chemical shift prediction errors for the protein CPAP-interacting epitope of Danio rerio STIL (BMRB ID 19318) (Hatzopoulos et al. 2013) studied at 0 °C are shown in Fig. 10. The main effect of neglecting temperature corrections was a downfield bias of 15N chemical shifts. Although the overestimation of order by the 15N chemical shifts (cyan data points) is evident from comparison of panels (b) and (a), this has a negligible effect on the derived total Z-score, when using all chemical shifts collectively, since the weight factor for 15N secondary chemical shifts is small.
Weighted errors for RCCS prediction for the protein CPAP-interacting epitope of Danio rerio STIL, (BMRB ID 19318) at T = 273 K. The signed scaled error, ε, between observed and predicted shifts and the CheZOD Z-score is shown along the sequence as in Fig. 8. Predictions when not using temperature correction are shown in panel b with the amino acid sequence shown for reference. 15N chemical shifts are shown using cyan dots
The accuracy of RCCS prediction was greatly improved in POTENCI by the introduction of both next-nearest neighbors and pairwise correlated amino acid pairs described by linear equations and corrections for dependence on pH and temperature as discussed above. The question remains whether the chemical shift prediction can be improved further by adding more sequence and condition features or by using a more sophisticated mathematical description. To address this question, we analyzed our database of 9810 disordered residues and searched for multiple occurrences of triplet and pentad amino acid segments both within the same, and across all, proteins in the database. For each reoccurring segment, S, we inspect the standard deviation, σ S , of the observed chemical shift within the group. The distribution of such standard deviations, representative of the “true variation” of the chemical shift, is analyzed and shown in Fig. 11. Among entries with assigned 15N shifts (8846 residues), there were 2032 and 419 reoccurring triplets across all protein sequences and within the same protein, respectively, and 127 and 58 pentads across and within protein(s), respectively. For segments across proteins we compare the chemical shifts corrected for temperature, pH and offset (Eq. 7). It is seen that the chemical shift variation for identical triplets across all protein sequences (black curve) is comparable to a normal distribution having the same errors as POTENCI predictions (magenta broken curve)—except for 1HN and 13C′, to a lesser extent—which show larger variation in the triplets. For comparison, the chemical shift variation for identical pentads across proteins (green curve) reduces to about half the value for the triplets, e.g. the median variation is 0.053 ppm for 13Cα among pentads (compared to 0.12 ppm among triplets). The observation that sharing five rather than three residues reduces chemical shift variation confirms that next-nearest neighbors are important for chemical shift prediction. These rather small variations among triplets and pentads compared to the POTENCI errors suggests that considering all three (or more) residues in a triplet simultaneously would improve chemical shift prediction. However, considering 20N combinations of N residues would require introducing an expansive number of adjustable parameters, and lead to over-fitting. It should, however, be noted that the analyzed variations correspond to residue types that occur more frequently in IDPs, and therefore might not be representative of the chemical shift variation for all possible combinations of residues. As another caveat, we point out that a fraction of the triplets would be part of longer repeat segments such as pentads whereas we have already removed the segments that only repeated within the same protein from the “across” analysis.
Distribution of the variations in observed chemical shifts among identical sequence motifs. Cumulative distribution function (cdf) for the standard deviation, σS, of observed chemical shifts for each reoccurring segment (see text); triplet (black and red curve)/pentad (green and blue), across (black and green curve) all proteins in the POTENCI database and within the same protein (red and blue); see also text. The theoretical cumulative distribution function of variation is shown with a broken magenta curve corresponding to the half-normal distribution with a standard deviation taken as the POTENCI error in the training set (Table 3). Segments only present in the same protein were excluded from the analysis across proteins. For the analysis across proteins, the chemical shifts were corrected for pH and temperature (Eq. 7, “methods”), and offset (which was determined during the fitting process)
