Abstract
The ubiquitous occurrence of microorganisms gives rise to continuous public concerns regarding their pathogenicity and threats to human environment, as well as potential engineering benefits in biotechnology. The development and wide application of environmental biotechnology, for example in bioenergy production, wastewater treatment, bioremediation, and drinking water disinfection, have been bringing us with both environmental and economic benefits. Strikingly, extensive applications of microscopic and molecular techniques since 1990s have allowed engineers to peep into the microbiology in “black box” of engineered microbial communities in biotechnological processes, providing guidelines for process design and optimization. Recently, revolutionary advances in DNA sequencing technologies and rapidly decreasing costs are altering conventional ways of microbiology and ecology research, as it launches an era of next-generation sequencing (NGS). The principal research burdens are now transforming from traditional labor-intensive wet-lab experiments to dealing with analysis of huge and informative NGS data, which is computationally expensive and bioinformatically challenging. This study discusses state-of-the-art bioinformatics and statistical analyses of 16S ribosomal RNA (rRNA) gene high-throughput sequencing (HTS) data from prevalent NGS platforms to promote its applications in exploring microbial diversity of functional and pathogenic microorganisms, as well as their interactions in biotechnological processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rapidly decreasing cost and continuous development (e.g., data throughput, base quality) of NGS in the past decade have largely promoted the widespread application of 16S ribosomal RNA (rRNA) gene HTS in exploring microbial diversity across multiple disciplines from medicine, biology, evolution, to ecology and environmental sciences. As opposed to traditional labor-intensive molecular methods used for microbial fingerprinting, such as fluorescent in situ hybridization (FISH), denaturing gradient gel electrophoresis (DGGE), PCR cloning, and terminal restriction fragment length polymorphism (T-RFLP), the most challenging as well as time-consuming experimentation of a NGS-based study usually not lies in the preliminary molecular experiments (e.g., DNA extraction, PCR, cloning) or sequencing (e.g., can be done by commercial companies), but rather in the subsequent tedious and rigid bioinformatics and statistical analyses, which are solid foundations to an effective way of revealing the underlying microbiological mechanisms behind efficient operations of biotechnological processes.
To meet the needs of processing diverse types of large 16S rRNA gene HTS data (that may differ in read length, sequencing depth, base quality, and error profiles) from various NGS platforms, a number of standardized or customized bioinformatics pipelines, platforms, and tools are proposed or developed, and kept regularly updated (Caporaso et al. 2010a, b; Schloss et al. 2009; Cole et al. 2009). The usage of these bioinformatics tools greatly improves our understandings of microbiology and microbial ecology in various natural habitats and engineered biological systems (e.g., bioenergy and wastewater treatment bioreactors) (Ibarbalz et al. 2013; Cai et al. 2013; Ju and Zhang 2014a, b; Peng et al. 2014; Guo and Zhang 2012; Xia et al. 2012; Ye et al. 2011).
This review paper is to technically guide the application of 16S rRNA gene HTS in investigating microbial diversity and interactions in biotechnological processes so as to reveal novel alternative microbial resources for use in environmental biotechnology (e.g., bioenergy production, pollution control), as well as to explore unaware microbial interactions that could be utilized to guide the manipulation of microbial community functioning. The state-of-the-art bioinformatics analysis tools or pipelines, including the most popular web-based or locally installed bioinformatics tools/platforms, reference databases, and the widely used ecological matrices, are sequentially introduced to promote their more extensive application. Moreover, popular statistical approaches and procedures for network analysis are elucidated by emphasizing on their application in mining huge NGS data from a large number of samples. Above all, most protocols introduced in this study are not only applicable to 16S rRNA gene-based analysis, but also to future NGS applications that target other marker genes.
Bioinformatics analysis
Bioinformatics analysis of HTS data includes three core aspects: (1) pretreatment of raw sequence data, (2) microbial diversity analysis, and (3) advanced data analysis and visualization. Several local or web-based software packages have been developed to trim, filter, analyze, and visualize large amplicon sequence data from NGS, including QIIME, mothur, RDP, VAMPS, and MEGAN (Table 1). The locally installed, command line-based QIIME and mothur are currently the two most popular platforms with complete pipelines to guide users through standardized or customized data analysis procedures. For users with limited computational resources, these tools are also available at Galaxy (Goecks et al. 2010) and CloVR wrappers (Angiuoli et al. 2011) via web-based cloud computing.
