Abstract
By assembling mesocosms and utilizing high-throughput sequencing, we aim to characterize the shifts of the bacterial community in freshwaters driven by two contrasting submerged macrophyte species, Ceratophyllum demersum L. and Vallisneria spiralis L. Although the microbe in both the water column and sediment were largely modulated by the macrophyte, the effect varied considerably depending on bacterial locations and macrophyte species. Actinobacteria was the most abundant taxa in the water column of all the three treatments, but its abundances were significantly higher in the two planted treatments. Moreover, Alphaproteobacteria showed high abundance only in the unplanted control. For bacterial taxa in the sediment, C. demersum significantly increased the relative abundance of Anaerolineae but reduced the relative abundance of Betaproteobacteria and Gammaproteobacteria, while V. spiralis increased the relative abundance of Deltaproteobacteria and Gammaproteobacteria. Additionally, in the C. demersum treatment, the water column bacterial community increased more dramatically in richness, alpha diversity, and the relative abundance of the dominant taxa than those in the V. spiralis treatment. Taken together, the findings from this study reveal that the two species of submerged macrophyte modified the bacterial community in waters, despite the obvious interspecific performance differences.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Submerged macrophytes are important components of aquatic ecosystems, occupy key interfaces, and therefore have major effects on productivity and biogeochemical cycles (Carpenter & Lodge, 1986; Srivastava et al., 2008; Baart et al., 2010). Since it was found that submerged macrophytes are crucial for the stabilization of the clear water state in shallow eutrophic and mesotrophic lakes (van Donk & van de Bund, 2002), numerous efforts have been made to address the underlying mechanisms related to the impact of submerged macrophytes in the aquatic ecosystem (Søndergaard et al., 2010; Vermaire et al., 2011).
With the development of culture-independent biology techniques, determining how macrophytes shape the bacterial community structure has become an active area of research (Brix & Schierup, 1989; Metzker, 2010; Zeng et al., 2012). In aquatic ecosystems, the presence of macrophytes could enhance the horizontal and vertical heterogeneity of the water environment by releasing substantial amounts of oxygen and organic carbon (Herrmann et al., 2009; Lewicka-Rataj et al., 2018) and by altering the pH and availability of the surrounding light and nutrients (Kirschner & Velimirov, 1997; Wu et al., 2007; Hilt, 2015), factors which could reasonably be expected to affect microbial communities. Additionally, different species of macrophytes characterized by dissimilar physiology and chemical compositions would result in the release of various types of substrates and thereby affect the availability of ecological niches for different bacterial strains (Brum & Esteve, 2001; Hempel et al., 2009; Pang et al., 2016). It is a common consensus among aquatic ecologists that plant species have a strong selective effect on rhizosphere microorganisms, epiphyton, and bacterioplankton (Marschner & Timonen, 2005; He et al., 2014; Fan et al., 2016). Compounds released from macrophytes could serve as selective agents that directly alter the bacterial community composition (BCC), and the species-specific influence of macrophytes on BCC is variable with the season and distance (Murray & Hodson, 1986; Zeng et al., 2012). Despite the rapid growth of knowledge concerning the effect of submerged macrophytes on modifying the microbial structure in aquatic systems, most studies have solely focused on water or sediment microorganisms, and the comparative analysis regarding the impact of submerged macrophytes on both parts has rarely been reported.
High-throughput sequencing (HTS), currently the most popular next-generation sequencing method, has been widely used to survey and improve our understanding of the diversity of prokaryotic communities in various environmental habitats and across multiple research disciplines (Peralta et al., 2012; Xie et al., 2015). Unlike the other approaches (e.g., PCR cloning, fluorescent in situ hybridization, denaturing gradient gel electrophoresis), in addition to providing comprehensive information without bias (overall error rates < 1%), Illumina’s HTS can generate up to billions of higher quality reads per flow cell at a far lower price. Despite the wide use of HTS, thus far, there is limited information about the microbial compositional structures modulated by macrophytes based on HTS technology.
Herein, we use Illumina’s HTS to comprehensively characterize the diversity and composition of the bacterial community in laboratory mesocosms with and without submerged macrophytes. Two species, Ceratophyllum demersum L. and Vallisneria spiralis L., were chosen in this study because they both grow fast and can easily develop into dense stands. Moreover, it has been documented that each could yield good results in improving and maintaining water quality (Qiu et al., 2001; Dai et al., 2012). More importantly, their morphologies are remarkably distinct: C. demersum is a free-floating submerged species and has fluffy, filamentous, bright-green leaves; V. spiralis is a rooted submerged species and has narrow, linear leaves. Therefore, in addition to evaluating the role of submerged macrophytes on the water column and sediment microbial community, we hypothesize that C. demersum has a relatively stronger effect on bacterioplankton in the water column, whereas V. spiralis plays a stronger role in regulating the bacterial community in sediment.