Restricting the above analysis to segments within the same protein, it is seen (Fig. 11) that the chemical shift variation is significantly lower (ca. half the magnitude for triplets, red curve) than the corresponding variation for the same segment size across all the proteins. e.g. for 13Cα the median variation is 0.06 ppm for triplets within proteins. There are several possible explanations for this decrease in chemical shifts variation. Firstly, segments within the same protein share the same sample conditions such as pH and temperature and it might be that, although POTENCI considers these effects, the parametric dependence might not be adequate. Secondly, other buffer conditions such as e.g. ionic strength and type of buffer component are not included in POTENCI, and might have an influence on the chemical shift in a way that is not currently captured by the offset correction parameter. Unfortunately, not all conditions are reported consistently in the BMRB data and, in particular, the ionic strength is only provided in some cases. A systematic empirical investigation of chemical shifts under varied conditions could prove valuable for resolving this issue. Thirdly, it should be noted that repeated segments within the same protein share features from the underlying sequence such as total charge and local polarity etc. and the segments might even be part of longer pseudo-repeats in sequence as is often seen for IDPs (Dunker et al. 2002; Simon and Hancock 2009). It appears from these observations, that there would be an underlying sample-specific effect that perturbs the conformational equilibrium very subtly in a way dependent on the central residue type and the sample conditions. These effects might be important, but are difficult to address without applying too many adjustable parameters with the risk of over-fitting of the data. Finally, the chemical shift variation among pentads within the same protein is very low (Fig. 11, blue curve) and approaches the experimental precision for the chemical shift, e.g. the median value is ca. 0.01 ppm for the heavy atoms and 0.001 ppm for protons. The true variation could be difficult to measure for experimentalists due to potential complete overlap on all chemical shift axes in a multi-dimensional NMR experiment for assignments, and hence, conservatively, the same chemical shifts would often be assigned to two different pentads although the actual chemical might differ slightly explaining the small kink in our curves. Therefore, the dispersion in multidimensional NMR spectra of IDPs are completely described by the local penta-peptide segments. This also means that predicted chemical shifts are not expected to improve with the addition of further neighboring amino acids beyond next-nearest neighbors in the parameterization. Rather, the chemical shift prediction accuracy is limited by the effects from sample conditions.
Validation of POTENCI
Statistical regression models can suffer from over-fitting. Therefore, in order to assess how our model would generalize to an independent dataset, the reliability of POTENCI predictions was assessed by cross-validation. Twelve representative proteins were selected and the chemical shifts were predicted with a leave-one-out procedure and compared to the observed shifts. Analysis of the prediction errors, following this cross-validation procedure, is a fair assessment of the accuracy of POTENCI. It was seen that POTENCI performs significantly better than the two currently most accurate predictors, ncIDP (Tamiola et al. 2010) and the QQXQQ library method of Kjaergaard et al. (2011), (Kjaergaard and Poulsen 2011), which show RMSDs higher by between 22.4% (Cα) and 83.7% (Hα) (Fig. 5). We also showed that the majority of the largest errors can be removed by adapted offset correction and by excluding residues with supposed residual order, and in this case POTENCI still performs better than the other methods with about the same ratios of improvement (Table S6). Furthermore, the RMSDs for POTENCI decreased by 1.7% on average without cross-validation, i.e. if the parameter set for the full training set of proteins including the protein to be predicted was used. This relatively small decrease in RMSD supports the conclusion that POTENCI was not over-fitted by our stochastic regression procedure. Akaikes information criterion (AIC) was used for deriving the optimal balance between the number of adjustable model parameters and goodness of fit (“methods”, Eq. 14). This procedure appears to be a suitable choice, since it prevents over-fitting of the data in an objective way and is asymptotically equivalent to minimizing the goodness of fit in leave-one-out cross-validation (Stone 1977). Indeed, POTENCI applies a relatively small ratio of number of parameters divided by number of data points, i.e. between 0.023 and 0.027 for all the heavy atoms (see Table 1). This number is low compared to the ratio of ca. 0.10 used in the linear regression applied to train the method of Tamiola et al. (2010). This ratio becomes as high as 0.144 for the method of Tamiola et al. for 1Hα, and we argue that over-fitting in this case might be responsible for the observation of downfield biases for 1Hα RCCS predictions for proteins not part of the training set for this method, as, for example, seen in Fig. 8 (see also Fig. S3). The very low standard error for 1Hα, measured in ppm, demands a very accurate offset setting of the chemical shift axes. If the offset is not set properly, it would lead to significant bias in the sign of the secondary chemical shift and distribution of errors as seen in Fig. S1. We foresee that the accuracy of POTENCI could be improved even further upon expansion of the database with future assigned IDPs that would allow for a larger number of adjustable parameters to be used according to AIC—in particular for the correlated contributions and next-nearest neighbor corrections. Such corrections would, of course, be much smaller than those presented here, and would have comparatively less impact.
