Abstract
Sediment environments harbor a repertoire of microorganisms that contribute to animal health and the microecosystem in aquaculture ecosystems, but their community diversity and the potential factors that control it remain unclear. Here, we applied 16S rRNA gene amplicon sequencing to investigate bacterial diversity and assembly mechanisms in the sediments of shrimp cultural ponds at the mesoscale. Our results showed that sediment bacterial communities contained 10,333 operational taxonomic units (OTUs) but had only 34 core OTUs and that the relative abundances of these core OTUs were significantly correlated with the physicochemical properties of the sediments. Proteobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Acidobacteria, Firmicutes, Actinobacteria, Ignavibacteriae, Spirochaetae and Planctomycetes were the ten most abundant bacterial phyla. Notably, some opportunistic pathogens (e.g. Vibrio and Photobacterium) and potential functional microbes (e.g. Nitrospira, Nitrosomonas, Desulfobulbus and Desulfuromusa) were widely distributed in shrimp cultural pond sediments. More importantly, we found that there was a significant negative but weak distance-decay relationship among bacterial communities in shrimp culture pond sediments at the mesoscale, and that the spatial turnover of these bacterial communities appeared to be largely driven by stochastic processes. Additionally, environmental factors, such as pH and total nitrogen, also played important roles in influencing the sediment bacterial structure. Our findings enhance our understanding of microbial ecology in aquatic ecosystems and facilitate sediment microbiota management in aquaculture.
Key points
• Core bacterial taxa in cultural pond sediments contributed to the shrimp health and element cycling.
• There was a significant negative distance-decay relationship among bacterial communities in shrimp culture pond sediments at the mesoscale, and its spatial turnover appeared to be largely driven by stochastic processes.
• Environmental factors (e.g. pH and total nitrogen) played important roles in influencing bacterial structure in shrimp cultural pond sediments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Natural environments teem with microorganisms that play crucial roles in ecosystem functions, such as biogeochemical cycling and maintaining macroorganismal health (Newton et al. 2011; Bárcena et al. 2019; Trivedi et al. 2020). Revealing the mechanisms that generate and maintain microbial diversity in the environment is critical to determine the links between community stability and ecosystem functions. Generally, there are two types of ecological processes (deterministic and stochastic processes) that determine microbial assemblages, but the relative contributions of these two processes have been debated (Zhou and Ning 2017). Some studies of microbial communities have explored diverse aquatic ecosystems in recent years and have shown that their microbial assemblies are often regulated by different ecological processes. One of the studies addressing this issue showed that stochastic processes dominated the microbial community variations in water environment in a lotic river (Chen et al. 2019). Additionally, strong stochastic assembly of microbial communities has been observed in other aquatic ecosystems, such as lakes (Zhao et al. 2017), reservoir (Liu et al. 2015) and wastewater (Ofiteru et al. 2010). In contrast, another study demonstrated that deterministic processes (e.g. environmental selection) played a key role in shaping the water microbial community in subtropical bays (Mo et al. 2018). Similarly, deterministic processes play a critical role in shaping the microbial variation in water and sediment environments in lakes (Wang et al. 2013; Yan et al. 2017). These studies have deepened our understanding of microbial diversity and assembly mechanisms in aquatic ecosystems.
Aquaculture is the third largest source of animal proteins worldwide, accounting for 17% of global protein consumption (FAO 2018), but the frequent occurrence of diseases in aquaculture settings restricts its development. Importantly, the microorganisms in water and sediment environments in aquaculture ecosystems are critical to cultured animal health, water quality and element cycling (Moriarty 1997; Zhou et al. 2009). Previous studies have found that several microorganisms can accelerate the decomposition of residual feed and faeces to improve water quality (Tan et al. 2016); others can convert toxic substances into low-toxicity or nontoxic forms to protect the cultured species (Su et al. 2016). Moreover, the characteristics of the microbial communities in farming environments are closely related to the occurrence of aquatic animal diseases (Li et al. 2017; Xiong et al. 2017). Given the importance of environmental microorganisms, revealing their microbial diversity and control factors can facilitate microbiota management to promote productivity and sustainability in aquaculture. A few studies have focused on microbial diversity in aquaculture ecosystems and have found that environmental factors and geographical distance both contribute to microbial assemblages in water and sediment environments (Li et al. 2017; Hou et al. 2017; Huang et al. 2018). Such information has provided the basis for our understanding of the microbial diversity in aquaculture ecosystems and the possible factors controlling it.
Sediment microorganisms are an important component of aquaculture ecosystems; they exhibit very high microbial diversity, which is often higher than that in water and in animal intestines (Al-Harbi and Uddin 2005; Del’Duca et al. 2015; Dabadé et al. 2016; Zhang et al. 2020). Among the microorganisms in aquaculture sediments, some are opportunistic pathogens (e.g. Vibrio) and functional microbes (e.g. Nitrospira and Desulfobulbus) (Hou et al. 2018; Fan et al. 2019) that are related to the health of the cultured animals and to element cycling. In particular, the microbial taxa in cultured animal intestines and in sediment environments in aquaculture ecosystems are mostly the same (Huang et al. 2018; Hou et al. 2018), and this close relationship is critical for host health (Schryver and Vadstein 2014). Taken together, these studies suggest that sediment microorganisms make important contributions to microecosystem and cultured animal health in aquaculture ecosystems, but their community diversity and assembly mechanism remain elusive.
