Abstract
The phylogenetic composition and physiological profiles of bacterial communities in freshwater springs were evaluated during the blooming and non-blooming stages of the invasive plant species, Hydrilla verticillata. Community-level physiological profiles (CLPPs) and pyrosequencing of 16S rRNA gene amplicons were used to study potential Hydrilla mediated shifts in the physiological potential and phylogenetic composition of the bacterial community in infested systems. The results of CLPP revealed that the microbes in the Hydrilla invaded sites utilized less substrates during blooming periods than during non-blooming periods of the plant. Spearman’s rank correlation analysis showed some relationships between the relative abundances of bacterial taxa and the Biolog substrate utilization pattern. The relative abundance of the identified taxa showed some striking differences based on the blooming status of Hydrilla and to a lesser extent on site variation. The relative abundance of Actinobacteria, Bacteriodetes, and Verrucomicrobia was generally higher during Hydrilla blooms, while Deltaproteobacteria was generally higher during non-blooming stages of Hydrilla. The detected genera also varied based on the blooming stages of the plant. Based on the findings, it appears that Hydrilla alters the phylogenetic composition and structure of the bacterial community during the blooming stage.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hydrilla verticillata, commonly called Hydrilla, is one of the most invasive and studied aquatic vascular plant species and is widely distributed in various countries (Langeland, 1996). Since its accidental introduction into Florida approximately 25 years ago, Hydrilla has caused major problems in water bodies in the state due to the intensity and vastness of its growth (Blackburn et al., 1969). In 1997, the plant was discovered in Wakulla Springs (Wakulla, FL, USA), classified as a natural treasure, and one of the longest and deepest known submerged freshwater springs and cave systems in the world (Cao et al., 1999). Historically, the Wakulla Spring, which is the centerpiece of a 6,000 acre preserve, has been known for its pristine water which has attracted divers and other visitors from many countries. However, for more than a decade, water quality in the spring has been on a decline due to increased nitrate levels, algal blooms, and the invasive aquatic plant species. Hydrilla has caused blockages in the waterways of the springs resulting in curtailment of boating tours and other recreational activities causing economic losses.
Hydrilla invasion also has been reported to have consequential ecological impacts, such as disruption of the dominant native species (Blackburn et al., 1969; Langeland, 1996) and the potential to alter the microbial community structure of invaded areas (Posey, 1988; Coles et al., 1999; Han et al., 2007). These changes can potentially alter ecosystem processes such as productivity, decomposition, nutrient cycling, and hydrology (D’Antonio & Vitousek, 1992; Vitousek et al., 1997; Hahn, 2003).
Although many studies (Kourtev et al., 2003; Costa et al., 2005; Garbeva et al., 2005) have focused on the impact of plant species on soil microbial communities, few have evaluated the microbial community of the water column in aquatic systems invaded by plant species such as Hydrilla. In one study (He et al., 2012), the researchers evaluated epiphytic microbial community on Hydrilla and eelgrass using the clone library procedure which provides limited data compared to current and widely used high throughput sequencing methods. Their study also did not evaluate bacteria communities in the water column. Thus, our objective in this study was to assess the bacterial community structure and function using the culture independent 454 pyrosequencing method in two Hydrilla invaded springs, the Wakulla Springs and the adjoining Sally Ward Springs, during the blooming and non-blooming stages of Hydrilla.
Materials and methods
Study sites
The sampling sites for this investigation were Wakulla and Sally Ward Springs, both infested with Hydrilla, and two underground aquifers which are the major sources of spring water into Wakulla Springs, Fig. 1. Three sub-sites were selected within the Wakulla Spring/River area: W1 (N30°14.101′, W084°18.160′), W2 (N30°14.155′, W084°17.747′), and W3 (N30°14.162′, W084°17.777′). Due to the smaller size of Sally Ward Spring, two sub-sites were selected: SW1 (N30°14.481′, W084°18.643′) and SW2 (N30°14.502′, W084°18.661′). The samples from the underwater aquifers were obtained from two channels (B and C) which join at a central point and feed water directly into the Wakulla Spring head.