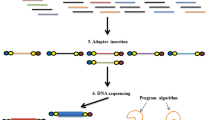
Regardless of which software or platform to employ, complete processing procedures of 16S rRNA gene amplicon data typically incorporate three parts (Fig. 1), that is, (1) data pretreatment, including de-multiplexing of barcoded sequences, quality filtering, denoise, chimera checking, and data normalization; (2) construction of operational taxonomic unit (OTU) table, including picking OTUs, picking representative sequences, aligning representative sequences, taxonomic assignment of representative sequences, and building of a phylogenetic tree of the OTUs; and (3) advanced data analysis and visualization, including the alpha- and beta-diversity analyses, clustering and coordinates analysis, and data visualization (e.g., heatmaps, Figs. 2b; 2D and 3D principal coordinates plots, Fig. 2d; and networks, Fig. 4b).
Bacterial composition (a, b), clustering analysis (c), and principal coordinate analysis (c) of 10 microbiomes of full-scale municipal sewage influent (1–2) and effluent (3–4) (Cai et al. 2013), and activated sludge (5–6) of an anoxic/oxic process (Ju et al. 2014), full-scale rotating biological contractor biofilms (7–8) (Peng et al. 2014), and lab-scale fresh and saline sewage EBPR activated sludge (9–10) (Mao et al. 2014), respectively. For each sample, 18,000 sequences of V3–V4 regions of 16S rRNA gene are assigned by web RDP classifier. a class level; b major genera (>2 % in at least one sample); c, d based on class-level abundance
Rarefaction curves of 10 microbiomes of full-scale municipal sewage influent (1–2) and effluent (3–4) (Cai et al. 2013), and activated sludge (5–6) of an anoxic/oxic process (Ju et al. 2014), full-scale rotating biological contractor biofilms (7–8) (Peng et al. 2014), and lab-scale fresh and saline sewage EBPR-activated sludge (9–10) (Mao et al. 2014), respectively. The rarefaction curve, plotting the number of observed OTUs (at a similarity cutoff of 97 %) as a function of the number of sequences, was computed using RDP pyrosequencing pipeline rarefaction tool
Flow diagram of network analysis for mining the huge NGS data (a) and its application in exploring strong (Spearman’s ρ ≥ 0.6 or ≤ −0.6) and significant (P value <0.01) correlations between activated sludge species (0.97 OTUs) and operational or quality parameters in a municipal wastewater treatment plant over 5 years (Ju et al. 2014). Abbreviation: T temperature (°C), SL salinity, HRT hydraulic retention time (day), SVI sludge volume index (mL/g), IC influent COD (mg/L); SRT sludge retention time (day), IN influent NH4-N (mg/L), EN effluent NH4-N (mg/L), NR NH4-N removal rate (%)
De-multiplexing and quality filtering
The initial step with handling raw barcoded sequence data of 16S rRNA gene amplicons is to de-multiplex the whole sequence set into individual subsets belonging to different samples based on sample-specific nucleotide barcodes. To avoid adverse effects in the downstream data analysis, all reads with a considerable proportion of either poor quality bases (e.g., low quality score, ambiguous base, or homopolymer) or mismatches in the primer or barcodes, must be removed before assigned to different samples (Caporaso et al. 2010a, b; Schloss et al. 2009).
QIIME and mothur have respective command lines for de-multiplexing and quality filtering of sequence data in several formats (e.g., FASTQ, FASTA, QUAL) generated by prevalent NGS platforms. Exactly, QIIME employs two python scripts, namely “split_libraries.py” and “split_libraries_fastq.py,” to perform coupled de-multiplexing and quality filtering of raw data generated by a single or multiple 454 runs and Illumina lanes, respectively; while mothur depends on “Trim.seqs” to screen and sort pyrosequences in a way similar to those implemented in web-based RDP pipeline (http://rdp.cme.msu.edu/). Both QIIME and mothur cannot de-multiplex barcoded sequences directly from a sff file (e.g., generated from 454 pyrosequencing), but they offer commands (“process_sff.py” in QIIME and “Sffinfo” in mothur) to transform sff into readable FASTA and QUAL files. For quality filtering of raw reads by whichever platforms or software, the following filtering criteria are typically applied, such as minimum average quality score allowed in a read, maximum number of ambiguous bases allowed, minimum and maximum sequence length, maximum length of homopolymer allowed, maximum mismatches in primer or barcode allowed, whether to truncate reverse primer, and so on.
Denoise and chimera checking
As PCR-based amplicon pyrosequencing and other HTS technologies revolutionized the study of microbial diversity, noise introduced during PCR amplification and pyrosequencing, such as sequencing errors (Table 2), PCR single-base errors, and PCR chimeras, can lead to inflated estimates of alpha diversity of microbial communities in a given habitat by orders of magnitude (Reeder and Knight 2010; Quince et al. 2011). Therefore, it is vital to remove noise from raw data to resolve true diversity.