In this study, we analysed the 16S rRNA gene sequences of 180 samples taken from 60 culture ponds across six regional sites in China and aimed to reveal the bacterial diversity and assembly mechanisms of the sediment environment in shrimp cultural ponds at the mesoscale (tens to thousands of kilometres). To expound on these questions, we explored (i) the sediment bacterial features and their biogeographic patterns; (ii) the relative importance of deterministic versus stochastic factors in regulating sediment bacterial assembly; and (iii) the effects of environmental drivers on the sediment bacterial community. Our findings can provide a reference for the study of microbial ecology in aquatic ecosystems and support the management of sediment microbiota in aquaculture.
Materials and methods
Sample collection and physicochemical analysis
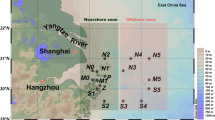
A total of 180 sediment samples were collected from 60 Litopenaeus vannamei cultural ponds (three parallel samples from each pond) in Qingzhou (QZ, n = 9 ponds * 3 samples), Zhuhai (ZH, n = 9*3), Zhangpu (ZP, n = 9*3), Maoming (MM, n = 15*3), Qionghai (QH, n = 8*3) and Tianjin (TJ, n = 10*3) cities in China (19.20°–39.29°N, 108.56°–117.93° E). The site locations were recorded by GPS (global positioning system; Garmin Vista HCx, Lenexa, USA). The geographical distances between sample sites ranged from 0.31 to 2063.35 km. All the sampled ponds had similar sizes (~ 3300 m2), water depths (~ 12 m), cultural period (about 60 days) and stocking densities (~ 200,000 shrimp larvae/pond) (Supplemental Table S1). For all ponds, no probiotics, disinfectants or other products were added within five days before sampling. During sampling, 5.0 g sediment was placed into a 15-mL sterile centrifugal tube for each sample. All samples were stored at – 80 °C until DNA extraction. The sediment pH was measured on-site using a pH meter (ZD-05, Beijing Century Euron Co, Ltd, Beijing, China). The concentrations of total nitrogen (TN) and total phosphorus (TP) of samples were measured using an auto-discrete analyser (Model CleverChem 380, DeChem-Tech.GmbH, Hamburg, Germany). The concentrations of total carbon (TC) and total organic carbon (TOC) of samples were measured using a total organic carbon analyser (Aurora 1030W, OI Analytical, College Station, TX, USA). The sediment physicochemical properties are described in Supplemental Table S2.
DNA extraction, PCR amplification, and sequencing
Sediment genomic DNA was extracted by the PowerSoil DNA Isolation Kit (Mobio, Carlsbad, San Diego, CA, USA). The 338F and 806R universal primer pair was used to amplify the V3-V4 regions of 16S rRNA gene PCR products from the sediment samples, which were equally combined and then sequenced using the Illumina MiSeq platform (Illumina, San Diego, USA) by Majorbio Bio-Pharm Technology Co Ltd. (Shanghai, China). Paired-end sequences were merged with FLASH (Magoč and Salzberg 2011), and merged sequences were processed with Quantitative Insights Into the Microbial Ecology pipeline (QIIME version 1.9.0) (Caporaso et al. 2010). In brief, sequences with ambiguous bases or truncated at any site of more than three consecutive bases receiving a Phred quality score Q < 20 were deleted. Chimeric sequences were discarded using the UCHIME algorithm (Edgar et al. 2011). Bacterial phylotypes were identified using UCLUST (Edgar 2010) and classified into operational taxonomic units (OTUs) at a 97% cut-off. The most abundant sequence from each OTU was selected as a representative and then taxonomically assigned against the Silva SSU database 128 using the RDP Classifier algorithm (http://rdpcmemsuedu/). This process allowed each identified OTU to have a close relative. The core OTUs in the shrimp cultural pond sediments were present at all regional sites, and in ≥ 80% of all samples with average relative abundances ≥ 0.2% (Wu et al. 2019).
Estimating stochasticity of sediment bacterial assembly
To determine which ecological processes shape sediment bacterial assembly, community assembly stochasticity was assessed using a null-model-based index. The stochasticity ratio has been described previously (Zhou et al. 2014). Since null-model algorithms usually require a high number of replicates, more than 8 pond samples were taken from each regional site. We calculated the stochasticity ratio using the following metrics: when using the Bray-Curtis (abundance- weighted and unweighted) model, the stochasticity ratio was calculated based on typical null-model algorithms for bacterial taxonomic metrics; when using weighted and unweighted UniFrac, the stochasticity ratio was calculated based on the typical null-model algorithms for phylogenetic metrics (Kembel 2009; Chase et al. 2011; Stegen et al. 2013). The samples within each site were considered to share the same regional species pool in the null-model algorithms.