Materials and methods
Experimental mesocosms
A total of 9 PVC tanks (0.6 m × 0.5 m × 0.8 m; length × width × height) were used for simulation of the lake system and comparative analyses. A 10-cm layer of sediment was placed in each tank, and each tank was then filled with water. The sediment used in these mesocosms was collected from the top 0–10 cm of sediment in a eutrophic landscape river flowing through Tongji University, Shanghai. The physical and chemical characteristics of water and sediment are shown in Table 1. This experiment manipulated three experimental treatment sets (planted with C. demersum; planted with V. spiralis; unplanted control), with three replicates for each treatment set. The two species of macrophytes were collected in June 2014 from Donghu Lake in Wuhan. They were pre-incubated for approximately 4 weeks in a large tank with the same water and sediment as the mimic systems. After washing and removing the adherent water on plants with a line wedge of bibulous paper, these two macrophyte species were evenly planted in three tanks (approximately 0.50 kg fresh weight per square meter; C. demersum: ~ 25 plants; V. spiralis: ~ 20 plants) immediately after filling the tanks with water. All mesocosms were exposed to natural sunlight in an open room with a transparent roof. During the experimental period, July 7 to October 3, 2014, the air temperature ranged from 20°C to 38°C. An appropriate amount of tap water was added periodically (every 2–4 days) to maintain the initial water level. For more details, please refer to Dai et al. (2017).
Physical and chemical analyses
Water and sediment samples were collected at the beginning and end of the experiment. Chemical variables included chemical oxygen demand (COD), total phosphorus (TP), soluble reactive phosphorus (SRP), total nitrogen (TN), and ammonia nitrogen (NH4+–N). TN, NH4+–N, and TP of water samples were analyzed according to Standard Methods in Environment Monitoring of China (National Bureau of Environment Protection, 2002). COD was measured using a spectrophotometer (DR/2800, Hach Co., Loveland, CO, USA). SRP was determined according to Murphy & Riley (1962).
Sediment samples were all naturally air-dried and sieved with a standard 100-mesh sieve. P fractions were determined using the SMT protocol (Ruban et al., 1999). The TP concentration in sediments was determined by the ascorbic acid method after combusting the sediment at 450°C for 3 h and extracting it by 20 mL of 3.5 M HCl for 16 h. Inorganic phosphorus in the supernatant (IP) was measured by the method described above after extracting the sediment with 20 mL of 1 M HCl for 16 h. Organic phosphorus (OP) was calculated by taking the difference between TP and IP. Organic matter (OM) was measured as the weight of the dry sediment after being heated in a muffle furnace at 550°C for 3 h. TN was measured as nitrate by alkaline persulfate oxidation digestion (Lachat Method 12-107-04-1-B, Milwaukee, WI).
Bacterial community analysis using high-throughput sequencing
Water column and surface sediment samples were collected at the termination of the experiment, taken to the laboratory, and processed immediately (within 10 min). Bacterioplankton samples in water (500 mL of water) were collected on 0.2-µm pore-sized filters after prefiltration through 5.0-µm pore-sized filters (diameter 45 mm; Whatman, UK). The filters containing microbes were cut into small pieces with a sterile scalpel, and bacterial genomic DNA was then extracted using a bacterial DNA kit (Omega Bio-Tek, Inc., Norcross, GA, USA) following the protocol. The sediment samples were collected at five random spots in each system and mixed in polyethylene bags. The DNA was then extracted from approximately 400 mg of sediment using a soil DNA kit (Omega) according to the manufacturer’s alternative protocol. All DNA extractions were purified with an AxyPrep™ DNA kit.
Next-generation sequencing library preparations and Illumina MiSeq sequencing were conducted at GENEWIZ, Inc. (Beijing, China). DNA samples were quantified using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA) and DNA quality was checked on a 0.8% agarose gel. Five to fifty ng DNA was used to generate amplicons using a MetaVx™ Library Preparation kit (GENEWIZ, Inc., South Plainfield, NJ, USA). A panel of proprietary primers was designed to anneal to the relatively conserved regions bordering the V3, V4, and V5 hypervariable regions. The V3 and V4 regions were amplified using forward primers containing the sequence “CCTACGGRRBGCASCAGKVRVGAAT” and reverse primers containing the sequence “GGACTACNVGGGTWTCTAATCC.” The V4 and V5 regions were amplified using forward primers containing the sequence” GTGYCAGCMGCCGCGGTAA” and reverse primers containing the sequence “CTTGTGCGGKCCCCCGYCAATTC.”
Quality filtering on joined sequences was performed, and sequences that did not fulfill the following criteria were discarded: sequence length < 200 bp, no ambiguous bases, mean quality score ≥ 20. The sequences were then compared with the reference database (RDP Gold database) using the UCHIME algorithm to detect chimeric sequences that were then removed. The sequence data are deposited into the NCBI’s Sequence Read Archive Database under accession numbers SAMN06212356.