There are several software alternatives for noise removal and chimera checking of amplicon sequences derived from NGS technologies. The software commonly used to remove or correct PCR and pyrosequencing errors/noise includes Denoiser (implemented in QIIME), AmpliconNoise (including PyroNoise and SeqNoise) (Quince et al. 2011), Acacia (Bragg et al. 2012), and Pre.cluster (in mothur). The command line-based Denoiser and AmpliconNoise are among the most popular tools, and both of them use raw flowgram file (.sff) as input, which contains all raw information from sequences, quality score, to flowgrams. The difference between PyroNoise and Denosier is that the former uses an expectation-maximization (EM) algorithm to identify most likely sequence for every read, while the latter uses a greedy-scheme viewed as an approximation to PyroNoise. As these two tools consider both base quality and flowgram, the denoising process is extremely computationally intensive and time-consuming. For example, Denosier may require 35 h to denoise half Titanium run reads (450,000 reads) on 200 CPUs. Other denoising tools including Acacia and Pre.cluster use FASTA as input without considering of flowgram signals, thus their operational time is much less. Strikingly, Acacia achieves equivalent sensitivity and specificity for homopolymer error-correction from FASTA files, but at speeds that are ~500× faster and 2000× faster than Denoiser and AmpliconNoise, respectively (Bragg et al. 2012). Therefore, Acacia is particularly suitable for rapid homopolymer error-correction for extremely large amplicon datasets.
For filtering of PCR chimeras, there are several types of available software, such as ChimeraSlayer (Haas et al. 2011), UCHIME (Edgar et al. 2011), Perseus (Quince et al. 2011), and DECIPHER (Wright et al. 2012). Among them, command lines of ChimeraSlayer, UCHIME (reference and de novo modes), and Perseus have been wrapped in mothur. ChimeraSlayer, which reads a FASTA file and a chimera-free reference database, is the recommended method for chimera checking in QIIME. However, these different methods often disagree with one another on the list of identified chimeras (Goodrich et al. 2014), probably because of their different mechanisms or algorithms. More efforts are required to evaluate these methods and coordinate their inconsistencies in chimera identification.
Noteworthy, a very small proportion of archaeal sequences may be generated for 16S rRNA gene amplicon datasets amplified with bacteria-specific primers. These unexpected sequences should be identified after denoising and chimera removal, and are advised to be discarded before subsequent data normalization (Zhang et al. 2012; Qian et al. 2010)
Data normalization
In general, raw sequence data post quality filtering, denoising, and chimera checking are often referred as “clean” or “effective” reads. The number of clean sequences obtained for each sample, that is sequencing depth, can significantly differ across different samples in the same sequencing run, which is mainly due to technical (sample-independent) rather than biological (sample-dependent) reasons. Uneven sequencing depth can affect diversity estimates in a single sample (i.e., alpha diversity), as well as comparisons across different samples (i.e., beta diversity), thus data normalization is required.
Two methods, i.e., relative abundance and random sampling (i.e., rarefaction), are commonly used to account for various sequencing depths across samples (Goodrich et al. 2014). However, relative abundance, calculated as normalizing sequence counts for a taxon against total sample sequence counts, is subjected to statistics pitfalls and can lead to correlation-based clustering of samples by sequencing depths (Friedman and Alm 2012). Such bias resulted from simply computing correlations using relative abundances of taxa across samples (with uneven sequencing depth) is known as “compositionality bias,” which can distort data interpretation. For instance, artefactual correlations may be observed between two non-correlated, rare community species in the presence of highly abundant species (Faust and Raes 2012). In the other method termed “rarefaction,” normalization is performed by random extraction of equal number of sequences from each sample, and this number is typically identified as the minimum sequence counts for all samples. The major drawback with rarefaction method is the loss of valuable sequence data from samples with relative high sequence counts, especially in the presence of large unevenness of sequencing depth across samples, leading to conservative diversity estimates.
In addition, z-score, calculated as the difference between observed value and mean value divided by the standard deviation, has also been commonly used to normalize and compare samples with different sequencing depths, although it has disadvantages, such as losing meaningfulness of raw data, and magnifying small differences (Oswald et al. 2011).