Statistical analysis
Non-metric multidimensional scaling (NMDS) and analysis of similarity (ANOSIM) were performed to evaluate the overall differences in sediment bacterial communities using the Bray-Curtis distance. Spearman’s correlation analyses were employed to test the correlations between the relative abundances of core OTUs and sediment properties. BioEnv and canonical correspondence analyses were used to identify the sediment properties and geographic distances that were the most important to bacterial structure, which were used to construct an environmental factor matrix for variation partitioning analysis (VPA) in R 3.3.2 with the vegan package (R Core Team 2015). The same subset of sediment properties was incorporated into a structural equation model (SEM) to evaluate their effect on the bacterial diversity and structure of shrimp cultural pond sediments (Bagozzi and Yi 2012). Meanwhile, distance decay curves were generated using the geographic distance and bacterial similarity among all sediment samples based on the beta nearest taxon index (Bray and Curtis 1957).
Results
Bacterial diversity and core OTUs of shrimp cultural pond sediments
Sequencing of 16S rRNA gene amplicons generated 8,730,698 quality sequences from all sediment samples, and there was an average of 48,503 ± 5894 sequences per sample (Supplemental Table S3). After subsampling to 20,002 sequences per sample, 3,600,360 sequences (10,333 bacterial OTUs with an average of 2141 ± 311 OTUs) were retained (Supplemental Table S3). Then, the α-diversity indices at the OTU level were calculated using rarefaction curves at OTU level at a sequencing depth of 20,002 with 1000 iterations, where the Shannon and Chao1 indices had stabilized (Supplemental Figure S1). Then, the α-diversity indices were calculated based on the OTUs of each library. Good’s coverage index was 0.96 ± 0.01 for each sample, indicating that each library of 16S rRNA sequences represented majority of the bacterial communities in all sediment samples. The Shannon and Chao1 indices were 6.37 ± 0.28 and 2,914 ± 472, respectively (Supplemental Table S3).
Despite the high diversity, only approximately 0.03% of the OTUs (34 out of 10,333 OTUs) constituted core OTUs that accounted for 13.45% of all the bacterial sequences in the sediment (Fig. 1a). These core OTUs were classified as Bacteroidetes (11 OTUs, relative abundance of 4.52%), Proteobacteria (8 OTUs, 3.59%), Acidobacteria (3 OTUs, 0.91%), Cyanobacteria (3 OTUs, 1.21%), Ignavibacteriae (3 OTUs, 1.14%), Chlorobi (2 OTUs, 0.64%), Planctomycetes (1 OTU, 0.54%), Chloroflexi (1 OTU, 022%), Tenericutes (1 OTU, 0.45%) and WS6 (1 OTU, 0.24%) (Fig. 1b and Supplemental Table S4). Some potential functional microbes, such as Robiginitalea OTU20074, Desulfomicrobium OTU6931 and Hydrogenophaga OTU4062, were core OTUs in the sediments (Supplemental Table S4). Spearman’s correlation analyses further revealed that the relative abundances of core OTUs were significantly (P < 0.05 in all cases) correlated with at least one of the sediment physicochemical properties (Fig. 1c and Supplemental Table S4). For example, the relative abundance of Robiginitalea OTU20074 was significantly positively correlated with the ratio of carbon to nitrogen (C/N) (r = 0.40, P < 0.01) and carbon to phosphorus (C/P) (r = 0.44, P < 0.01), but negatively associated with TN (r = − 0.40, P < 0.01), TP (r = − 0.16, P < 0.05) and the ratio of nitrogen to phosphorus (N/P) (r = − 0.16, P < 0.05).
The abundance and composition of core bacterial OTUs in shrimp cultural pond sediments. a Percentage and relative abundance of core versus other OTUs. b Taxonomic composition of core OTUs at the phylum level. c Correlations between the relative abundances of core OTUs and sediment properties. The colour gradient on the right represents the Spearman’s rank correlation coefficients, with more positive values (dark blue) indicating stronger positive correlations and more negative values (dark red) indicating stronger negative correlations. The sizes of the coloured boxes indicate the correlation strengths. The asterisks denote the significance levels of the Spearman’s rank correlation coefficients. ** P < 0.01, * P < 0.05
Taxonomic composition of bacterial communities in shrimp cultural pond sediments
All sediment sequences were affiliated with at least 61 or 1189 bacteria divisions at the phylum or genus level (Supplemental Table S3). Proteobacteria (relative abundance was 31.43%), Bacteroidetes (25.38%), Chloroflexi (8.32%), Cyanobacteria (3.03%), Acidobacteria (2.95%), Actinobacteria (2.88%), Firmicutes (2.72%), Ignavibacteriae (2.53%), Spirochaetae (2.49%) and Planctomycetes (2.49%) were the ten most abundant phyla in the shrimp cultural pond sediments (Fig. 2), while Spirochaeta (1.40%), Robiginitalea (1.05%), Acholeplasma (0.94%), Ignavibacterium (0.91%), Candidatus Thiobios (0.75%), Nitrospira (0.72%), Ilumatobacter (0.72%), Algoriphagus (0.68%), Caldithrix (0.67%) and Desulfobulbus (0.64%) were the ten most abundant genera (Supplemental Table S5). Notably, some opportunistic pathogens, such as Vibrio and Photobacterium, and potentially functional microbes, such as Nitrospira, Nitrosomonas, Desulfobulbus and Desulfuromusa, were detected in most sediment samples (Supplemental Table S5).