Sample collection and processing
To analyze the water column bacterial communities, water samples were collected during the blooming (April 2012) and non-blooming (December 2011) stages of Hydrilla. Approximately, 2 l of water sample was collected in duplicate at each sampling site using a Vertical Point Water Sampler (Aquatic Research Instrument, Hope, ID) and transferred into sterilized bottles. Water parameters such as conductivity, salinity, and temperature were measured (YSI 30®46-YSI, St. Petersburg, FL) along with pH (AR15-Fisher Scientific, Pittsburg, PA) prior to sample collection (Table 1). In addition, water samples for nutrient analysis were collected and transported on ice to the City of Tallahassee Water Quality Laboratory (Tallahassee, FL, USA) for analyses using various US Environmental Protection Agency methods. Chlorophyll-a by EPA spectrophotometric method SMI0200H (AWWA, 1998), total nitrogen by EPA ammonia, unionized method 350.1, total phosphorus as P by EPA calorimetric, automated block digester AAII method 365.4 and dissolved organic carbon by EPA high temperature combustion method SM5310B (EPA, 1986).
In the laboratory, each of the duplicate samples was combined yielding a 4 l composite sample from each site. Three hundred milliliters from each site was set aside for determining bacterial cell abundance and community-level physiological profile (CLPP). In addition, 3 l of each water sample were filtered through 0.2-μm Nuclepore track-etched membrane filters (Whatman laboratory, NJ, catalog # 111106) to capture bacteria. The filters were stored at −20°C for subsequent DNA extraction and analysis.
Bacterial abundance
Bacterial cell abundance was determined by DAPI (4′,6-diamino-2-phenylindole) (Molecular Probes, Inc., Eugene, OR) staining (Pernthaler et al., 1998). Briefly, formalin-fixed water subsamples of 3–5 ml in triplicate were stained in the dark for 30 min with DAPI (final concentration 10 µg/ml). The samples were then filtered through pre-wetted black polycarbonate membrane filters (diameter, 25 mm, pore size, 0.22 mm, GE Osmonics Inc., Minnetonka, MN). Bacterial counts were done at 1,000× magnification using a Zeiss Axioskop epifluorescence microscope (Carl Zeiss Inc., Thornwood, NY). One-way ANOVA was used to test for significant differences between cell counts at each site during the non-blooming and blooming cycles.
CLPPs analysis
CLPPs were assessed by the Biolog EcoPlate™ system (BIOLOG Inc., Hayward, CA) as previously reported (Insam, 1997). 120 µl of each water sample was inoculated into each well of the Biolog EcoPlate™ and incubated at 25°C for 192 h. Substrate utilization (absorbance at 590 nm) was measured at zero time-point and every 24 h (Yao et al., 2000) thereafter for 168 h using a BIOTEK UQuant Microplate Spectrophotometer (BIO-TEK Instruments Inc., Winooski, USA). The measurements of individual substrates were corrected for background absorbance by subtracting the absorbance of the control (no carbon source) samples. Well optical density values that were negative or <0.06 were manually set to zero (Classen et al., 2006). The 31 carbon substrates contained in each ecoplate were designated into six respective guilds; carbohydrates, carboxylic acids, amino acids, amides/amines, polymers, and esters. The utilization pattern of each guild by the bacterial communities was determined by averaging the OD of the substrates within the respective guilds after 192 h. The CLPP AWCD Biolog data were analyzed using principal component analysis (PCA) to identify sample groupings based on a simplification of the dataset into its most variable axes. PCA was done using the PAST program (Hammer et al., 2001) and the results visualized by constructing ordination plots of the first two axes, which represent the greatest proportion of variation.
DNA extraction and pyrosequencing
Community DNA was extracted from cells captured from water samples onto 0.2-μm Nuclepore track-etched membrane filters using the MoBio PowerWater® DNA Isolation Kit (MoBio, Carlsbad, CA) following manufacturer’s instructions. DNA yield and purity were measured using a ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). In order to check quality of the extracted DNA, universal eubacterial primers 27F and 1492R (Rohwer et al., 2001) were used to amplify 16S rRNA gene fragments. The polymerase chain reaction (PCR) mixture was prepared using Taq mastermix (Denville Scientific, Metuchen, NJ) with 2 μl template DNA, PCR grade sterile water, and 5 pmol/μl of each primer in a total reaction of 25 μl. Sterile water was used as negative controls in each batch of PCR reactions. The PCR reactions were performed in a Biorad thermocycler (Hercules, CA) under the following conditions: initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, annealing at 55°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 10 min. Product size and purity were confirmed by electrophoresis in 1% agarose gels using ethidium bromide. Once DNA quality was confirmed to meet the standards of the pyrosequencing facility, the DNA samples were shipped to MR DNA (www.mrdnalab.com) in Shallow Water, TX for pyrosequencing analysis following protocols originally described by Dowd et al. (2008). Briefly, the 16S universal Eubacterial primers 27Fmod (5′-AGRGTTTGATCMTGGCTCAG-3′) and 519Rmodbio (5′-GTNTTACNGCGGCKGCTG-3′) were used to amplify the V1–V3 region of the 16S rRNA gene using 30 cycles of PCR. Amplification was done using HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA) under the following conditions: 94°C for 3 min, followed by 28 cycles of 94°C for 30 s; 53°C for 40 s and 72°C for 1 min after which a final elongation step at 72°C for 5 min was performed. Following PCR, all amplicon products from different samples were mixed in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, MA, USA). Samples were sequenced utilizing Roche 454 FLX titanium instruments and reagents following manufacturer’s guidelines.