Picking OTUs and representative sequences
After initial quality filtering, denoising, and chimera checking, clean sequences are clustered into OTUs (i.e., referred as phylotypes when a recognized reference database is used). The OTUs are picked based on sequence identity, and various identity cutoffs of 16S rRNA gene have been used for different taxonomic ranks. For example, identity cutoffs recommended by MEGAN are 99 % for species, 97 % for genus, 95 % for family, and 90 % for order level, respectively (Huson et al. 2007). The OTU picking strategy and algorithms have significant effects in the downstream data interpretation. Based on whether to use a reference database, OTU picking strategies are classified into three categories: de novo, closed reference, and open reference (Caporaso et al. 2010a, b). De novo OTU picking clusters sequences among themselves without a reference database, whereas closed reference OTU picking matches sequences against a reference database and those unmatched at given identity cutoffs are discarded. In an open-reference OTU picking process of QIIME, all sequences are first picked for closed reference OTUs, and any unmatched reads are subsequently clustered for de novo OTUs. In particular for OTU picking from 16S rRNA gene sequences of different hypervariable regions, only closed reference OTU picking is applicable, and publicly available GreenGenes, SILVA SSU, and RDP are commonly employed as sources of reference databases.
There are many clustering or alignment tools available for OTU picking, such as Uclust, cd-hit, BLAST, mothur, usearch, and prefix/suffix. These tools are implemented in QIIME. Among them, the mothur method contains three clustering algorithms to pick de novo OTUs, namely, nearest neighbor, furthest neighbor, or average neighbor. These algorithms are also available by calling “Cluster” command in mothur platform. Upon accomplishment of OTUs picking, the next consideration is which sequence to choose as a representative of an OTU cluster. In general, a representative sequence can be a random, the longest, the most abundant (as default in QIIME), or the first sequence in an OTU cluster. In particular, the distance method in mothur identifies the sequence with the smallest maximum distance to the other sequences as the representative sequence.
Taxonomic assignment
The methods for taxonomic assignment of representative OTU sequences contain three strategies, i.e., word match, best hit, and Lowest Common Ancestor (LCA; (Huson et al. 2007)). The RDP classifier (Wang et al. 2007), either run as a web-based or a local tool, employs a word-matching strategy and does not require alignment, thus the speed is high (Fig. 2a, b). By contrast, both best-hit and LCA strategies require identity search (i.e., alignment) of query sequences against a 16S rRNA reference databases (e.g., RDP, GreenGenes, SILVA SSU) using an alignment tool, thus the efficiency of annotation is determined by the speed, sensitivity, and accuracy of alignment software, apart from sizes and quality of reference databases. The difference between best-hit annotation and LCA algorithm for taxonomic assignment lies in that whether a sequence read is assigned by its best hit (e.g., alignment with the highest score), or by LCA of multiple hits in a given reference database. At present, MEGAN (Huson et al. 2007) and web-based SINA Alignment Service (http://www.arb-silva.de/aligner/) both support taxonomic assignment by LCA algorithm.
Pairwise alignment tools include BLAST (Altschul et al. 1990), BLAT (Kent 2002), Usearch/Uclust (Edgar 2010), mothur (“Classify.seqs” command), SINA aligner (Pruesse et al. 2012), and RTAX (Soergel et al. 2012). Among them, the most-cited tool has been BLAST, due to its high sensitivity and accuracy with acceptable computational costs. Other tools, such as BLAT, have increased speeds by at least two orders of magnitudes than BLASTN, but at the cost of decreased sensitivity (e.g., BLAT allows short, high-similarity alignments, but almost no gapped search). Usearch has been evaluated as 30–200 times faster than BLASTN while keeping almost equal sensitivity and error rates in searching low (0–70 %), medium (70–80 %), and high (80–100 %) similarity nucleotide sequences, as described at http://www.drive5.com/usearch/perf/. This powerful tool, in combination with a condensed reference database (e.g., OTUs sets of public 16S rRNA gene databases, customized sub-databases of functional genes (Yu and Zhang 2013)), is especially suitable for rapid screening of marker (e.g., 16S rRNA) or functional (e.g., antibiotic resistance) genes from huge short-gun metagenomic datasets, as demonstrated by Albertsen et al. (2013) and Yang et al. (2014).