Geographic patterns of the sediment bacterial community
The NMDS analysis showed that there were differences in sediment bacterial community structure among regional sites in different (Fig. 3a). This result was further corroborated by the ANOSIM, which revealed that the bacterial community structure significantly (r = 0.8115, P < 0.001) differed between any two of the compared regional sites (Fig. 3a). To further explore the geographic distribution of the sediment bacterial community, we generated the distance decay curves using the geographic distance and bacterial similarity and found that the sediment bacterial community decreased in slightly similarity but significantly (P < 0.001) with increasing geographic distance (Fig. 3b). These results suggest that there was an obvious but weak distance-decay pattern in the bacterial community in shrimp cultural pond sediments at the mesoscale.
Ecological processes contribute to sediment bacterial assembly
We used a null model-based approach to calculate the stochastic ratios of sediment bacterial assembly with taxonomic and phylogenetic metrics. The average stochastic ratios based on these metrics were all higher than 80% (Fig. 4a), suggesting that stochastic factors can play more important roles than deterministic factors in influencing the bacterial assembly in shrimp cultural pond sediments at the mesoscale. To discern the relative importance of the various factors contributing to sediment community variation, we performed VPA based on phylogenetic diversity metrics. Eight sediment physicochemical properties that provided the highest Pearson correlation with sediment communities were selected by the BioEnv procedure. The VPA results showed that the sediment properties explained 16.73% (P < 0.001), and geographic distance alone explained 6.27% (P < 0.001) bacterial variations with 4.30% interaction effect detected, respectively, leaving 72.70% of the variation unexplained (Fig. 4b). These results support those inferred from the stochastic ratio analysis.
Ecological processes contributing to the bacterial assembly of shrimp cultural pond sediments. a Ecological stochasticity of sediment bacterial assembly estimated by the stochasticity ratio, which was calculated for each pair of samples based on taxonomic diversity (Bray-Curtis, BC) and phylogenetic diversity (Phyl, UniFrac) weighted by abundance (Wt) or not (Uw) Boxes and whiskers indicate quartiles. b VPA of the effects of geographic distance and environmental factors on sediment bacterial structure. *** P < 0.01
Environmental drivers of sediment bacterial community
Because the sediment physicochemical properties were determined to be important, we performed a more in-depth analysis using an SEM and revealed the effects of environmental drivers on the sediment bacterial community (Fig. 5). This model was assessed using the Goodness-of-Fit (GoF) statistic, and the GoF value was 0.158. The results showed that sediment properties significantly affected the bacterial diversity (r = 0.17, P < 0.01) and structure (r = − 0.73, P < 0.01). Among specific sediment physicochemical variables, pH (r = 0.33, P < 00.1), TN (r = − 0.33, P < 0.05) and C/P (r = − 0.24, P < 0.05) significantly affected the sediment bacterial diversity, while TN (r = 0.48, P < 0.01) and N/P (r = 0.18, P < 0.05) significantly affected the bacterial structure. Thus, sediment properties were important factors in shaping the bacterial assembly in shrimp cultural pond sediments.
SEM showing the environmental drivers of the bacterial community in shrimp culture pond sediments. Directed graph of SEM, and GoF statistic value was 0.158. Each box represents an observed variable or latent variable. Path coefficients are reflected in the width of the arrow, with blue solid and red dashed arrows indicating significantly positive and negative effects. ** P < 0.01, * P < 0.05
Discussion
Sediment microorganisms contribute to animal health and microecosystem in aquaculture systems, and their community diversity and assembly mechanisms are fundamental but rarely investigated topics. In this study, we aimed mainly to investigate the bacterial diversity in shrimp cultural pond sediments and to explore the relative contributions of ecological processes on its community assembly. Extensive studies have focused on sediment microorganisms in aquaculture ecosystems and found that they can vary widely at the OTU or genus level but tend to be dominated by Proteobacteria, Bacteroidetes and Chloroflexi (Al-Harbi and Uddin 2005; Del’Duca et al. 2015; Hou et al. 2018; Zhao et al. 2020a). These studies revealed a common conclusion about sediment bacterial compositions in aquaculture, but the core taxa of aquaculture systems remain unknown. Our study found that despite its high diversity, sediment communities in shrimp cultural ponds had only small core OTUs. These sediment core taxa might serve as a ‘most wanted’ list for future experimental efforts to understand their genetic, biochemical, physiological and ecological traits. For example, Robiginitalea OTU20074 and Desulfomicrobium OTU6931 are known to enhance sulfur cycling, and Robiginitalea is thought to have the ability to produce hydrogen sulfide (Liesack and Finster 1994; Zhang et al. 2018). A Hydrogenophaga OTU (OTU4062) was also identified as core taxa, reflecting its importance for denitrification (Liu et al. 2019). In fact, hydrogen sulfide, ammonia nitrogen and nitrite nitrogen are the commonest toxicants and are harmful to shrimp health and growth (Lin and Chen 2001; Suo et al. 2017). Thus, these core taxa in culture pond sediments might play vital roles in water quality and shrimp health.