Unfortunately, the studied training set of chemical shifts inevitably contains outliers, both due to errors in the chemical shifts due to human assignment mistakes and experimental noise, but also due to systematic biases caused by effects not included in the model definition, such as small fractions of residual structure or differences in buffer conditions. Since outliers have large impact on the model parameters in regression models, it is important to remove the outliers prior to fitting. IDPs often contain ordered segments of varying length and degrees order as recently discussed (Nielsen and Mulder 2016), and therefore we did not include such putative ordered segments and only included apparent completely disordered residues according to our Z-score definition of local order (Table S1). It was indeed identified by cross-validation that local residual structure leads to over-dispersed error distributions (Fig. S1). However, other types of outliers cannot be distinguished from rare cases related to true data points prior to model building and therefore need to be identified a posteriori. We applied our procedure of iteratively decreasing the weight, r VIF , (Eq. 14) on the number of parameters in the definition of AIC, followed by removal of outliers (ca. 1% of data points) based on the principle of quantile matching (“methods”, Eq. 19). A high value of r VIF in the first iteration ensured that the data points were not fitted too heavily, so that outliers could be distinguished and removed. The principle of quantile matching was applied here to remove the correct number of over-dispersed points. While removing too few erroneous points is bad for reasons discussed above, too excessive outlier stripping would risk removing true data points with high information content. Our choice of limiting quantile, Q = 3.0 (Eq. 19), is of course not universal, but with this choice we observed that the errors followed a normal distribution quite well (Fig. 2) as required for linear regression models. After the final cycle of model optimization followed by outlier stripping, the procedure appeared to have converged as, on average, the number of outliers did not increase in the final cycle (between 8.6% more and 15.7% less than in the previous cycle were found). We note that the inclusion of neighbor correlated contributions (Eq. 10) allowed us to account for data, which appeared as outliers without this contribution.
POTENCI was optimized in steps of standard least squares fitting imbedded in a stochastic procedure (see “methods” and Supplementary Methods) to optimize the subset definition (Eq. 11), which cannot be varied exhaustively or with an analytical gradient. Stochastic methods are not necessarily guaranteed to converge, but the success of the convergence will depend on how efficiently the solution space was searched with the purpose of identifying the global minimum. It is, in principle, impossible to assess whether the global minimum was indeed found, but the success can be estimated from the apparent precision judged by the convergence of multiple parallel solutions to the optimization. We analyzed the optimized parameters and chemical shift predictions for the individual optimizations based on the 12 different proteins in the cross-validation set (Table 4). Some small and homogenous variations for the parameters are observed (data not shown), but we argue the most important property is the impact on the chemical shift predictions. Therefore, we derived the predicted chemical shifts by each of the 12 sets for Hsp12 (cf. Fig. 8) and show the scaled chemical shift errors in Fig. 12. It is seen that the variation in prediction is about one order of magnitude lower than the prediction RMSD related to the accuracy of the POTENCI method, is evenly distributed across the sequence, and is independent of error size.
Variations in predicted chemical shifts using 12 different parameter sets. The scaled chemical shift difference as in Fig. 8 shown as a function of the position in the sequence using the scalings representative of the accuracy of POTENCI taken as the RMSDs from the cross-validation set of 12 proteins (Table 3). Values for the prediction are superimposed for parameter sets based on each protein from the cross-validation set (Table 4). The plot window is zoomed and shows all predictions except T107 15N
The mechanism of neighbor effects