Phylogenetic analysis
Phylogenetic relationships between aligned representative OTU sequences can be visualized and explored in a phylogenetic tree. Methods used for multiple DNA and/or protein sequence alignments include ClustalW, MUSCLE (Edgar 2004), Clustal Omega (Sievers et al. 2011), Kalign (Lassmann and Sonnhammer 2005), T-COFFEE (Notredame et al. 2000), COBALT (Papadopoulos and Agarwala 2007), and FastTree (Price et al. 2010). Many software or packages have been developed for inferring phylogenies and building trees for multiple sequence alignments, such as MEGA (Tamura et al. 2011), RAxML (Stamatakis 2014), MRBAYES (Huelsenbeck and Ronquist 2001), PhyML (Guindon et al. 2010), TreeView (Page 2001), Clearcut (Evans et al. 2006), FigTree (Morariu et al. 2009), and ARB (Ludwig et al. 2004).
MEGA is a popular and versatile tool because of its user-friendly graphical user interface and manuals, as well as plentiful alternative resources, such as alignment tools (e.g., ClustalW, MUSCLE), ways of building trees (e.g., maximum-likelihood, neighbor-joining) from sequence data and distances, evolutionary distance estimation (e.g., pairwise, overall mean), and substitution models. However, RAxML and PhyML are the most widely used programs for maximum-likelihood phylogenetic analysis, probably because they are specifically designed and optimized for such purpose. Web servers of these two programs make them more easily accessible by researchers. Notably, ARB is a useful graphical software package equipped with a variety of tools for sequence alignment and editing, phylogenetic analyses, design and evaluation of hybridization probe or PCR primer, etc. Nonetheless, it may not be suitable for directly handling large NGS data.
Many independent alignment and tree building tools are accessible via several NGS data analysis pipelines. QIIME by default uses PyNAST to align sequences and multiple programs (e.g., FastTree, Clearcut, ClustalW, MUSCLE) to build a Newick format tree file which can be later visualized with programs, such as FigTree (Caporaso et al. 2010a, b). By contrast, mothur employs multiple approaches (e.g., Needleman-Wunsch, Gotoh, BLASTN) within the command “Align.seqs” to align sequences and builds a relaxed neighbor-joining tree in Newick format by calling “Clearcut” command (Schloss et al. 2009).
Alpha- and beta-diversity analyses
For alpha-diversity analysis within one sample, mothur provides lots of diversity metrics (e.g., Shannon, Berger-Parker, Simpson, Q statistic; observed richness, Chao1, ACE, and jackknife), while QIIME by default uses phylogenetic diversity (PD)-whole tree, chao1, and observed species. Another way to compare species richness is by rarefaction curve, i.e., a plot of number of taxon/OTU as a function of number of randomly sampled sequences (Fig. 3). The slope at the end point of a rarefaction curve denotes fraction of unexplored taxon/OTU in a sample at current sequencing depth, thus a smaller slope represents a better reflection of community richness. Tools for rarefaction analysis are available in QIIME (“single_rarefaction.py” and “multiple_rarefaction.py”), mothur (“Rarefaction.single” and “Rarefaction.shared” commands), and RDP web server.
For analysis of beta diversity between samples, that is the degree to which the samples differ from one another, several distance metrics, such as Unifrac, Bray-Curtis, Euclidean, Jaccard index, Yue & Clayton, and Morisita-Horn, have been often employed. In general, beta-diversity metrics, unlike alpha diversity metrics, are remarkably robust to issues, such as sequencing noise and presence of rare sequences (e.g., singletons) (Gobet et al. 2010). Based on whether to consider sequence abundance, beta-diversity metrics can be classified into two categories: quantitative (e.g., Bray-Curtis, weighted Unifrac) and qualitative (e.g., binary Jaccard index) (Goodrich et al. 2014). Noteworthy, weighted and unweighted Unifrac distances, as implemented in QIIME, take into consideration phylogenetic relationships between taxon/OTU sequences. This superiority of Unifrac distance make it outperfom other metrics in community comparison, especially in the case of no apparent difference in abundance profiles of taxon/OTU between samples.
Statistical and network analysis
High throughput of NGS technologies and sample multiplexing enable parallel sequencing of a large number of samples with factorial design and adequate replicates to allow rigorous statistical analysis. Here, popular approaches used for statistics analysis, modeling, and visualization of diversity or function profiles in microbiota data are briefly introduced, such as two-sample/group tests, clustering, classification, ordination, and network analysis. The commonly used tools for statistics and visualization of biological/ecological data include open-source free software or packages, such as R packages (e.g., vegan, ade4), PAST (http://folk.uio.no/ohammer/past/), STAMP (http://kiwi.cs.dal.ca/Software/STAMP), and commercial graphical software, such as CANOCO, PRIMER-E, SPSS, and EXCEL. Free open-source software packages for network visualization and exploration include Cytoscape, Gephi, and R-WGCNA package.