Analysis of pyrosequencing-derived data
Pyrosequencing data was analyzed using the Mothur pipeline by Schloss et al. (2011). Briefly, adapters, barcodes, primers, and sequences <200 bp and containing ambiguous ‘N’ were removed from the raw sequences. Cleaned sequences were aligned using the Silva reference database via the Mothur alignment command. Aligned sequences were denoised using the “pre.cluster” command to merge sequence counts that were within 2 bp of a more abundant sequence (Huse et al., 2010; Roeselers et al., 2011). Chimeric sequences were removed using UCHIME algorithm (http://drive5.com/uchime) (Edgar et al., 2011). In addition, sequences that were suspected to be mitochondria–chloroplast–Archaea–Eukarya–unknown were removed from further analysis. DNA distance matrices were calculated and used to define the number of operational taxonomic units (OTUs) at sequence divergences of 3% (Wu et al., 2010). All OTUs were classified using the 16S rRNA training set nine database from RDP (Wang et al., 2007).
Diversity richness was calculated using rarefaction curves, ACE (Chao & Lee, 1992) and Chao 1 (Chao, 1984) non-parametric richness estimators, while community diversity was determined using Invsimpson and Shannon diversity indices. Dendrogram using the thetaYC tree was generated to determine similarities among treatments and the unweighted unifrac test of significance was used to determine whether the clustering within the tree was statistically significant. To identify relationships between the identified bacterial taxa and the Biolog substrate utilization pattern, Spearman’s rank correlations were determined using SPSS 19.0 software (SPSS Inc.).
Nucleotide sequence accession number
The 16S rRNA gene sequences derived from pyrosequencing have been deposited in the NCBI Sequence Read Archive under accession number SAMN01993801.
Results
Bacteria abundances
Bacterial abundances based on DAPI counts are shown in Figs. S1 and S2. Although the bacterial cell counts from Wakulla Springs were not significantly different between the blooming and non-blooming (one-way ANOVA, P > 0.05) periods, the counts were typically higher during non-blooming. This was in contrast to the Sally Ward Springs where cell counts during the non-blooming stages were significantly higher (P < 0.05) than the blooming stages (Fig. S1). At all of the springs, there were significant differences (P < 0.001) in cell counts among the sub-sites on each of the sample collection dates (Fig. S2). Cell counts from the Wakulla Spring sites were typically higher than those from the Sally Ward sites, with the exception of site 1 which had similar counts to those of the Wakulla sites during the non-blooming stages. The underground aquifers had significantly lower bacterial cell counts than either Wakulla or Sally Ward Springs in all cases.
CLPPs analysis
The results of CLPP revealed that the microbes in the Hydrilla invaded sites utilized less substrates during blooming periods than during non-blooming periods of the plant (Fig. S3). Substrate utilization by the bacteria in the Wakulla and Sally Ward Springs was much higher than that in the aquifers. To determine the substrates most preferred by the microbial community in the invaded systems, the sum of all the substrate utilization values for each of the respective guilds from all samples was used to calculate a mean value for each of the substrate guilds. The results showed esters, followed by carbohydrates, as the most utilized substrates by the microbial community during the blooming stages and polymers, followed by carbohydrates, were most utilized during the non-blooming stage. The other substrates were utilized at similar rates during blooming and non-blooming periods (Fig. 2).
Community-level physiological profile (CLPP) of microbial communities in the water column of springs during blooming and non-blooming stages of Hydrilla. The CLPP is based on microbial community utilization patterns of carbon substrates in the Biolog EcoPlate™ system. a Non-blooming, b blooming, W Wakulla Spring, SW Sally Ward Spring, B and C underground conduits
The PCA (Fig. 3a) and cluster analysis (Fig. 3b) were performed on the combined substrate utilization values from all sub-sites at both sampling dates. The bacterial community physiological profiles from the invaded springs did not show any specific clustering pattern based on the presence or absence of the plant or among the different sub-sites. The analyses did show that the bacterial community in samples collected from the non-invaded aquifer sites clustered together and separately from those from the invaded springs. Principal components 1 and 2 explained 99.7% of the total variance in the data of CLPP, with 98.8% being explained by PC1 (Fig. 3a). This indicates that the differences along PC1 were more influential than along PC2. Shannon diversity indices were calculated on average the AWCD after 192 h (Table S1) and ranged from 2.50 to 2.76 while evenness ranged from 0.72 to 0.93. Neither the Shannon index nor the evenness differed greatly among the sub-sites.