The filtered sequence data were further processed to reduce sequencing and PCR errors using the mothur Miseq SOP (http://www.mothur.org/wiki/MiSeq_SOP). Next, the clean sequences were imported into Quantitative Insights into Microbial Ecology (QIIME, version 1.8.0) for the open-reference operational taxonomic units (OTUs) picking and core diversity analyses. The OTUs were assigned using usearch61 alignment and taxonomically assigned by Ribosomal Database Program (RDP) classifier at a confidence threshold of 0.8 using the latest version of Greengenes database. The normalized sequences from all samples with clean sequences between 44,427 and 115,606 were clustered into OTUs at identity thresholds of 0.97. An OTU-based rarefaction curve, alpha diversity metrics (i.e., observed species, Chao1 richness, and Shannon’s diversity index H), and beta diversity metrics (weighted UniFrac) were calculated for all samples that had clean sequences at an even sequencing depth of 44,000 sequences (Ju & Zhang, 2015).
Statistical analysis
We generated ordination plots for visualizing the community shifts, including rarefaction curves (RC), abundance-based coverage estimators (ACE), Chao richness estimator, coverage, Shannon diversity, and Simpson diversity using the Vegan package in R (Oksanen et al., 2013; R Development Core Team, 2015). A one-way ANOVA or a Kruskal–Wallis test (if the data could not be transformed to follow the assumption) was performed on the target variables to test the difference between water column and sediment samples as well as between the three treatments in terms of physical and chemical variables, bacterial 16S rRNA gene copy number, observed species, and Shannon–Wiener indices.
We conducted the principal coordinate analysis (PCoA) to compare similarities among samples based on beta diversity metrics using weighted UniFrac distances (considering both species phylogeny and abundance). To further evaluate the significant differences in bacterial communities between the samples, the randomization/permutation procedure analysis of similarities was used. Complete separation is indicated by R = 1, whereas R = 0 suggests no separation (Clarke & Gorley, 2001). We also present the linear discriminant analysis (LDA) effect size (LEfSe) analysis on the website http://huttenhower.sph.harvard.edu/galaxy under the following conditions: (1) the alpha value for the factorial Kruskal–Wallis test among classes is < 0.05; and (2) the threshold on the logarithmic LDA score for discriminative features is > 2.0 (Zhang et al., 2013). The differential features were identified on the OTU level. The treatment groups were used as the class of subjects (no subclass). Additionally, QIIME scripts were used to detect significant differences between the OTU abundance in the different sample groups (http://qiime.org/scripts/group_significance.html). All the analyses and graphing were accomplished with R version 3.2.1 (R Development Core Team, 2015).
Results
Properties of water and sediment
At the end of the experiment, the concentrations of TN, NH4+, TP, SRP, and COD in the vegetated water column were all significantly lower than those in the control (P < 0.05). Moreover, significantly lower concentrations of TP and SRP were detected in the V. spiralis group compared to the C. demersum group. For sediment, the unplanted control had significantly higher concentrations of TP and OP than the vegetated groups and significantly higher concentrations of OM than the V. spiralis group (P < 0.05). No significant difference was found between the vegetated groups.
Bacterial richness and diversity
The appearance of a plateau in the rarefaction curve for each sample indicates that the bacterial community diversity had been well captured at the sequencing depth used in the present study (Fig. 1A, B). The observed OTUs in the water columns and sediment samples of the three different systems were 1194 and 4724 in the C. demersum group, 917 and 4285 in the V. spiralis group, and 812 and 4581 in the control. In all groups, the bacterial richness in sediment was significantly higher than the richness in the water column (P < 0.05). However, no significant difference in bacterial community richness was detected among the groups either in the water column or in the sediment (Fig. 1C).
The Shannon–Wiener diversity index of bacterial communities in water columns was 6.39 (C. demersum), 4.70 (V. spiralis), and 4.65 (control); in sediments it was 9.77, 9.27 and 9.64, respectively. In addition to the significant difference between the water column and sediment, a significantly higher Shannon–Wiener diversity index was observed in the water column of the C. demersum group compared to the V. spiralis group and the control group (P < 0.05; Fig. 1D).