Two-sample/group tests
A common metagenomic experimentation is to compare taxa/OTUs or functional categories between two samples, or each of them across two groups of samples (e.g., different treatments, treatment vs. control, before vs. after treatment) (Ju et al. 2014). Many statistical tests are available to decide whether observed differences between two groups are statistically significant or merely raised by chance (e.g., natural variations during measurement). The most widely used one is two-sample t-test, also known as “Student’s t-test.” This test, either it is independent or paired, is based on an assumption of normal distribution of each of the two data sets being compared, although their sample sizes are not necessarily the same. The data normality can be pretested by a normality test, such as Kolmogorov-Smirnov, Shapiro-Wilk, or Anderson-Darling normality test. Particularly for independent two-sample t-test, independence and equal variances (which can be tested by F-test, Levene’s test, etc.) of two populations are required. In the case of non-normal distribution of data sets, nonparametric two-sample tests robust to data non-normality, such as Wilcoxon signed-rank test, and Mann-Whitney U test are applicable for significance testing of difference between group medians.
Multiple-sample/group tests
To test significant differences between means of multiple samples (two or more), analysis of variance (ANOVA) is often applied based on three assumptions, i.e., normality of response variable, sample independence, and equality of population variance. Typically, one-way ANOVA is applicable to compare three or more levels of one factor/variable, while two-way ANOVA is more popular when two factors/variables, each with multiple levels, are involved in the experiments (Ju et al. 2013). Compared with a one-way mode, two-way ANOVA enables tests of effects of two factors/variables at the same time, as well as tests of independence between two factors (i.e., whether interaction effect is present or not).
In the case of non-normal distribution of populations, it is feasible to transform your data to make it follow a normal distribution, or to choose a nonparametric test (e.g., Kruskal-Wallis H test, PerANOVA) that does not require an assumption of normality. However, one-way ANOVA appears to be robust to moderate violations of normality assumption (with false positive rate not much affected) (Lix et al. 1996). Moreover, in the case of failures to meet equal variance assumption, there are several alternatives to one-way ANOVA, such as Welch test, Brown and Forsythe test, and Kruskal-Wallis H test.
Clustering and classification
Clustering and classification are two contrasting methods widely used for statistical mining of taxonomic or functional entities in microbiota data. The unsupervised clustering is the process of grouping a set of instances (e.g., samples, genes, and taxa) into clusters based on how “close” (similar) they are to one another. For example, hierarchical clustering has been a popular method to compare and visualize as much as differences between multiple samples (i.e., as measured by beta-diversity metrics) as possible via a small number of dimensions and in a resulting tree (Fig. 2c). By contrast, supervised classification is to learn which instances (e.g., taxa, genes, proteins) discriminate between categories (e.g., predefined groups of samples) based on a predefined training set of data points, and to build models using these discriminatory instances so as to predict which categories (or classes) novel instance inputs belong to. Readers may refer to Knights et al. (2011) for more applications and examples of supervised classification methods of microbial communities.
Notably, both taxonomic marker (e.g., 16S rRNA gene) and functional genes have been used for classification of microbial communities. However, what they can do (function), in comparison with who is there (taxonomy), is expected to provide far more discriminatory power for classification of biologically meaningful groups of samples (Xu et al. 2014), since functionally similar communities may show quite different taxonomic structures. It has been reported that PICRUSt, a powerful tool capable of gene content prediction of microbial community from environmental 16S rRNA gene sequences with the Random Forest classifier (Langille et al. 2013), can provide robust information as good as functional genes for classifying samples from environments with enough existing microbial reference genomes in the database (Xu et al. 2014).
Ordination analysis
Once ecological similarities or distances (e.g., beta-diversity metrics) between samples have been computed, the entire data set can be explored by multivariate ordination methods (apart from aforementioned clustering methods), such as principal component analysis (PCA), principal coordinates analysis (PCoA, also known as metric multidimensional scaling) (Fig. 2d), Nonmetric multidimensional scaling (NMDS), canonical correspondence analysis (CCA), linear discriminant analysis (LDA), and redundancy analysis (RDA). Among them, PCA is to create a set of new linearly uncorrelated variables (principal components) via orthogonal transformation so as to account for as much as of the variability of the original data as possible, while PCoA is a rotation of inter-sample distance matrix to project as much distance as possible to low dimensional ordination space (Knights et al. 2011). NMDS is similar to PCoA, except that it seeks to rank the pairwise distances between samples, and uses these ranks (rather than the actual distances in PCoA) to map samples nonlinearly into simplified low-dimensional ordination space to represent their ranked differences. CCA is perhaps the most widely used ordination method today. This method has been widely used by ecologists to relate abundances of species to environmental variables (i.e., sample metadata) (Zhang et al. 2012; Ju et al. 2013).