Principal component analysis of community-level physiological profiles of microbial communities. N non-blooming stages of Hydrilla, B blooming stages of Hydrilla, B and C underground aquifers (non-infested), SW Sally Ward (infested), W Wakulla Spring (infested). Shown are: a PCA analyses and b dendrogram (Euclidean distance) analysis of microbial responses to Biolog substrates
Diversity and composition of bacteria communities in freshwater springs invaded with Hydrilla
Although rarefaction curves did not reach saturation at a 3% genetic distance indicating that the full extent of taxonomic diversity was not surveyed, the Good’s coverage estimate ranged from 67 to 96% (Table 2, Figs. S4 and S5) showing that the majority of the bacterial community was covered. The Shannon diversity indices did not vary greatly between the blooming and non-blooming stages of Hydrilla being 6.03 and 6.29, respectively. The average Shannon index for the non-invaded aquifers was 5.80 in April samples and 5.73 in December samples. The Chao 1 richness estimate for the samples collected from the invaded systems was generally higher in December than in April and averaged 8,154 and 7,518, respectively. Contrastingly, for the samples collected from the non-invaded aquifers, the Chao 1 richness estimate was generally higher in April (4,452) compared to December (3,349). The ACE richness estimator showed similar findings to the Chao 1 estimator (Table 2, Fig. S4).
The pyrosequencing-based analysis of the V1–V3 region of the 16S rRNA genes resulted in 256,352 sequences. Following filtering, denoising, chimera removal and clustering, a total of 209,420 high quality sequences with read lengths ≥200 bp (average read length was 298 bp) across all 14 water samples were used to classify the bacterial community structure in Wakulla and Sally Ward Springs and the aquifers.
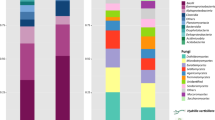
The 209,420 sequences classified below domain level were affiliated to 24 bacterial phyla including five Proteobacterial classes (Fig. 4, Fig. S6). The Proteobacteria represented approximately 49.8% of the total sequences with Betaproteobacteria (19.5%), Alphaproteobacteria (7.6%), and Gammaproteobacteria (6.1%) representing the most abundant. The most abundant bacterial taxa identified in the invaded systems at the blooming stage of Hydrilla were Betaproteobacteria (12.1%), Actinobacteria (11.8%), unclassified Proteobacteria (14.8%), and Alphaproteobacteria (4.6%). In contrast, the most abundant taxa during non-blooming were Betaproteobacteria (7.4%), Alphaproteobacteria (3.1%), and Gammaproteobacteria (2.7%). The most abundant taxa at the non-invaded sites were Actinobacteria (2.7%), Betaproteobacteria (2.6%), Gammaproteobacteria (1.3%), and Alphaproteobacteria (1.2%).
Relative abundance of dominant bacterial phyla and proteobacterial classes in water samples from freshwater springs infested with Hydrilla. OTUs were classified using the 16S rRNA training set nine database from RDP at a 3% genetic distance. N non-blooming stages of Hydrilla, B blooming stages of Hydrilla, B and C underground aquifers (non-infested), SW Sally Ward (infested), W Wakulla Spring (infested)
At the genus level, 37,296 OTUs from all water bodies were classified from the more than 209,000 sequences based at a 3% genetic distance. To assess the most abundant OTUs, only those with abundances >100 sequences across all samples were classified (Table S3). Of these, the most dominant (>2% of the genera classified) were Opitutus (8.4% from the phylum Verrucomicrobia), Limnohabitans (7.1% from the class Betaproteobacteria), Luteibacter (7.0% from the class Gammaproteobacteria), Acinetobacter (6.5% from the class Gammaproteobacteria), Flavobacterium (6.1% from the class Flavobacteria), and Rheinheimera (4.4% from the class Gammaproteobacteria).