Additionally, our results showed that some opportunistic pathogens, such as Vibrio species, which become enriched in shrimp or fish intestines and are generally associated with host disease outbreaks (Li et al. 2017; Huang et al. 2020), were also present in sediment. Indeed, the activities of benthonic animals (e.g. shrimp, shellfish, crab) are closely related to sediments; specifically, L. vannamei has the characteristics of feeding from sediment and ingesting particulate matter into their intestine. Presumably, opportunistic pathogens in sediment environments may enter benthic animal intestines through the feeding behaviors of these animals thus increasing the risk of host disease.
We further found that there was significant negative but weak distance-decay relationship (DDR) among bacterial communities in shrimp cultural pond sediments at the mesoscale. The DDR is a fundamental pattern in ecology (Morlon et al. 2011), and over the past few decades, microbial biogeographic patterns have been reported in a variety of environments (Fierer et al. 2009; Griffiths et al. 2011; Delgado-Baquerizo et al. 2018). Consistent with those in other human-managed ecosystems, such as agricultural soil (e.g. wheat soil and alkaline agricultural soil at the mesoscale; Zhao et al. 2020b) and engineering systems (e.g. activated sludge at the regional, continental and global scales) (Wu et al. 2019), the spatial turnover rates of bacterial communities also presented obvious but weak DDRs. Thus, ‘scale-dependency’ is an important consideration, different approaches for microbial community management in human-managed systems should be applied at different scales. Moreover, we found that the bacterial spatial turnover rates in human-managed systems were lower than those observed in some natural environments, especially in nonflowing ecosystems (e.g. soil and sediment) (Fierer and Jackson 2006; Martiny et al. 2011).
Generally, the spatial turnover of the microbial community is driven both by deterministic and stochastic processes (Hanson et al. 2012), but there is an ongoing debate regarding the relative contributions of these two processes. Thus, our study raises an important question: what is the relative importance of deterministic versus stochastic processes in regulating the bacterial assembly in shrimp cultural pond sediments at the mesoscale? To address this concern, we calculated the stochastic ratios of sediment bacterial assembly and found that its spatial turnover appeared to be largely driven by stochastic processes. Consistent with that in other human-managed ecosystems, the bacterial spatial turnover of activated sludge in wastewater treatment plants was also largely driven by stochastic processes (Wu et al. 2019). Like in natural ecosystems, such as river-bay systems and cool temperate forests, stochasticity factors play a relatively important role in controlling bacterial variation in sediments (Lu et al. 2021) and soils (Bahram et al. 2016). In contrast, deterministic processes were the primary factors controlling the bacterial community assemblages of wheat field soil (Shi et al. 2018), as well as of natural deserts and soils (Caruso et al. 2011; Schmidt et al. 2014). Thus, the microbial community assemblies of different environments are often regulated by different ecological processes.
We also found that sediment properties and geographic distance are both important factors in shaping sediment bacterial assembly in shrimp cultural ponds. Our VPA results showed that sediment bacterial variables explained only 27.3% by sediment properties and geographic distance, leaving 72.7% of the variation unexplained. Similarly, the bacterial variables of lake sediment (Xiong et al. 2012), wheat field soil (Shi et al. 2018) and forest soil (Zhou et al. 2008) explained only 22.5%, 44.8% and 41.1% by environmental factors and spatial parameters, respectively. Similar trends were observed in the above studies and in our study, with environmental variables explaining < 50% of the bacterial variation; the reasons for the unexplained variation also include stochastic processes (e.g. dispersal, ecological drift) (Zhou et al. 2014). Thus, the VPA results also showed that bacterial assembly in shrimp cultural pond sediments appears highly likely to be driven by stochastic processes. Moreover, sediment properties played important roles in influencing the sediment bacterial structure and core taxa. These findings could be important for developing operating strategies to maintain the sediment microbial diversity that promotes stable system performance. Perhaps one could alternate the sediment properties (e.g. pH, TN, C/P, N/P, which are associated with feed inputs and other operations in shrimp culture) to manipulate the sediment microbiota and select for microorganisms that have the desired functions in shrimp cultural ponds.
In conclusion, we systematically investigated the bacterial diversity and assembly mechanism of shrimp cultural pond sediments within an ecological framework (Fig. 6). Our findings highlight how little we know about the microbial ecology of the sediment environment in aquaculture ecosystems, which are some of the most common human-managed systems. Despite its high bacterial diversity, the important sediment core community consists of only a small number of taxa whose relative abundances were strongly linked to sediment physicochemical properties. Importantly, some opportunistic pathogens and potential functional microbes were widely distributed in the sediment. Furthermore, there was a significant negative but weak DDR among the sediment bacterial communities, and their spatial turnover appeared to be largely driven by stochastic processes, although sediment physicochemical properties also played important roles. These results greatly enhance our mechanistic understanding of aquatic microbial ecology and facilitate sediment microbiota management for healthy aquaculture.
Data availability
All the raw reads in the current study have been deposited at the Sequence Read Archive of the NCBI under the accession number PRJNA545396.