Bacterial community composition
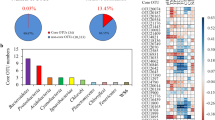
Over 95% and 87% of the annotated sequences were affiliated with 31 bacterial classes and 1 archaeal class of 17 phyla in the water column samples and sediment samples, respectively. Figure 2 shows that there were distinct structural differences between bacterial communities in water columns and sediments as well as among the three treatments (Fig. 2). On average, in the water columns, Actinobacteria represented the most abundant taxa in all three treatments, but significantly higher relative abundance was observed in the two planted treatments (LSD: P < 0.05), compared to the control. Other dominant taxa following Actinobacteria (accounting for over 10%) included Sphingobacteriia and Cytophagia in the C. demersum treatment, Flavobacteriia and Sphingobacteriia in the V. spiralis treatment, and Alphaproteobacteria, Flavobacteriia, and Saprospirae in the control. Additionally, for these taxa, there were significant differences among the three groups. Meanwhile, compared with the relative microbial abundance in the control, C. demersum significantly enhanced the Anaerolineae (16.2% VS 15.8%) but reduced the abundance of Betaproteobacteria (15.9% VS 17.1%) and Gammaproteobacteria (13.4% VS 14.3%) in the sediment. In the V. spiralis group, Deltaproteobacteria (17.3% VS 15.5%) and Gammaproteobacteria (17.3% VS 14.3%) were significantly enhanced (P < 0.05).
The relative abundance of dominating bacterial phyla and classes in different systems. Relative abundances were calculated as the number of sequences assigned to each bacterial group divided by the total number of 16S rRNA gene in each sample and expressed as percentages. Only bacterial groups of high relative abundance (> 5% in all systems) were shown in the graph. Bacterial groups of low abundance were grouped as Others
Bacterial community structure comparison
The results reveal that the variability of microbial communities could be mainly explained by the first principle components (PC1), which accounts for up to 76% of the variability. Microflora in the water column and sediment were significantly separated at P = 0.01 with the R value of 0.630, together with 0.327 and 0.105 for the two parts among the three groups. Moreover, remarkably larger variability of microflora among the water columns was observed (Fig. 3).
Three-dimensional principal coordinate analysis (PCoA) plot (A) and the distance values show the differences of bacterial community in water column and sediment among the three different systems (B), and pairwise community distances were determined using the weighted UniFrac algorithm. The smallest observation, first quartile, median, third quartile, and largest observation are indicated in (B)
To further explore the bacterial community differences among the three groups, QIIME scripts and LEfSe were employed, and both indicated that there were significant differences among water samples but none among sediment samples. QIIME scripts compare each OTU based on the past sample groupings to see if it is differentially represented. In this study, each system had its own dominant OTUs. In the C. demersum and V. spiralis groups, each had 7 OTUs significantly higher than in the other groups, while in the control, there were 8 OTUs (Table 2). Most of the OTUs belonged to Bacteroidetes and Actinobacteria: one OTU in the V. spiralis group belonged to Cyanobacteria, and two OTUs in the control group were affiliated with Proteobacteria. LEfSe can identify differentially abundant taxa by combining tests for statistical significance, biological consistency, and effect relevance (Segata et al., 2011). A total of 9 bacterial clades were detected as high-dimensional biomarkers for separating water microbiota among the three treatments (Fig. 4A). Seven were in the C. demersum group, and one was in the V. spiralis and control group, respectively. The most differentially abundant phyla in the system with C. demersum were Gammaproteobacteria (belonging to Proteobacteria), followed by Mb-NB09, BS119 TM7-1, TM7, and OM190 (belonging to Planctomycetes); the predominant phylum in the V. spiralis and control group was BSV26 (belonging to Chlorobi) and Oscillatoria psychidae (belonging to Cyanobacteria), respectively.
Differentially abundant taxa identified using LEfSe analysis. The histogram A shows the LDA scores computed for features (OTU level) differentially abundant in water columns among different systems. Taxonomic cladogram. B reports the taxa (highlighted by small circles and by shading) showing significant different abundance values (according to LEfSe) in the three different systems; for each taxon, the color denotes the class with higher median for both the small circles and the shading
Discussion
Similar to previous findings (Zeng et al., 2012; Waters et al., 2015), there were differentiated microbial communities between the present and absent-plant treatments. In addition to finding a higher abundance of the most dominant taxon in water columns due to the growth of macrophytes, all of the proceeding dominant groups in planted systems belonged to Bacteroidetes. These results support the finding of a previous study demonstrating a positive relationship between specific bacterial phyla and macrophytes (Gordon-Bradley et al., 2014). Van der Gucht et al. (2005) inferred that the die-off submerged macrophytes could contribute to higher concentrations of the high molecular weight fraction of dissolved organic matter (DOC) in clear water lakes and hence facilitated the proliferation of the Bacteroidetes (Kirchman, 2002). Additionally, consistent with the previous study, which showed a high numerical importance at phytoplankton-dominated sites in the large and shallow subtropical lake (Wu et al., 2007), we found that Alphaproteobacteria dominated only in the unplanted control, where the nutrient concentration and phytoplankton density were remarkably higher in the systems with macrophyte-absent control than in the planted systems (Table 1; Dai et al., 2017). The supply of phytoplankton-derived DOC could contribute to the phenomenon (Stepanauskas et al., 2003; Bunse et al., 2016), because the abundance of Alphaproteobacteria is inferred almost exclusively controlled by resources and DOM concentrations and DOM properties are among the most important ones (Amaral et al., 2016). Compared with the community of epibiotic bacteria and surrounding bacterioplankton of Potamogeton crispus in freshwater lakes, He et al. (2014) found that Alphaproteobacteria was the most dominant group among epibiotic bacterial taxa, whereas Actinobacteria dominated bacterioplankton. In our study, Actinobacteria showed significantly higher relative abundance in the planted water columns, rather than Alphaproteobacteria. These indicated that DOC released by submerged macrophytes has limited impacts on the bacterioplankton in lentic waters.