With POTENCI a set of chemical shift correction terms, Δ k (n), for the amino acid neighbors (Eqs. 2 and 9) were derived. This set of corrections quantifies the effect of local sequence on local conformational sampling through chemical shift perturbation. A few interesting trends were observed (Fig. 3). For example, aromatic residues produce upfield shifts on protons revealing a clear effect of ring-currents. Nuclei closest to the neighboring amino acid are the most affected, i.e. 15N/1HN by neighbor i − 1, 13C′ by neighbor i + 1 while 13Cα and 13Cβ are the least influenced, indicating that the chemical shift is perturbed through space by local magnetic fields. Furthermore, there is a clear trend that amino acid correction matrices are correlated in pairs corresponding to bonded atoms (13Cα/13Cβ, 15N/1HN) and also 1Hα/1Hβ. Since secondary chemical shifts are utilized to identify residual order, such as partial helix formation, it is important to assess whether the nature of the neighbor amino acid directly leads to shifts in the local backbone angle sampling for the segment, as discussed previously (Ting et al. 2010; Kjaergaard et al. 2011; Kjaergaard and Poulsen 2011). However, this is difficult to address directly from the correction matrices, since these represent the sum of all the sources of amino acid perturbations, such as, for example, ring-current shifts. In order to separate the effects, we performed a principal component analysis (PCA) (Wold et al. 1987) of all the corrections, where all 20 amino acids where represented by the 7 × 4 = 28 correction terms, and the linear combination of these constituents explaining most of the variation were derived by the PCA procedure. The first four loadings (correction term combinations) and scores (grouping of amino acids) are visualized in Fig. 13. First, it is seen that the most important component (explaining 46.2% of the variation) is defined by a loading having the largest weight on the proton terms, with a positive sign. This large negative shift in the scores for peptides containing aromatic residues most likely arises due to ring-current effects. Second, the next-largest component (together with the first component explaining 67.9% of the variation) is defined by a loading with positive sign for 13C′ and 13Cα (and small positive weights for 1Hβ) and a negative sign for the remaining nuclei. Strikingly, this sign combination matches exactly the well-known secondary chemical shifts for alpha-helix formation (Wishart et al. 1991). Indeed, analysis of the scores for the second component reveals that residues that increase the population of local helical structure (Ting et al. 2010) (Asp and Asn) display the highest value of component 2, whereas helix dis-favoring residues (Ile and Val) show the lowest value. We chose to exclude Pro from this analysis, since it resulted in slightly more noisy parameters, but the same analysis including Pro revealed the same trends for the first two components with an extreme value of − 10.1 for the second component, indicating that Pro disfavors helical local conformations more than Ile and Val, a result consistent with theoretical values from Ting et al. (2010). Altogether, our results support the findings by Kjaergaard et al. (2011), Kjaergaard and Poulsen (2011) in the context of peptide libraries, where changes in the Ramachandran distribution were shown to contribute to the sequence correction factors. The systematic trends in the loadings and scores for components 3 and 4 are less clear (data not shown) and the interpretation would therefore be largely speculative.
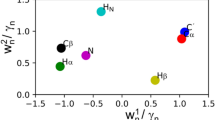
Principal component analysis of neighbor corrections. Visualization of the two first principal components, PCi, with rank, i = 1, 2 of neighbor contributions (see text). \(P{C_i}(X)=\sum\nolimits_{{n,k}} {{w_i}(n,k)\Delta _{k}^{n}(X)}\) where X is the amino acid type and n denotes the nucleus type. Neighbor contributions were scaled to zero mean and unit variance prior to PCA analysis. Pictograms visualizing the loadings, \({w_i}(n,k)\), in panels (a, b) for i = 1, 2, respectively, with color-ramps as in Fig. 3. c 2D Scatter plot showing the scores, \(P{C_i}(X)\), annotating each point with its corresponding amino acid one-letter code
POTENCI includes corrections for correlated effects of neighbors, \(\omega _{{l,m}}^{k}(n)\) (Eq. 10), meaning that it accounts for the different effects a certain amino acid has on another specific amino acid using groups of amino acids. Several significant correlation corrections were observed (see Fig. 4; Table 3). The largest effects are observed when Gly and Pro are involved, most likely reflecting the special sampling of local backbone conformation for these residues. For example, Gly followed by Pro (xxGPx segment, see legend to Fig. 4) has a large correlation correction of 1.3 ppm for Gly 13Cα and large negative corrections for 15N (− 0.97 ppm) and 1HN (− 0.14 ppm). The number for 13Cα is well reflected in the different neighbor correction factors of − 0.79 and − 2.25 ppm in the context of the penta-peptide libraries, GGXGG and QQXQQ, respectively (Kjaergaard et al. 2011; Kjaergaard and Poulsen 2011) and similarly well-matched differences for 15N (− 1.56 ppm) and 1HN (− 0.20 ppm). Another important correlation correction, − 0.35 ppm for 1HN, is needed when Gly has an aromatic residue as next-nearest preceding neighbor (rxGxx segment). This suggests that ring-current effects are stronger for Gly as a consequence of a missing side-chain. Between the two different peptide libraries, GGXGG and QQXQQ, we find differences of − 0.34, − 0.25, and − 0.29 ppm for Trp, Phe, and Tyr, respectively, matching well with our corrections.