When applied to exploring microbial ecology, linear ordination methods, such as PCA, LDA, or RDA that are generally meant to be designed for continuous data, may be sometimes limited by the nonlinear or nonmonotone species-species interactions or environment-species responses. Moreover, as opposed to PCA which mainly employs a Euclidean distance, PCoA or NMDS works with any dissimilarity/distance measure or other association coefficients that is specifically designed to deal with data sparsity (e.g., presence of many double zeros) and compositionality bias (e.g., Bray-Curtis dissimilarity). Readers may refer to Ramette (2007) for more advice on how to choose an appropriate ordination method for multivariate analyses of ecological data.
Network-based modeling
16S rRNA gene HTS enables intensive community-wide investigation of microbial structure. Network-based modeling is increasingly applied to predict microbial community assembly (e.g., co-occurrence and co-exclusion patterns), gene co-expression or regulatory patterns, metabolic associations, or protein-protein interactions from huge NGS data, posing novel insights into complex ecological (e.g., cooperative and competitive relationships) or biological associations in various natural or engineering habitats (Doncheva et al. 2012; Steele et al. 2011; Ju et al. 2014). Moreover, time-dependent responses of microbial communities to changes of environmental variables have been traced in interactive environment-species association networks (Ju and Zhang 2014a, b), thanks to developments in both barcoded NGS technologies and bioinformatics software (Ruan et al. 2006). The prediction of environment-species associations has a great significance in the manipulation of microbial communities for selective enrichment of beneficial species and elimination of detrimental microorganisms in anaerobic bioreactors operated for waste and wastewater treatment or bioenergy production (Vanwonterghem et al. 2014).
Network analysis is typically launched by creating an incidence (presence and absence) or abundance matrix of entities (e.g., OTUs, gene or protein), across a range of temporal (time) and/or spatial (location) scales (Fig. 4a). As an association between entities is assumed as one-to-one (pairwise) or one-to-many, it then can be well modeled by computing pairwise correlation/distance measures (e.g., Pearson’s or Spearman’s coefficients) or sparse multiple regression (Faust and Raes 2012). The above computing step is repeated a large number of times by permutation tests to generate P value as an indicator of statistical significance of a predicted relationship. Finally, all statistically significant (P value below the threshold, typically 0.05 or 0.01) relationships are visualized in a network interface, where a node represents an entity and an edge stands for a robust relationship between two entities (Fig. 4b).
In addition, topological and statistical properties of a microbial association network, such as modularity, clustering coefficients, node degree, network diameter, and density, may carry biological or ecological meanings. For example, modules and hubs (nodes with highest degree) in an ecological network are widely interpreted as niches and keystone species, respectively (Fig. 4b). Readers may refer to Proulx et al. (2005) for network thinking in ecology and evolution; Doncheva et al (2012) for topological analysis and interactive visualization of biological networks, and Ju and Zhang (2014a, b) for a demonstration of a correlation-based statistical method for predicting microbial co-occurrence and assembly patterns in a biological wastewater treatment system from large time-series 16S rRNA gene pyrosequencing datasets.
Concluding remarks
The wide application of 16S rRNA gene HTS allows community-wide characterization of microbial diversity and composition in various natural or engineering habitats. With the assistance of highly multiplexed barcoded sequencing and the state-of-the-art bioinformatics and statistical tools, it is technically feasible and economically affordable to simultaneously sequence up to hundreds of to a thousand of sample amplicons in a single NGS run equipped with biological and technical replicates to allow rigid statistical analyses of microbial community dynamics over adequate temporal and/or spatial environmental gradients. This is often required to obtain thorough understandings of environmental impacts on microbial community structure and completely resolve species-species interactions (e.g., by network analysis). Moreover, there is an inevitable tradeoff between data throughput and read length for currently prevalent NGS platforms. Generally, larger data size (i.e., higher sequencing depth) is more beneficial to investigate complex or diverse microbial communities and rare species, while longer read length provides higher taxonomic resolution and may resolve highly similar species (e.g., microdiversity). Therefore, as to which NGS platform to employ highly depends on the research purpose. Better still, 16S rRNA gene HTS is encouraged to work more in complementary with other techniques, such as other metagenomic approaches (e.g., metagenome, metatranscriptome), bioimaging (e.g., FISH), and isotope labeling, to explore metabolic potentials and gene expressions of active microorganisms.