In order to identify relationships between the relative abundances of bacterial taxa and the Biolog substrate utilization pattern, Spearman’s rank correlation analysis was done. Among all the substrates; amino acids (r 2 = 0.619, P = 0.05), polymers (r 2 = 0.582, P = 0.05), and esters (r 2 = 0.780, P = 0.01) were positively correlated to Betaproteobacteria, while carbohydrates (r 2 = 0.596, P = 0.05) were only correlated to Bacteriodetes. Polymers (r 2 = 0.569, P = 0.05) were positively correlated to Deltaproteobacteria (Table S2).
Similarity among microbial communities
In order to describe the phyologenetic similarities among the microbial communities in samples based on sampling dates and sites, a dendrogram was generated using the thetaYC calculators in the Mothur platform (Fig. 5). The unweighted unifrac commands were used to determine whether the clustering within the tree was statistically significant or not. There was significant differences in clustering of the communities based on the blooming stages of Hydrilla (unweighted unifrac significance = 0.03) and also the sites (non-invaded versus invaded) (unweighted unifrac significance = 0.009). Bacteria in water samples collected during the non-blooming stages clustered more closely together while those from the blooming stages were more closely related. In addition, bacteria in the water samples from the Hydrilla invaded sites were more closely related to each other than to the samples from the non-invaded aquifers.
ThetaYC neighbor joining tree based on a 3% genetic distance from samples collected from freshwater springs during blooming (B, red) and non-blooming (N, blue) stages of Hydrilla. The thetaYC tree was constructed using the Mothur platform. C and B (yellow) underground aquifers (non-infested), W Wakulla Spring (infested), SW Sally Ward Spring (infested)
Discussion
Invasive plant species have become a serious problem in various ecosystems. Once established they can alter or displace the native communities including the structure of microbial communities and their role in nutrient cycling (Angeloni et al., 2006). Results of previous studies (Kourtev et al., 2003; Windham & Ehrenfeld, 2003) have shown that invading plants can alter microbially driven nitrogen cycling processes in terrestrial ecosystems (Kourtev et al., 2003) and freshwater wetlands (Windham & Ehrenfeld, 2003). Such plants have also been shown to possibly alter the microbial community structure in terrestrial soils (Duda et al., 2003; Angeloni et al., 2006) and brackish marsh sediments (Ravit et al., 2003; Angeloni et al., 2006).
In this study, the microbial community activities and structure in Hydrilla invaded and non-invaded springs were investigated, using both community physiological (CLPPs) and 16S rRNA gene profiles. For the invaded sites, this approach permitted detection of shifts in the activities and structure of water column microbial communities between the blooming and non-blooming cycles of Hydrilla.
Among the differences observed between the invaded and non-invaded sites were in bacterial abundances. The invaded sites, Wakulla and Sally Ward Springs, were found to have significantly higher abundances of bacteria (P < 0.05) (even when the plant was dormant) than the non-invaded underground aquifers sites (Fig. S2). These differences could be due to the different nature of the systems, although the measurements of water parameters for the springs and aquifers were similar (Table 1). However, the underground aquifers are in a protected environment devoid of light and lacking exposure to the many elements in nature.
Sole-carbon-source utilization tests have been previously used to monitor differences in metabolic profiles in microbial communities in soil and other natural environments (Garland & Mills, 1991; Campbell et al., 1997; Gomez et al., 2004). Using the Biolog EcoPlate™ system, we observed that the bacterial community substrate utilization was generally higher in the Hydrilla invaded sites, than in the non-invaded aquifers. These differences are likely related to variations in bacterial numbers and community structure in the different water bodies which may be influenced, at least in part, by the presence or absence of Hydrilla. In the Hydrilla invaded sites, substrate utilization by the bacteria was higher during the non-blooming stage of the plant compared to the blooming stages. The bacterial cell counts were also higher during the non-blooming stages of the plant in the invaded sites. Several explanations may account for this. For example, during the blooming period, the Hydrilla may be adding some nutrient exudates that the bacteria have a preference for over the Biolog substrates. It has been reported that Hydrilla contains compounds such as loliolide, thymidine, octadecanedioic acid (Xiao et al., 2007), and caffeic acid ester (Hipskind et al., 1992). Whether the bacteria can utilize these carbon compounds as energy sources is not fully understood; however, this could be an important area for future investigation to evaluate whether these compounds can be utilized by bacteria or have a detrimental effect on them. When the Hydrilla and their products are reduced during non-blooming periods, the bacteria may preferentially turn again to metabolism of the Biolog substrates. Another possible scenario is that the bacteria community in the spring that are able to more effectively utilize the Biolog substrates may be reduced by the Hydrilla, thus the lower substrate utilization pattern.