Applications of POTENCI
The sequence-specific random-coil chemical shift is the core of many methods for inferring structural and dynamical properties (Cornilescu et al. 1999; Berjanskii and Wishart 2005; Cavalli et al. 2007; Shen et al. 2008, 2009; Camilloni et al. 2012; Tamiola and Mulder 2012; Nielsen and Mulder 2016). The improvement in accuracy for POTENCI relative to other methods means that subtler deviations from complete disorder can be detected, as discussed above. A perspicacious standard for deriving “exact” protein-specific RCCSs has been to denature a protein artificially (e.g. using a denaturant and/or low pH) in order to obtain the “intrinsic random coil (IRC) shifts”. The IRC shifts can be obtained through the additional sequence-specific assignment of the denatured state or a denaturant titration series (Modig et al. 2007; Kjaergaard et al. 2010). The POTENCI procedure is compared to the IRC approach for the C-terminal domain of the protein TDP-43 (Chen et al. 2016) in Fig. 14. Differences between experimental and POTENCI-predicted chemical shifts reveal larger fluctuations (higher CheZOD Z-scores) for residues 65–79, consistent with partial helix formation, and very small secondary chemical shifts (and very low Z-scores) for other parts of the sequence, with the further exception for residues 80–92, which again display slightly larger values. Exactly the same trends are observed for the IRC approach, with the only exceptions being the acidic residues Glu17 and Asp152, which are protonated under the acidic conditions (pH 2.5) employed to enforce the denatured state in the IRC approach. The similar conclusions drawn from the two chemical shift approaches are mirrored in the similar sequence profiles for the derived CheZOD Z-score (Fig. 14a, b, black curves). In contrast, the same chemical shift differences derived using the QQXQQ or ncIDP methods reveal much noisier profiles, having much larger average chemical shift fluctuation and higher background values for the Z-score (Fig. 14c, d). In conclusion, with the introduction of POTENCI it is no longer necessary to perform the additional complete resonance assignments in the denatured state, but rather the empirical POTENCI predictions can be directly applied to deduce important information on the deviation from complete disorder. In addition, spikes related to problems with chemical shift differences for the IRC approach due to titratable side chains are avoided when using POTENCI. Since pH and temperature corrections are incorporated, POTENCI can be applied as well to study pH or temperature induced structural changes to reveal details of protein folding.
Weighted errors for RC chemical shift prediction and comparison with denatured chemical shifts for the C-terminal domain of TDP-43 (BMRB ID 26728) (pH 6.5) (Chen et al. 2016). Signed, scaled differences between observed and predicted shifts and the CheZOD Z-score are shown along the sequence as in Fig. 8. For comparison, in panel b, we show the differences between the observed chemical shifts and another set of corresponding assignments for the same protein sequence derived under denaturing conditions [pH 2.5; 8 M Urea (Chen et al. 2016) (BMRB ID 26816)]. The titratable amino acids Glu17, Glu108 and Asp152 are highlighted using yellow boxes. Note that the experimental 13Cα and 13Cβ chemical shifts are not available for Glu108. Residue numbering starts with 1 for the first residue in the protein sequence (The first 11 residues have no assignments and are not shown)
Automatic resonance assignments are at the heart of fast protein structure-determination pipelines, such as structural genomics projects (Bartels et al. 1997; Burley 2000; Montelione et al. 2000; Moseley et al. 2001; Oezguen et al. 2002; Jung and Zweckstetter 2004; Williamson and Craven 2009; Rosato et al. 2012; Schmidt and Guntert 2012; Zhang et al. 2014). Sequence-specific RCCSs predicted by POTENCI would be instrumental for the performance of (automatic) assignment procedures for IDPs (Verdegem et al. 2008; Tamiola and Mulder 2011; Isaksson et al. 2013; Lee et al. 2015; Piai et al. 2016). More specifically, for a completely disordered protein, the spectral region that needs to be searched for assignment candidates is proportional to the product of the prediction RMSDs of RCCS for the applied method. For an HNCO correlation spectrum, for example, this spectral region is 3.8- and 3.6-times smaller when applying POTENCI rather than the QQXQQ and ncIDP methods, respectively (based on the RMSDs as presented in Fig. 5). When analyzing modern high-dimensional experiments specialized for assigning IDPs (Zawadzka-Kazimierczuk et al. 2012; Bermel et al. 2013; Piai et al. 2014) this factor would even be larger than ten-fold. This would mean that fewer multi-dimensional experiments would be needed for the assignments and the process could be considerably faster. Fast sequential assignments combined with analysis of secondary chemical shifts based on the POTENCI predictions and sequence-specific CheZOD Z-score calculations, as in Figs. 8a and 14a, form an effective procedure for the accurate quantification of dynamics for IDPs. We propose an even faster approach based on assignment-free assessment of sequence-specific disorder using POTENCI predictions and unassigned multidimensional NMR correlation spectra. POTENCI could be applied to predict the multidimensional spectra of IDPs when used in conjunction with spectrum simulation programs such as Virtual Spectrum (Nielsen and Nielsen 2014) where observed peaks not matching any predicted positions would be indicative of residual order. Conversely, the difference between a predicted peak position and the position of the nearest observed peak could be used to estimate a probability of residual order. We foresee a future where NMR spectroscopy combined with POTENCI predictions and automated-analysis methods can be used for large scale classification of IDPs in “dynamical genomics projects”, complementing and extending the structural genomics of folded proteins (Baker and Sali 2001; Simons et al. 2001; Chandonia and Brenner 2006) to dramatically increase our panorama of the protein universe.