References
Al-Harbi AH, Uddin N (2005) Bacterial diversity of tilapia (Oreochromis niloticus) cultured in brackish water in Saudi Arabia. Aquaculture 250(3-4):566–572. https://doi.org/10.1016/j.aquaculture.2005.01.026
Bagozzi RP, Yi Y (2012) Specification, evaluation, and interpretation of structural equation models. J ACAD Market Sci 40(1):8–34. https://doi.org/10.1007/s11747-011-0278-x
Bahram M, Kohout P, Anslan S, Harend H, Abarenkov K, Tedersoo L (2016) Stochastic distribution of small soil eukaryotes resulting from high dispersal and drift in a local environment. ISME J 10(4):885–896. https://doi.org/10.1038/ismej.2015.164
Bárcena C, Valdés-Mas R, Mayoral P, Garabaya C, Durand S, Rodriguez F, Fernández-García MT, Salazar N, Nogacka AM, Garatachea N, Bossut N, Aprahamian F, Lucia A, Kroemer G, Freije JMP, Quirós PM, López-Otín C (2019) Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat Med 25(8):1234–1242. https://doi.org/10.1038/s41591-019-0504-5
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr 27(4):326–349. https://doi.org/10.2307/1942268
Caporaso JG, Kuczynski J, Stombaugh J (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Caruso T, Chan YK, Lacap DC, Lau MCY, Mckay CP, Pointing SB (2011) Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J 5(9):1406–1413. https://doi.org/10.1038/ismej.2011.21
Chase JM, Kraft NJ, Smith KG, Vellend M, Inouye BD (2011) Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2(2):24–art24. https://doi.org/10.1890/ES10-00117.1
Chen W, Ren K, Isabwe A, Chen H, Liu M, Yang J (2019) Stochastic processes shape microeukaryotic community assembly in a subtropical river across wet and dry seasons. Microbiome 7(1):138. https://doi.org/10.1186/s40168-019-0749-8
Dabadé DS, Wolkers-Rooijackers JC, Azokpota P, Hounhouigan DJ, Zwietering MH, Nout MJ, den Besten HMW (2016) Bacterial concentration and diversity in fresh tropical shrimps (Penaeus notialis) and the surrounding brackish waters and sediment. Int J Food Microbiol 218:96–104. https://doi.org/10.1016/j.ijfoodmicro.2015.11.013
Del’Duca A, Cesar DE, Abreu PC (2015) Bacteiral community of pond’s water, sediment and in the guts of tilapia (Oreochromis niloticus) juveniles characterized by fluorescent in situ hybridization technique. Aquac Res 46(3):707–715. https://doi.org/10.1111/are.12218
Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-Gonzalez A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N (2018) A global atlas of the dominant bacteria found in soil. Science 359(6373):320–325. https://doi.org/10.1126/science.aap9516
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Fan L, Wang Z, Chen M, Qu Y, Li J, Zhou A, Xie S, Zeng F, Zou J (2019) Microbiota comparison of pacific white shrimp intestine and sediment at freshwater and marine cultured environment. Sci Total Environ 657(20):1194–1204. https://doi.org/10.1016/j.scitotenv.2018.12.069
FAO (2018) Fishery and Aquaculture Statistics. Food and Agriculture Organization of the United Nations, Rome
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103(3):626–631. https://doi.org/10.1073/pnas.0507535103
Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC (2009) Global patterns in below ground communities. Ecol Lett 12(11):1238–1249. https://doi.org/10.1111/j.1461-0248.2009.01360.x
Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS (2011) The bacterial biogeography of British soils. Environ Microbiol 13(6):1642–1654. https://doi.org/10.1111/j.1462-2920.2011.02480.x
Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH (2012) Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10(7):497–506. https://doi.org/10.1038/nrmicro2795
Hou D, Huang Z, Zeng S, Liu J, Wei D, Deng X, Weng S, He Z, He J (2017) Environmental factors shape water microbial community structure and function in shrimp cultural enclosure ecosystems. Front Microbiol 8:2359. https://doi.org/10.3389/fmicb.2017.02359
Hou D, Huang Z, Zeng S, Liu J, Weng S, He J (2018) Comparative analysis of the bacterial community compositions of the shrimp intestine, surrounding water and sediment. J Appl Microbiol 125(3):792–799. https://doi.org/10.1111/jam.13919
Huang F, Pan L, Song M, Tian C, Gao S (2018) Microbiota assemblages of water, sediment, and intestine and their associations with environmental factors and shrimp physiological health. Appl Microbiol Biotechnol 102(19):8585–8598. https://doi.org/10.1007/s00253-018-9229-5
Huang Z, Zeng S, Xiong J, Hou D, Zhou R, Xing C, Wei D, Deng X, Yu Y, Wang H, Deng Z, Weng S, Kriengkrai S, Ning D, Zhou J, He J (2020) Microecological Koch’s postulates reveal that intestinal microbiota dysbiosis contributes to shrimp white feces syndrome. Microbiome 8(1):32. https://doi.org/10.1186/s40168-020-00802-3