Pyrosequencing analysis of the V1–V3 regions of the 16S rRNA genes from water samples collected from all sites yielded 209,420 clean sequences and 37,296 OTUs. The invaded sites had 15,134 and 15,030 OTUs during non-blooming and blooming stages of Hydrilla, respectively. The non-invaded sites had 3,218 and 3,914 OTUs during the same periods of sampling. The 37,296 OTUs were classified into 24 known bacterial taxa including some rare taxa (Fig. 4, Figs. S6 and S7). All taxa are known freshwater bacteria (Zwart et al., 2002; Newton et al., 2011). Although the dominant phyla and Proteobacteria classes were found at all the sub-sites in the invaded springs at both blooming and non-blooming stages of the plant, there were some variations in their relative proportions (Fig. S7).
One difference was the greater abundance of the phylum Actinobacteria during the blooming stages of Hydrilla compared to the non-blooming stages. However, this pattern was also seen in the non-invaded sites and, therefore, may be linked to other factors such as seasonal influences. Further study that includes greater sampling frequency may better clarify this issue. Actinobacteria are traditionally associated with terrestrial ecosystems (Rheims et al., 1999), but are also found in freshwater (Nielsen et al., 2006) and marine environments (Du et al., 2006). They are primarily saprophytic and reported to contribute significantly to the recycling of complex biopolymers such as lignocellulose, hemicellulose, pectin, and chitin in sediments (Goodfellow & Williams, 1983; Stackebrandt et al., 1997; Du et al., 2006). However, their ecological functions in limnetic environments are not fully understood (Allgaier & Grossart, 2006).
The relative abundance of the phyla Bacteriodetes and Verrucomicrobia was higher during the blooming stage of Hydrilla in comparison to the non-blooming period. For the non-invaded systems, their abundance was similar in proportion on both sample collection dates. These collective results show a positive relationship between these bacterial phyla and Hydrilla blooms. Both Bacteriodetes and Verrucomicrobia are common and important phyla in freshwater ecosystems (Zwart et al., 2002; Newton et al., 2011).
There were also some minor variations in the bacterial phyla between the different sampling sites. For example, a greater abundance of Betaproteobacteria and Gammaproteobacteria was detected in Wakulla Spring irrespective of the presence or absence of a Hydrilla bloom than in Sally Ward Spring and the aquifers (Fig. S7).
At the genus level, there were some similarities and differences among the detected genera based on site to site variations and blooming versus non-blooming stages of the plant. The genus Opitutus of the phylum Verrucomicrobia and the family Opitutaceae was the most abundant among all the sites combined and represented 8.4% of the total identified genera. Their abundance did not appear to be altered by the absence or presence of Hydrilla since they were distributed relatively equally among both the invaded and non-invaded sites. The ecological functions of this genus are not fully understood, however, according to Newton et al. (2011), some members of this phylum seem to be associated with high-nutrient environments or algal blooms.
The greatest differences in bacterial community structure between Hydrilla blooming and non-blooming stages in the invaded springs were observed at the genus level (Table S3). The genera Propionibacterium, Sediminibacterium, Flavobacterium, Brevundimonas, Polynucleobacter, Limnohabitans, Acinetobacter, and Methylophagia were present in higher abundance during the Hydrilla blooming period than non-blooming. Other genera (Hydrogenophaga and Massilia) were more abundant during the non-blooming stages of Hydrilla. Differences were also observed in the relative abundances of bacterial genera between the invaded vs. non-invaded systems. While the genus Opitutus was found among all the sites, they were the most dominant group among the aquifers, while Luteibacter was most abundant in the invaded systems (Table S3).
The phylogenetic results appear to support the hypothesis that Hydrilla invasion does impact the bacterial community structure of invaded systems. The impact of plants on environmental microbial communities has been reported by other investigators as well. Wieland et al. (2001) reported that the type of plant species (clover, bean, or alfalfa) selects for the microbial communities on their roots. Plant species are known to produce specific exudates (Söderberg et al., 2002), different leaching metabolites and proportions of chemicals (Fiehn et al., 2000) which can impact the bacterial community structure. These findings support previous results in our laboratory in which the epiphytic bacterial community structure of Hydrilla was found to be distinct from that on the native plant species eelgrass (Gordon-Bradley et al., 2013).