Conclusions
We have presented here a method, POTENCI, for predicting RCCSs for proteins from amino acid sequence. The cross-validation performance of POTENCI on 12 very unstructured proteins result in chemical shift RMSD values of 0.1861, 0.1677, 0.1862, 0.5341, 0.0735, and 0.0319 ppm for 13C′, 13Cβ, 13Cα, 15N, 1HN and 1Hα, respectively, while 1Hβ is predicted with an RMSD of 0.0187 ppm. POTENCI exhibits a significantly improved accuracy compared to current best methods, with decreased RMSD values between 25 and 78%, and is at least as accurate as “intrinsic random coil” referencing. We attribute the improved accuracy of our method to two important assets. First, pH and temperature corrections are applied. When neglecting these corrections, one would observe rogue errors in predicted RCCSs for titratable residue at non-neutral pH and systematically biased 15N chemical shifts at low temperatures. Second, data-mining of a very large database of chemical shifts for validated, completely unstructured protein segments allowed us to take more sequence features into account. At current, we believe that adding further sequence features will not affect the prediction significantly, but, rather, the interplay between local sequence and sample conditions has become limiting to the accuracy of RCCS prediction. We have demonstrated the use of POTENCI-derived secondary chemical shifts together with the CheZOD Z-score method to detect very subtle signs of residual structure in IDPs that cannot be separated from the noise with other methods. We envisage that POTENCI may become the standard for RCCSs, and will be applied for the characterization of IDPs at large. POTENCI is available for download and as a web server implementation from http://www.protein-nmr.org.
References
Akaike H (1974) New look at statistical-model identification. IEEE Trans Autom Control AC19:716–723
Akaike H (1985) Prediction and entropy. A celebration of statistics. Atkinson ACF, SE New York, Springer, pp 1–24
Baker D, Sali A (2001) Protein structure prediction and structural genomics. Science 294:93–96
Bartels C, Guntert P, Billeter M, Wuthrich K (1997) GARANT—a general algorithm for resonance assignment of multidimensional nuclear magnetic resonance spectra. J Comput Chem 18:139–149
Berjanskii MV, Wishart DS (2005) A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc 127:14970–14971
Bermel W et al (2013) High-dimensionality C-13 direct-detected NMR experiments for the automatic assignment of intrinsically disordered proteins. J Biomol NMR 57:353–361
Braun D, Wider G, Wuethrich K (1994) Sequence-corrected 15N “random coil” chemical shifts. J Am Chem Soc 116:8466–8469
Brutscher B et al (2015) NMR methods for the study of instrinsically disordered proteins structure, dynamics, and interactions: general overview and practical guidelines. Adv Exp Med Biol 870:49–122
Bundi A, Wüthrich K (1979) 1H-nmr parameters of the common amino acid residues measured in aqueous solutions of the linear tetrapeptides H-Gly-Gly-X-L-Ala-OH. Biopolymers 18:285–297
Burley SK (2000) An overview of structural genomics. Nat Struct Biol 7:932–934
Camilloni C, De Simone A, Vranken WF, Vendruscolo M (2012) Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry 51:2224–2231
Cavalli A, Salvatella X, Dobson CM, Vendruscolo M (2007) Protein structure determination from NMR chemical shifts. Proc Natl Acid Sci USA 104:9615–9620
Chandonia J-M, Brenner SE (2006) The impact of structural genomics: expectations and outcomes. Science 311:347–351
Chen TC, Hsiao CL, Huang SJ, Huang JR (2016) The nearest-neighbor effect on random-coil nmr chemical shifts demonstrated using a low-complexity amino-acid sequence. Protein Pept Lett 23:967–975
Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13:289–302
De Simone A et al (2009) Accurate random coil chemical shifts from an analysis of loop regions in native states of proteins. J Am Chem Soc 131:16332–16333
Dunker AK et al (2002) Intrinsic disorder and protein function. Biochemistry 41:6573–6582
Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208
Eliezer D et al (2005) Residual structure in the repeat domain of tau: echoes of microtubule binding and paired helical filament formation. Biochemistry 44:1026–1036
Felli IC, Pierattelli R (2012) Recent progress in NMR spectroscopy: toward the study of intrinsically disordered proteins of increasing size and complexity. IUBMB Life 64:473–481
Georgiev AG (2009) Interpretable numerical descriptors of amino acid space. J Comput Biol 16:703–723