Regarding how macrophytes affect the bacterial diversity in the water column, there are conflicting results in the literature, as Wu et al. (2007) observed a higher bacterial diversity in the macrophyte-absent areas compared with the macrophyte-dominated sites. Gordon-Bradley et al. (2015) found that the Shannon diversity indices did not vary greatly between the blooming and nonblooming stages of Hydrilla. The study from Declerck et al. (2005) indicates that the richness of bacteria was positively correlated to submerged vegetation in shallow lakes. In our study, we found a significantly higher Shannon–Wiener diversity index only in the water column with C. demersum. These results imply that different species of macrophytes could have distinctive effects on the bacterioplankton community. This conclusion can be further supported by the particular microbial taxa that were identified via QIIME scripts and LEfSe. In general, there are limited results and conclusions about the bacterial richness and Shannon–Wiener diversity related to macrophyte growth. Therefore, more studies are recommended to confirm the exact effect and uncover the underlying mechanisms, and we urge further investigation of detailed taxonomic compositions and ecological features, e.g., metabolism, growth, and mortality.
Although variations of sediment microbes were also observed among the treatments, compared to bacterioplankton, the two species of submerged macrophyte displayed a much weaker effect on the sediment microbes. Ahn et al. (2007) came to the analogous results that no significant effects of the presence of macrophyte on sediment microbial communities were detected in the wetland system, but they attributed this to the low growth of plants and sampling anomalies. Other studies have demonstrated differences in the species composition and biomass of sediment bacteria between macrophyte and unvegetated areas (Boschker et al., 2000; Kurtz et al., 2003; Shao et al., 2011). However, their data from small sample sizes did not allow them to conduct the statistical analysis. Using terminal restriction fragment length polymorphism (T-RFLP) and LIBSHUFF analysis, Zhao et al. (2013) detected significant differences in bacterial communities between unvegetated and rhizosphere sediments. Moreover, they found significantly lower diversity of the bacterial community associated with the rhizosphere sediments of submerged macrophytes, compared to that of the bulk sediment (zhao et al., 2017). The discrepancy among these studies could be attributed to the different analysis approaches used, methods of sample collection, and research systems (mesocosm and large, shallow lake).
As we hypothesized, C. demersum showed a stronger effect on the richness and alpha diversity of bacterioplankton. Moreover, the abundance of seven bacterial taxa was significantly higher in the C. demersum group, whereas in the V. spiralis system, only one taxon was identified. It has been documented that characteristics of the leaf structure play a crucial role in dictating the types of microbes that thrive on them (Redford et al., 2010). In our study, C. demersum has long stems with numerous side shoots and thread-like leaves that are produced in whorls of six to twelve, which makes the single specimen appear as a large, bushy mass, while the leaves of V. spiralis are flat, linear and the surface can range up to 19 mm in width. Therefore, C. demersum has the potential to provide a more protective habitat for bacterial colonizers. Although both species of submerged macrophytes showed inhibitory effects on cyanobacteria (Gross et al., 2003; Xian et al., 2006a), one OTU with significantly higher frequencies belonging to Cyanobacteria was identified in the V. spiralis group. These results imply that as the distinct allelopathy on phytoplankton between the two species (Xian et al., 2006b; Dai et al., 2017), C. demersum has a stronger inhibitory effect on Cyanobacteria than V. spiralis. Additionally, the differences in plant exudates, oxygen release, pH alteration, light and nutrient availability, decomposition products of dissolved organic matter, etc., are reasonably expected to affect microbial communities differently (Boschker et al., 1999; Gordon-Bradley et al., 2014; Levi et al., 2017). Contrary to our expectation, no high-dimensional biomarker and a significantly different frequency higher OUT was identified in the planted systems, although the relative abundance of Anaerolineae in the system of C. demersum, along with Deltaproteobacteria and Gammaproteobacteria in the system of V. spiralis, exhibited an obvious increment compared to the sediment dominant taxon in the control system. More studies, especially long-term experiments, are necessary to verify the results.
In summary, we employed C. demersum and V. spiralis as the representative submerged macrophyte in freshwater and reported a comprehensive characterization of the bacterial communities shaped by macrophytes in mimic ecosystems.