Kembel SW (2009) Disentangling niche and neutral influences on community assembly: assessing the performance of community phylogenetic structure tests. Ecol Lett 12(9):949–960. https://doi.org/10.1111/j.1461-0248.2009.01354.x
Li T, Li H, Gatesoupe FJ, She R, Lin Q, Yan X, Li J, Li X (2017) Bacterial signatures of “Red-Operculum” disease in the gut of Crucian Carp (Carassius auratus). Microb Ecol 74(10):510–521. https://doi.org/10.1007/s00248-017-0967-1
Liesack W, Finster K (1994) Phylogenetic analysis of five strains of gram-negative, obligately anaerobic, sulfur-reducing bacteria and description of Desulfirornusa gen nov, including Desulfuromusa kysingii sp nov, Desulfurornusa bakii sp nov, and Desulfuromusa succinoxidans sp nov. Aust Vet J 44(4):753–758. https://doi.org/10.1099/00207713-44-4-753
Lin Y, Chen J (2001) Acute toxicity of ammonia on Litopenaeus vannamei Boone juveniles at different salinity levels. J Exp Mar Biol Ecol 259(1):109–119. https://doi.org/10.1016/S0022-0981(01)00227-1
Liu L, Yang J, Yu Z, Wilkinson DM (2015) The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J 9(9):2068–2077. https://doi.org/10.1038/ismej.2015.29
Liu X, Xu J, Huang J, Huang M, Wang T, Bao S, Tang W (2019) Bacteria-supported iron scraps for the removal of nitrate from low carbon-to-nitrogen ratio wastewater. RSC Adv 9(6):3285–3293. https://doi.org/10.1039/c8ra09091b
Lu Z, Liu Z, Zhang C, Wei Q, Zhang S, Li M (2021) Spatial and seasonal variations of sediment bacterial communities in a river-bay system in South China. Appl Microbiol Biotechnol 105(7):1979–1989. https://doi.org/10.1007/s00253-021-11142-z
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Martiny JB, Eisen JA, Penn K, Allison SD, Horner-Devine MC (2011) Drivers of bacterial beta-diversity depend on spatial scale. Proc Natl Acad Sci U S A 108(9):7850–7854. https://doi.org/10.1073/pnas.1016308108
Mo Y, Zhang W, Yang J, Lin Y, Yu Z, Li S (2018) Biogeographic patterns of abundant and rare bacterioplankton in three subtropical bays resulting from selective and neutral processes. ISME J 12(9):2198–2210. https://doi.org/10.1038/s41396-018-0153-6
Moriarty DJW (1997) The role of microorganisms in aquaculture ponds. Aquaculture 151(1):333–349. https://doi.org/10.1016/S0044-8486(96)01487-1
Morlon H, Schwilk DW, Bryant JA, Marquet PA, Rebelo AG, Tauss C, Bohannan BJM, Green JL (2011) Spatial patterns of phylogenetic diversity. Ecol Lett 14(2):141–149. https://doi.org/10.1111/j.1461-0248.2010.01563.x
Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsso S (2011) A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75(1):14–49. https://doi.org/10.1128/MMBR.00028-10
Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA, Sloan WT (2010) Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci U S A 107(35):15345–15350. https://doi.org/10.1073/pnas.1000604107
R Core Team (2015) A language and environment for statistical computing Vienna. The R Foundation for Statistical Computing, Austria. isbn:3-900051-07-0 http://www.R-project.org/
Schmidt SK, Nemergut DR, Darcy JL, Lynch R (2014) Do bacterial and fungal communities assemble differently during primary succession? Mol Ecol 23(2):254–258. https://doi.org/10.1111/mec.12589
Schryver PD, Vadstein O (2014) Ecological theory as a foundation to control pathogenic invasion in aquaculture. ISME J 8(12):2360–2368. https://doi.org/10.1038/ismej.2014.84
Shi Y, Li Y, Xiang X, Sun R, Yang T, He D, Zhang K, Ni Y, Zhu Y, Adams JM, Chu H (2018) Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome 6(1):27. https://doi.org/10.1186/s40168-018-0409-4
Stegen JC, Lin X, Fredrickson JK, Chen X, Kennedy DW, Murray CJ, Rockhold ML, Konopka A (2013) Quantifying community assembly processes and identifying features that impose them. ISME J 7(11):2069–2079. https://doi.org/10.1038/ismej.2013.93
Su Z, Li Y, Pan L, Xue F (2016) An investigation on the immunoassays of an ammonia nitrogen-degrading bacterial strain in aquatic water. Aquaculture 450(1):17–22. https://doi.org/10.1016/j.aquaculture.2015.07.001
Suo Y, Li E, Li T, Jia Y, Qin J, Gu Z, Chen L (2017) Response of gut health and microbiota to sulfide exposure in Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immun 63:87–96. https://doi.org/10.1016/j.fsi.2017.02.008
Tan LT, Chan KG, Lee LH, Goh BH (2016) Streptomyces bacteria as potential probiotics in aquaculture. Front Microbiol 7:79. https://doi.org/10.3389/fmicb.2016.00079
Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK (2020) Plant-microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18(11):607–621. https://doi.org/10.1038/s41579-020-0412-1
Wang J, Shen J, Wu Y, Tu C, Soininen J, Stegen JC, He J, Liu X, Zhang L, Zhang E (2013) Phylogenetic beta diversity in bacterial assemblages across ecosystems: deterministic versus stochastic processes. ISME J 7(7):1310–1321. https://doi.org/10.1038/ismej.2013.30