The collective results from this study show that Hydrilla invasion in an aquatic system can alter the bacterial community structure and metabolic activities in the water column possibly causing shifts in nutrient cycling and other bacterial environmental services. The pyrosequencing of the 16S rRNA gene provided a comprehensive overview of the bacterial community structure of the systems invaded with Hydrilla and the Biolog system showed differences in profiles of the metabolic activities of bacteria communities during Hydrilla blooms vs. when Hydrilla was dormant. Pyrosequencing results revealed that the predominant bacteria during blooming were Actinobacteria, Bacteriodetes, and Verrucomicrobia, whereas Deltaproteobacteria was predominant during the non-blooming stages of the plant. Further studies on the long term impact of Hydrilla on aquatic bacterial communities in multiple systems are warranted to further understand the environmental and ecological disturbances associated with this invasive plant.
References
Allgaier, M. & H.-P. Grossart, 2006. Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Applied and Environmental Microbiology 72: 3489–3497.
Angeloni, N. L., et al., 2006. Effects of an invasive cattail species (Typha × glauca) on sediment nitrogen and microbial community composition in a freshwater wetland. FEMS Microbiology Letters 263: 86–92.
AWWA, APHA, 1998. Standard Methods for the Examination of Water and Wastewater, 20th ed. APHA, Washington, DC.
Blackburn, R. D., et al., 1969. Identification and distribution of certain similar-appearing submersed aquatic weeds in Florida. Hyacinth Control Journal 8: 17–21.
Campbell, C. D., S. J. Grayston & D. J. Hirst, 1997. Use of rhizosphere carbon sources in sole carbon source tests to discriminate soil microbial communities. Journal of Microbiological Methods 30: 33–41.
Cao, H., J. B. Cowart & J. K. Osmond, 1999. Water sources of Wakulla Springs, Wakulla County, Florida: physical and uranium isotopic evidences. Southeastern Geology 39: 51–55.
Chao, A., 1984. Nonparametric estimation of the number of classes in a population. Scandinavian Journal of Statistics 11: 265–270.
Chao, A. & S. M. Lee, 1992. Estimating the number of classes via sample coverage. Journal of the American Statistical Association 87: 210–217.
Classen, A. T., et al., 2006. Community-level physiological profiles of bacteria and fungi: plate type and incubation temperature influences on contrasting soils. FEMS Microbiology Ecology 44: 319–328.
Coles, S. L., et al., 1999. Historical and recent introductions of non-indigenous marine species into Pearl Harbor, Oahu, Hawaiian Islands. Marine Biology 135: 147–158.
Costa, R., et al., 2005. Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiology Ecology 56: 236–249.
D’Antonio, C. M. & P. M. Vitousek, 1992. Biological invasions by exotic grasses, the grass/fire cycle, and global change. Annual Review of Ecology and Systematics 23: 63–87.
Dowd, S. E., et al., 2008. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 3: e3326.
Du, H., et al., 2006. Diversity and distribution of pigmented heterotrophic bacteria in marine environments. FEMS Microbiology Ecology 57: 92–105.
Duda, J. J., et al., 2003. Differences in native soil ecology associated with invasion of the exotic annual chenopod, Halogeton glomeratus. Biology and Fertility of Soils 38: 72–77.
Edgar, R. C., et al., 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200.
EPA, 1986. Test Methods for Evaluating Solid Waste, Physical/Chemical Methods. US EPA, Cincinnati, OH, USA.
Fiehn, O., et al., 2000. Metabolite profiling for plant functional genomics. Nature Biotechnology 18: 1157–1161.
Garbeva, P., et al., 2005. Effect of above-ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3. Environmental Microbiology 8: 233–246.
Garland, J. L. & A. L. Mills, 1991. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Applied and Environmental Microbiology 57: 2351–2359.
Gomez, E., J. Garland & M. Conti, 2004. Reproducibility in the response of soil bacterial community-level physiological profiles from a land use intensification gradient. Applied Soil Ecology 26: 21–30.
Goodfellow, M. & S. T. Williams, 1983. Ecology of actinomycetes. Annual Reviews in Microbiology 37: 189–216.
Gordon-Bradley, N., D. Lymperopoulo & H. N. Williams, 2013. Differences in bacterial community structure on Hydrilla vertlicillata and Vallisnera americana in a Freshwater Spring. Microbes and Environment 29: 67–73.
Hahn, D. R., 2003. Alteration of microbial community composition and changes in decomposition associated with an invasive intertidal macrophyte. Biological Invasions 5: 45–51.
Hammer, Ø., D. A. T. Harper & P. D. Ryan, 2001. PAST-Palaeontological Statistics. www.uv.es/~pardomv/pe/2001_1/past/pastprog/past.pdf. Accessed on 25.7.2009.
Han, X., et al., 2007. Effects of vegetation type on soil microbial community structure and catabolic diversity assessed by polyphasic methods in North China. Journal of Environmental Sciences 19: 1228–1234.