In summary, the two species of submerged macrophyte shaped the water bacterioplankton community at the end of the experiment, whose diversity and representative populations significantly differed from those in the unplanted control system but presented limited impact on the sediment bacterial community. Additionally, obvious interspecific performance differences were observed in this study. Habitat-related species sorting and physical and chemical properties in the water column could be the drivers of bacterial community formation and modulation. Hence, we argue that macrophytes play an important role in structuring bacteria community composition, but their effects may differ significantly, because their various physiological and morphological traits could provide different heterogeneous physical structures and nutrient flows.
References
Ahn, C., P. M. Gillevet & M. Sikaroodi, 2007. Molecular characterization of microbial communities in treatment microcosm wetlands as influenced by macrophytes and phosphorus loading. Ecological Indicators 7: 852–863.
Amaral, V., D. Graeber, D. Calliari & C. Alonso, 2016. Strong linkages between DOM optical properties and main clades of aquatic bacteria. Limnology and Oceanography 61: 906–918.
Baart, I., C. Gschöpf, A. P. Blaschke, S. Preiner & T. Hein, 2010. Prediction of potential macrophyte development in response to restoration measures in an urban riverine wetland. Aquatic Botany 93: 153–162.
Boschker, H. T. S., J. F. C. De Brouwer & T. E. Cappenberg, 1999. The contribution of macrophyte-derived organic matter to microbial biomass in salt-marsh sediments: stable carbon isotope analysis of microbial biomarkers. Limnology and Oceanography 44: 309–319.
Boschker, H. T. S., A. Wielemaker, B. E. M. Schaub & M. Holmer, 2000. Limited coupling of macrophyte production and bacterial carbon cycling in the sediments of Zostera spp. meadows. Marine Ecology Progress Series 203: 181–189.
Brix, H. & H. H. Schierup, 1989. The use of aquatic macrophytes in water-pollution control. Ambio Stockholm 18: 100–107.
Brum, P. R. & F. A. Esteves, 2001. Changes in abundance and biomass of the attached bacterial community throughOTU the decomposition of three species of aquatic macrophytes. In: Faria, B.M.; Farjalla, V.F. & Esteves, F.A. (eds). Aquatic Microbial Ecology in Brazil. Series Oecologia Brasiliensis, vol. IX. PPGE-UFRJ: 77-96.
Bunse, C., M. Bertos-Fortis, I. Sassenhagen, S. Sildever, C. Sjöqvist, A. Godhe, S. Gross, A. Kremp, I. Lips, N. Lundholm & K. Rengefors, 2016. Spatio-temporal interdependence of bacteria and phytoplankton during a Baltic Sea spring bloom. Frontiers in Microbiology 7: 517.
Carpenter, S. R. & D. M. Lodge, 1986. Effects of submersed macrophytes on ecosystem processes. Aquatic Botany 26: 341–370.
Clarke, K. R. & R. N. Gorley, 2001. PRIMER v5: User Manual/Tutorial. PRIMER-E: PlymOTUh: UK.
Dai, Y. R., C. R. Jia, W. Liang, S. H. Hu & Z. B. Wu, 2012. Effects of the submerged macrophyte Ceratophyllum demersum L. on restoration of a eutrophic waterbody and its optimal coverage. Ecological Engineering 40: 113–116.
Dai, Y. R., J. Wu, X. H. Ma, F. Zhong, N. X. Cui & S. P. Cheng, 2017. Increasing phytoplankton-available phosphorus and inhibition of macrophyte on phytoplankton bloom. Science of Total Environment 579: 871–880.
Fan, Z., R. M. Han, J. Ma & G. X. Wang, 2016. Submerged macrophytes shape the abundance and diversity of bacterial denitrifiers in bacterioplankton and epiphyton in the Shallow Fresh Lake Taihu, China. Environmental Science and Pollution Research 23: 14102–14114.
Declerck, S., J. Vandekerkhove, L. Johansson, K. Muylaert, J. M. Conde-Porcuna, K. Van Der Gucht, C. Pérez-Martíez, T. Lauridsen, K. Schwenk, G. Zwart, W. Rommens, J. López-Romos, E. Jeppesen, W. Vyverman, L. Brendonck & L. De Meester, 2005. Multi-group biodiversity in shallow lakes along gradients of phosphorus and water plant cover. Ecology 86: 1905–1915.
Gordon-Bradley, N., D. S. Lymperopoulou & H. N. Williams, 2014. Differences in bacterial community structure on Hydrilla verticillata and Vallisneria americana in a freshwater spring. Microbes and environments 29: 67–73.
Gordon-Bradley, N., N. Li & H. N. Williams, 2015. Bacterial community structure in freshwater springs infested with the invasive plant species Hydrilla verticillata. Hydrobiologia 742: 221–232.