Han B, Liu YF, Ginzinger SW, Wishart DS (2011) SHIFTX2: significantly improved protein chemical shift prediction. J Biomol NMR 50:43–57
Hatzopoulos GN et al (2013) Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure 21:2069–2077
Isaksson L et al (2013) Highly efficient NMR Assignment of intrinsically disordered proteins: application to B- and T cell receptor domains. PLoS ONE 8:e62947
Jung YS, Zweckstetter M (2004) Mars—robust automatic backbone assignment of proteins. J Biomol NMR 30:11–23
Kjaergaard M, Poulsen FM (2011) Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J Biomol NMR 50:157–165
Kjaergaard M, Poulsen FM (2012) Disordered proteins studied by chemical shifts. Prog Nucl Magn Reson Spectrosc 60:42–51
Kjaergaard M et al (2010) Temperature-dependent structural changes in intrinsically disordered proteins: formation of alpha-helices or loss of polyproline II? Protein Sci 19:1555–1564
Kjaergaard M, Brander S, Poulsen FM (2011) Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J Biomol NMR 49:139–149
Kohlhoff KJ et al (2009) Fast and accurate predictions of protein NMR chemical shifts from interatomic distances. J Am Chem Soc 131:13894–13895
Kragelj J, Ozenne V, Blackledge M, Jensen MR (2013) Conformational propensities of intrinsically disordered proteins from NMR chemical shifts. Chemphyschem 14:3034–3045
Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–1327
Marsh JA, Singh VK, Jia Z, Forman-Kay JD (2006) Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci 15:2795–2804
Meiler J (2003) PROSHIFT: Protein chemical shift prediction using artificial neural networks. J Biomol NMR 26:25–37
Merutka G, Dyson HJ, Wright PE (1995) ‘Random coil’ 1H chemical shifts obtained as a function of temperature and trifluoroethanol concentration for the peptide series GGXGG. J Biomol NMR 5:14–24
Modig K et al (2007) Detection of initiation sites in protein folding of the four helix bundle ACBP by chemical shift analysis. FEBS Lett 581:4965–4971
Montelione GT et al (2000) Protein NMR spectroscopy in structural genomics. Nat Struct Biol 7:982–985
Moseley HNB, Monleon D, Montelione GT (2001) Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Nucl Magn Reson Biol Macromol Pt B 339:91–108
Mukrasch MD et al (2005) Sites of tau important for aggregation populate (beta)-structure and bind to microtubules and polyanions. J Biol Chem 280:24978–24986
Neal S, Nip AM, Zhang HY, Wishart DS (2003) Rapid and accurate calculation of protein H-1, C-13 and N-15 chemical shifts. J Biomol NMR 26:215–240
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Molec Biol 48:443–453
Nielsen JT, Mulder FAA (2016) There is diversity in disorder—“In all chaos there is a cosmos, in all disorder a secret order”. Front Mol Biosci 3:4
Nielsen JT, Nielsen NC (2014) VirtualSpectrum, a tool for simulating peak list for multi-dimensional NMR spectra. J Biomol NMR 60:51–66
Nielsen JT, Eghbalnia HR, Nielsen NC (2012) Chemical shift prediction for protein structure calculation and quality assessment using an optimally parameterized force field. Progr Nucl Magn Reson Spectrosc 60:1–28
Nielsen JT et al (2016) In situ high-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum. Nat Commun 7:12454
Oezguen N et al (2002) Automated assignment and 3D structure calculations using combinations of 2D homonuclear and 3D heteronuclear NOESY spectra. J Biomol NMR 22:249–263
Perez Y, Gairi M, Pons M, Bernado P (2009) Structural characterization of the natively unfolded N-terminal domain of human c-Src kinase: insights into the role of phosphorylation of the unique domain. J Mol Biol 391:136–148
Piai A et al (2014) “CON-CON’’ assignment strategy for highly flexible intrinsically disordered proteins. J Biomol NMR 60:209–218
Piai A et al (2016) Amino acid recognition for automatic resonance assignment of intrinsically disordered proteins. J Biomol NMR 64:239–253
Platzer G, Okon M, McIntosh LP (2014) pH-dependent random coil (1)H, (13)C, and (15)N chemical shifts of the ionizable amino acids: a guide for protein pK a measurements. J Biomol NMR 60:109–129
Ramachandran GN, Ramakrishnan C, Sasisekharan V (1963) Stereochemistry of polypeptide chain configurations. J Mol Biol 7:95–99
Richarz R, Wüthrich K (1978) Carbon-13 NMR chemical shifts of the common amino acid residues measured in aqueous solutions of the linear tetrapeptides H-Gly-Gly-X-L-Ala-OH. Biopolymers 17:2133–2141
Romero P et al (2001) Sequence complexity of disordered protein. Proteins 42:38–48
Rosato A et al (2012) Blind testing of routine, fully automated determination of protein structures from NMR data. Structure 20:227–236