Wu L, Ning D, Zhang B, Li Y, Zhang P, Shan X, Ling F, Xiao N, Zhang Y, Vierheilig J, Wells GF, Yang Y, Deng Y, Tu Q, Wang A, Global Water Microbiome Consortium, Zhang T, He Z, Keller J, Nielsen PH, PJJ A, Criddle CS, Wagner M, Tiedje JM, He Q, Curtis TP, Stahl DA, Alvarez-Cohen L, Rittmann BE, Wen X, Zhou J (2019) Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat Microbiol 4(7):1183–1195. https://doi.org/10.1038/s41564-019-0426-5
Xiong J, Liu Y, Lin X, Zhang H, Zeng J, Hou J, Yang Y, Yao T, Knight R, Chu H (2012) Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ Microbiol 14(9):2457–2466. https://doi.org/10.1111/j.1462-2920.2012.02799.x
Xiong J, Zhu J, Dai W, Dong C, Qiu Q, Li C (2017) Integrating gut microbiota immaturity and disease-discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ Microbiol 19(4):1490–1501. https://doi.org/10.1111/1462-2920.13701
Yan Q, Stegen JC, Yu Y, Deng Y, Li X, Wu S, Dai L, Zhang X, Li J, Wang C, Ni J, Li X, Hu H, Xiao F, Feng W, Ning D, He Z, Van Nostrand JD, Wu L, Zhou J (2017) Nearly a decade-long repeatable seasonal diversity patterns of bacterioplankton communities in the eutrophic Lake Donghu (Wuhan, China). Mol Ecol 26(14):3839–3850. https://doi.org/10.1111/mec.14151
Zhang J, Han J, Chen G, Du Z (2018) Robiginitalea sediminis sp nov, isolated from a sea cucumber culture pond. Antonie Van Leeuwenhoek 111(6):905–911. https://doi.org/10.1007/s10482-017-0989-1
Zhang X, Li X, Lua J, Qiu Q, Chen J, Xiong J (2020) Quantifying the importance of external and internal sources to the gut microbiota in juvenile and adult shrimp. Aquaculture 531(30):735910. https://doi.org/10.1016/j.aquaculture.2020.735910
Zhao D, Xu H, Zeng J, Cao X, Huang R, Shen F, Yu Z (2017) Community composition and assembly processes of the freeliving and particle-attached bacteria in Taihu Lake. FEMS Microbiol Ecol 93(6):fix062. https://doi.org/10.1093/femsec/fix062
Zhao Z, Jiang J, Pan Y, Dong Y, Chen Z, Zhang G, Gao S, Sun H, Guan X, Wang B, Xiao Y, Zhou Z (2020a) Temporal dynamics of bacterial communities in the water and sediments of sea cucumber (Apostichopus japonicus) culture ponds. Aquaculture 528(15):735498. https://doi.org/10.1016/j.aquaculture.2020.735498
Zhao S, Liu J, Banerjee S, Zhou N, Zhao Z, Zhang K, Hu M, Tian C (2020b) Biogeographical distribution of bacterial communities in saline agricultural soil. Geoderma 361(1):114095. https://doi.org/10.1016/j.geoderma.2019.114095
Zhou J, Ning D (2017) Stochastic community assembly: does it matter in microbial ecology. Microbiol Mol Biol R 81(4):e00002–e00017. https://doi.org/10.1128/mmbr.00002-17
Zhou J, Kang S, Schadt CW, Garten CT (2008) Spatial scaling of functional gene diversity across various microbial taxa. Proc Natl Acad Sci USA 105(22):7768–7773. https://doi.org/10.1073/pnas.0709016105
Zhou Q, Li K, Bo L (2009) Role and functions of beneficial microorganisms in sustainable aquaculture. Bioresour Technol 100(16):3780–3789. https://doi.org/10.1016/j.biortech.2008.12.037
Zhou J, Deng Y, Zhang P, Xue K, Liang YT, Van Nostrand JD, Yang Y, He Z, Wu L, Stahl DA, Hazen TC, Tiedje JM, Arkini AP (2014) Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci U S A 111(9):836–845. https://doi.org/10.1073/pnas.1324044111
Funding
This work was financially supported by National Natural Science Foundation of China (31902392); China Agriculture Research System of MOF and MARA; the China Agriculture Research System (CARS-48); China- ASEAN Maritime Cooperation Fund, China-ASEAN Center for Joint Research and Promotion of Marine Aquaculture Technology; Natural Science Foundation of Guangdong Province, China (2019A1515011557); Fundamental Research Funds for the Central Universities (19lgpy103); and Key Research and Development Projects in Guangdong Province (2020B0202010009).
Author information
Authors and Affiliations
Contributions
DW Hou, RJ Zhou, SZ Zeng, DD Wei, XS Deng and CG Xing performed the sample collections and experiments. DW Hou and RJ Zhou analysed the data. DW Hou, ZJ Huang SP Weng and JG He contributed the conception of this work and wrote the paper. ZJ Huang was primarily responsible for the final content.
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain any studies with human participants performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 626 kb)
Rights and permissions
About this article
Cite this article
Hou, D., Zhou, R., Zeng, S. et al. Stochastic processes shape the bacterial community assembly in shrimp cultural pond sediments. Appl Microbiol Biotechnol 105, 5013–5022 (2021). https://doi.org/10.1007/s00253-021-11378-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11378-9