Gross, E. M., D. Erhard & E. Iványi, 2003. Allelopathic activity of Ceratophyllum demersum L. and Najas marina ssp. intermedia (Wolfgang) Casper. Hydrobiologia 506: 583–589.
He, D., L. Ren & Q. L. Wu, 2014. Contrasting diversity of epibiotic bacteria and surrounding bacterioplankton of a common submerged macrophyte, Potamogeton crispus, in freshwater lakes. FEMS microbiology ecology 90: 551–562.
Hempel, M., H. P. Grossart & E. M. Gross, 2009. Community composition of bacterial biofilms on two submerged macrophytes and an artificial substrate in a pre-alpine lake. Aquatic Microbial Ecology 58: 79–94.
Herrmann, M., A. M. Saunders & A. Schramm, 2009. Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Applied and Environmental Microbiology 75: 3127–3136.
Hilt, S., 2015. Regime shifts between macrophytes and phytoplankton – concepts beyond shallow lakes, unravelling stabilizing mechanisms and practical consequences. Limnetica 34: 467–480.
Ju, F. & T. Zhang, 2015. 16S rRNA gene high-throughput sequencing data mining of microbial diversity and interactions. Applied Microbiology and Biotechnology 99: 4119–4129.
Kirchman, D. L., 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiology Ecology 39: 91–100.
Kirschner, A. K. T. & B. Velimirov, 1997. A seasonal study of bacterial community succession in a temperate backwater system, indicated by variation in morphotype numbers, biomass, and secondary production. Microbial Ecology 34: 27–38.
Kurtz, J. C., D. F. Yates, J. M. Macauley, R. L. Quarles, F. J. Genthner, C. A. Chancy & R. Devereux, 2003. Effects of light reduction on growth of the submerged macrophyte Vallisneria americana and the community of root-associated heterotrophic bacteria. Journal of Experimental Marine Biology and Ecology 291: 199–218.
Levi, P. S., P. Starnawski, B. Poulsen, A. Baattrup-Pedersen, A. Schramm & T. Riis, 2017. Microbial community diversity and composition varies with habitat characteristics and biofilm function in macrophyte-rich streams. Oikos 126: 398–409.
Lewicka-Rataj, K., A. Świątecki & D. Górniak, 2018. The effect of Lobelia dortmanna L. on the structure and bacterial activity of the rhizosphere. Aquatic Botany 145: 10–20.
Marschner, P. & S. Timonen, 2005. Interactions between plant species and mycorrhizal colonization on the bacterial community composition in the rhizosphere. Applied Soil Ecology 28: 23–36.
Metzker, M. L., 2010. Sequencing technologies - the next generation. Nature Reviews Genetics 11: 31–46.
Murphy, J. & J. P. Riley, 1962. A modified single solution method for the determination of phosphate in natural waters. Analytica Chimica Acta 27: 31–36.
Murray, R. E. & R. E. Hodson, 1986. Influence of macrophyte decomposition on growth rate and community structure of Okefenokee Swamp bacterioplankton. Applied and environmental microbiology 51: 293–301.
National Bureau of Environment Protection Editorial Committee of Water and Wastewater Monitoring Analytical Methods, 2002. Water and wastewater monitoring analysis method, 4th ed. Chinese Environment Science Press, Beijing, China.
Oksanen, J., F. G. Blanchet, R. Kindt, P. Legendre, P. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, E. Szoecs & H. Wagner, 2013. Vegan: Community Ecology Package.
Pang, S., S. Zhang, X. Lv, B. Han, K. Liu, C. Qiu, C. Wang, P. Wang, H. Toland & Z. He, 2016. Characterization of bacterial community in biofilm and sediments of wetlands dominated by aquatic macrophytes. Ecological Engineering 97: 242–250.
Peralta, R. M., C. Ahn & P. M. Gillevet, 2012. Characterization of soil bacterial community structure and physicochemical properties in created and natural wetlands. Science of Total Environment 443: 725–732.
Qiu, D. R., Z. B. Wu, B. Y. Liu, J. Q. Deng, G. P. Fu & F. He, 2001. The restoration of aquatic macrophytes for improving water quality in a hypertrophic shallow lake in Hubei Province, China. Ecological Engineering 18: 147–156.
R Development Core Team, 2015. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria.
Redford, A. J., R. M. Bowers, R. Knight, Y. Linhart & N. Fierer, 2010. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environmental Microbiology 12: 2885–2893.
Ruban, V., J. F. Lópea-Sánchez, P. Pardo, G. Rauret, H. Muntau & P. Quevauviller, 1999. Selection and evaluation of sequential extraction procedures for the determination of phosphorus forms in lake sediment. Journal of Environmental Monitoring 1: 51–56.
Segata, N., J. Izard, L. Waldron, D. Gevers, L. Miropolsky, W. S. Garrett & C. Huttenhower, 2011. Metagenomic biomarker discovery and explanation. Genome Biology 12: R60.