Schmidt E, Guntert P (2012) A new algorithm for reliable and general NMR resonance assignment. J Am Chem Soc 134:12817–12829
Schwarzinger S et al (2000) Random coil chemical shifts in acidic 8 M urea: implementation of random coil shift data in NMRView. J Biomol NMR 18:43–48
Schwarzinger S et al (2001) Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc 123:2970–2978
Shen Y, Bax A (2007) Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J Biomol NMR 38:289–302
Shen Y et al (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acid Sci USA 105:4685–4690
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS plus: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223
Simon M, Hancock JM (2009) Tandem and cryptic amino acid repeats accumulate in disordered regions of proteins. Gen Biol 10:R59-R59
Simons KT, Strauss C, Baker D (2001) Prospects for ab initio protein structural genomics. J Mol Biol 306:1191–1199
Singarapu KK et al (2011) Structural characterization of Hsp12, the heat shock protein from Saccharomyces cerevisiae, in aqueous solution where it is intrinsically disordered and in detergent micelles where it is locally alpha-helical. J Biol Chem 286:43447–43453
Spera S, Bax A (1991) Empirical correlation between protein backbone conformation and C. alpha. and C. beta. 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc 113:5490–5492
Stone M (1977) Asymptotics for and against cross-validation. Biometrika 64:29–35
Tamiola K, Mulder FAA (2011) ncIDP-assign: a SPARKY extension for the effective NMR assignment of intrinsically disordered proteins. Bioinformatics 27:1039–1040
Tamiola K, Mulder FAA (2012) Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. Biochem Soc Trans 40:1014–1020
Tamiola K, Acar B, Mulder FAA (2010) Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc 132:18000–18003
Tamiola K, Scheek RM, Meulen P, Mulder FAA (2018) PepKalc-scalable and comprehensive calculation of electrostatic interactions in random coil polypeptides. Bioinformatics. https://doi.org/10.1093/bioinformatics/bty033
Theil H, Theil H (1971) Principles of econometrics
Ting D et al (2010) Neighbor-dependent Ramachandran probability distributions of amino acids developed from a hierarchical Dirichlet process model. PLoS Comput Biol 6:e1000763
van der Lee R et al (2014) Classification of intrinsically disordered regions and proteins. Chem Rev 114:6589–6631
Verdegem D, Dijkstra K, Hanoulle X, Lippens G (2008) Graphical interpretation of Boolean operators for protein NMR assignments. J Biomol NMR 42:11–21
Wang G, Dunbrack RL Jr (2003) PISCES: a protein sequence culling server. Bioinformatics 19:1589–1591
Wang Y, Jardetzky O (2002) Investigation of the neighboring residue effects on protein chemical shifts. J Am Chem Soc 124:14075–14084
Wang L, Eghbalnia HR, Bahrami A, Markley JL (2005) Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J Biomol NMR 32:13–22
Ward JJ et al (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337:635–645
Wilk MB, Gnanadesikan R (1968) Probability plotting methods for the analysis of data. Biometrika 55:1–17
Williamson MP (1990) Secondary-structure dependent chemical shifts in proteins. Biopolymers 29:1423–1431
Williamson MP, Craven CJ (2009) Automated protein structure calculation from NMR data. J Biomol NMR 43:131–143
Wishart DS, Sykes BD, Richards FM (1991) Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol 222:311–333
Wishart DS et al (1995) 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR 5:67–81
Wold S, Esbensen K, Geladi P (1987) Principal component analysis. Chemom Intell Lab Syst 2:37–52
Wright PE, Dyson HJ (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16:18–29
Zawadzka-Kazimierczuk A, Kozminski W, Billeter M (2012) TSAR: a program for automatic resonance assignment using 2D cross-sections of high dimensionality, high-resolution spectra. J Biomol NMR 54:81–95
Zhang HY, Neal S, Wishart DS (2003) RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR 25:173–195
Zhang ZY, Porter J, Tripsianes K, Lange OF (2014) Robust and highly accurate automatic NOESY assignment and structure determination with Rosetta. J Biomol NMR 59:135–145
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nielsen, J.T., Mulder, F.A.A. POTENCI: prediction of temperature, neighbor and pH-corrected chemical shifts for intrinsically disordered proteins. J Biomol NMR 70, 141–165 (2018). https://doi.org/10.1007/s10858-018-0166-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-018-0166-5