He, D., L. Ren & Q. Wu, 2012. Epiphytic bacterial communities on two common submerged macrophytes in Taihu Lake: diversity and host-specificity. Chinese Journal of Oceanology and Limnology 30: 237–247.
Hipskind, J., et al., 1992. A fungitoxic phenolic compound in Hydrilla verticillata. Biological Control 2: 51–58.
Huse, S. M., et al., 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environmental Microbiology 12: 1889–1898.
Insam, H., 1997. A new set of substrates proposed for community characterization in environmental samples. In Insam, H. & A. Rangger (eds), Microbial Communities, Functional versus Structural Approaches. Springer, Berlin: 259–260.
Kourtev, P. S., J. G. Ehrenfeld & M. Häggblom, 2003. Experimental analysis of the effect of exotic and native plant species on the structure and function of soil microbial communities. Soil Biology and Biochemistry 35: 895–905.
Langeland, K. A., 1996. Hydrilla verticillata (LF) Royle (Hydrocharitaceae), “The perfect aquatic weed”. Castanea 61: 293–304.
Newton, R. J., et al., 2011. A guide to the natural history of freshwater lake bacteria. Microbiology and Molecular Biology Reviews 75: 14–49.
Nielsen, J. L., et al., 2006. Detection of activity among uncultured actinobacteria in a drinking water reservoir. FEMS Microbiology Ecology 55: 432–438.
Pernthaler, J., et al., 1998. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Applied and Environmental Microbiology 64: 4299–4306.
Posey, M. H., 1988. Community changes associated with the spread of an introduced seagrass, Zostera japonica. Ecology 69: 974–983.
Ravit, B., J. G. Ehrenfeld & M. M. Haggblom, 2003. A comparison of sediment microbial communities associated with Phragmites australis and Spartina alterniflora in two brackish wetlands of New Jersey. Estuaries 26: 465–474.
Rheims, H., et al., 1999. Molecular monitoring of an uncultured group of the class Actinobacteria in two terrestrial environments. Journal of Microbiological Methods 36: 65–75.
Roeselers, G., et al., 2011. Evidence for a core gut microbiota in the zebrafish. The ISME Journal 5: 1595–1608.
Rohwer, F., et al., 2001. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20: 85–91.
Schloss, P. D., D. Gevers & S. L. Westcott, 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PloS One 6: e27310.
Söderberg, K. H., P. A. Olsson & E. Bååth, 2002. Structure and activity of the bacterial community in the rhizosphere of different plant species and the effect of arbuscular mycorrhizal colonisation. FEMS Microbiology Ecology 40: 223–231.
Stackebrandt, E., F. A. Rainey & N. L. Ward-Rainey, 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. International Journal of Systematic Bacteriology 47: 479–491.
Vitousek, P. M., et al., 1997. Human domination of Earth’s ecosystems. Science 277: 494–499.
Wang, Q., et al., 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 73: 5261–5267.
Wieland, G., R. Neumann & H. Backhaus, 2001. Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Applied and Environmental Microbiology 67: 5849–5854.
Windham, L. & J. G. Ehrenfeld, 2003. Net impact of a plant invasion on nitrogen-cycling processes within a brackish tidal marsh. Ecological Applications 13: 883–896.
Wu, J. Y., et al., 2010. Effects of polymerase, template dilution and cycle number on PCR based 16 S rRNA diversity analysis using the deep sequencing method. BMC Microbiology 10: 255.
Xiao, Y., et al., 2007. Chemical composition of Hydrilla verticillata (L. f.) Royle in Taihu Lake. Chinese Journal of Chemistry 25: 661–665.
Yao, H., et al., 2000. Microbial biomass and community structure in a sequence of soils with increasing fertility and changing land use. Microbial Ecology 40: 223–237.
Zwart, G., et al., 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquatic Microbial Ecology 28: 141–155.
Acknowledgments
This project was supported by funding from the National Science Foundation HBCU-RISE Program (NSF Grant No. HRD-0932137). We are grateful to Scott Savery from the Wakulla Spring State Park and Scott Barrett Dyer from the Florida Department of Environmental Protection for their help with sample collection from the Wakulla Spring State Park.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Stefano Amalfitano.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gordon-Bradley, N., Li, N. & Williams, H.N. Bacterial community structure in freshwater springs infested with the invasive plant species Hydrilla verticillata . Hydrobiologia 742, 221–232 (2015). https://doi.org/10.1007/s10750-014-1988-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-014-1988-0