Shao, K. Q., G. Gao, B. Q. Qin, X. M. Tang, Y. P. Wang, K. X. Chi & J. Y. Dai, 2011. Comparing sediment bacterial communities in the macrophyte-dominated and algae-dominated areas of eutrophic Lake Taihu, China. Candian Journal of Microbiology 57: 263–272.
Srivastava, J., A. Gupta & H. Chandra, 2008. Managing water quality with aquatic macrophytes. Reviews in Environmental Science Biotechnology 7: 255–266.
Stepanauskas, R., M. A. Moran, B. A. Bergamaschi & J. T. Hollibaugh, 2003. Covariance of bacterioplankton composition and environmental variables in a temperate delta system. Aquatic Microbial Ecology 31: 85–98.
Søndergaard, M., L. S. Johansson, T. L. Lauridsen, T. L. Jørgensen, L. Liboriussen & E. Jeppesen, 2010. Submerged macrophytes as indicators of the ecological quality of lakes. Freshwater Biology 55: 893–908.
Van der Gucht, K., T. Vandekerckhove, V. Nele, S. Cousin, K. Muylaet, K. Sabbe, M. Gillis, S. Declerk, L. De Meester & W. Vyverman, 2005. Characterization of bacterial communities in four freshwater lakes differing in nutrient load and food web structure. FEMS Microbiology Ecology 53: 205–220.
van Donk, E. & W. J. van de Bund, 2002. Impact of submerged macrophytes including charophytes on phyto- and zooplankton communities: allelopathy versus other mechanisms. Aquatic Botany 72: 261–274.
Vermaire, J. C., Y. T. Prairie & I. Gregory-Eaves, 2011. The influence of submerged macrophytes on sedimentary diatom assemblages. Journal of Applied Phycology 47: 1230–1240.
Waters, M. N., C. L. Schelske & M. Brenner, 2015. Cyanobacterial dynamics in shallow Lake Apopka (Florida, U.S.A.) before and after the shift from a macrophyte-dominated to a phytoplankton-dominated state. Freshwater Biology 60: 1571–1580.
Wu, Q. L., G. Zwart, J. F. Wu, M. P. Kamst-van Agterveld, S. J. Liu & M. W. Hahn, 2007. Submersed macrophytes play a key role in structuring bacterioplankton community composition in the large, shallow, subtropical Taihu Lake, China. Environmental Microbiology 9: 2765–2774.
Xian, Q., H. Chen, H. Liu, H. Zou & D. Yin, 2006a. Isolation and Identification of Antialgal Compounds from the Leaves of Vallisneria spiralis L. by Activity-Guided Fractionation (5 pp). Environmental Science and Pollution Research 13: 233–237.
Xian, Q., H. Chen, H. Zou & D. Yin, 2006b. Allelopathic activity of volatile substance from submerged macrophytes on Microcystin aeruginosa. Acta Ecologica Sinica 26: 3549–3554.
Xie, W. Y., J. Q. Su & Y. G. Zhu, 2015. Phyllosphere bacterial community of floating macrophytes in paddy soil environments as revealed by Illumina High-Throughput sequencing. Applied and Environmental Microbiology 81: 522–532.
Zeng, J., Y. Q. Bian, P. Xing & Q. L. Wu, 2012. Macrophyte species drive the variation of bacterioplankton community composition in a shallow freshwater lake. Applied and Environmental Microbiology 78: 177–184.
Zhang, C. H., S. F. Li, L. Yang, P. Huang, W. J. Li, S. Y. Wang, G. P. Zhao, M. H. Zhang, X. Y. Pang, Z. Yan, Y. Liu & L. P. Zhao, 2013. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nature Communication 4: 2163.
Zhao, D. Y., P. Liu, C. Fang, Y. M. Sun, J. Zeng, J. Q. Wang, T. Ma, Y. H. Xiao & Q. L. Wu, 2013. Submerged macrophytes modify bacterial community composition in sediments in a large, shallow, freshwater lake. Canadian Journal of Microbiology 59: 237–244.
Zhao, D., S. Wang, R. Huang, J. Zeng, F. Huang & Z. Yu, 2017. Diversity and composition of bacterial community in the rhizosphere sediments of submerged macrophytes revealed by 454 pyrosequencing. Annals of Microbiology 67: 313–319.
Acknowledgements
This study was financed by the National Natural Science Foundation of China (51609238 and 51578395). We declare no conflicts of interest. We thank the editor and reviewers for their constructive comments that improved this paper.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Handling editor: André Padial
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dai, Y., Wu, J., Zhong, F. et al. Macrophyte identity shapes water column and sediment bacterial community. Hydrobiologia 835, 71–82 (2019). https://doi.org/10.1007/s10750-019-3930-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-019-3930-y