References
Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH (2013) Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31(6):533–538
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Angiuoli SV, Matalka M, Gussman A, Galens K, Vangala M, Riley DR, Arze C, White JR, White O, Fricke WF (2011) CloVR: a virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinforma 12(1):356–370
Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW (2012) Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods 9(5):425–426
Cai L, Ju F, Zhang T (2013) Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl Microbiol Biotechnol 98(7):3317–3326
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010a) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26(2):266–267
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI (2010b) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Cole J, Wang Q, Cardenas E, Fish J, Chai B, Farris R, Kulam-Syed-Mohideen A, McGarrell D, Marsh T, Garrity G (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37(1):141–145
Doncheva NT, Assenov Y, Domingues FS, Albrecht M (2012) Topological analysis and interactive visualization of biological networks and protein structures. Nat Protoc 7(4):670–685
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200
Evans J, Sheneman L, Foster J (2006) Relaxed neighbor joining: a fast distance-based phylogenetic tree construction method. J Mol Evol 62(6):785–792
Faust K, Raes J (2012) Microbial interactions: from networks to models. Nat Rev Microbiol 10(8):538–550
Friedman J, Alm EJ (2012) Inferring correlation networks from genomic survey data. PLoS Comput Biol 8(9):e1002687
Gobet A, Quince C, Ramette A (2010) Multivariate Cutoff Level Analysis (MultiCoLA) of large community data sets. Nucleic Acids Res 38(15):e155–e155
Goecks J, Nekrutenko A, Taylor J (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11(8):R86
Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R, Ley RE (2014) Conducting a microbiome study. Cell 158(2):250–262
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59(3):307–321
Guo F, Zhang T (2012) Profiling bulking and foaming bacteria in activated sludge by high throughput sequencing. Water Res 46(8):2772–2782
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21(3):494–504
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17(3):377–386
Ibarbalz FM, Figuerola EL, Erijman L (2013) Industrial activated sludge exhibit unique bacterial community composition at high taxonomic ranks. Water Res 47(11):3854–3864
Ju F, Zhang T (2014a) Bacterial assembly and temporal dynamics in activated sludge of a full-scale municipal wastewater treatment plant. ISME J 9:683–695
Ju F, Zhang T (2014b) Novel microbial populations in ambient and mesophilic biogas-producing and phenol-degrading consortia unraveled by high-throughput sequencing. Microb Ecol 68(2):235–246
Ju F, Guo F, Ye L, Xia Y, Zhang T (2013) Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ Microbiol Rep 6(1):80–89
Ju F, Xia Y, Guo F, Wang Z, Zhang T (2014) Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ Microbiol 16(8):2421–2432
Kent WJ (2002) BLAT-the BLAST-like alignment tool. Genome Res 12(4):656–664
Knights D, Costello EK, Knight R (2011) Supervised classification of human microbiota. FEMS Microbiol Rev 35(2):343–359
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31(9):814–821
Lassmann T, Sonnhammer EL (2005) Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinforma 6(1):298–306
Lix LM, Keselman JC, Keselman H (1996) Consequences of assumption violations revisited: a quantitative review of alternatives to the one-way analysis of variance F test. Rev Educ Res 66(4):579–619
Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ (2012) Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30(5):434–439
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Buchner A, Lai T, Steppi S, Jobb G, Förster W (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32(4):1363–1371
Mao Y, Yu K, Xia Y, Chao Y, Zhang T (2014) Genome reconstruction and gene expression of “Candidatus Accumulibacter phosphatis” clade IB performing biological phosphorus removal. Environ Sci Technol 48(17):10363–10371
Minoche AE, Dohm JC, Himmelbauer H (2011) Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol 12(11):R112
Morariu VI, Srinivasan BV, Raykar VC, Duraiswami R, Davis LS (2009) Automatic online tuning for fast Gaussian summation. In: Advances in neural information processing systems, 1(1):1113-1120
Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302(1):205–217
Oswald ES, Brown LM, Bulinski JC, Hung CT (2011) Label-free protein profiling of adipose-derived human stem cells under hyperosmotic treatment. J Proteome Res 10(7):3050–3059
Page RD (2001) TreeView. Glasgow University, Glasgow, UK
Papadopoulos JS, Agarwala R (2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23(9):1073–1079
Peng X, Guo F, Ju F, Zhang T (2014) Shifts in the microbial community, nitrifiers and denitrifiers in the biofilm in a full-scale rotating biological contactor. Environ Sci Technol 48(14):8044–8052
Price MN, Dehal PS, Arkin AP (2010) FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490
Proulx SR, Promislow DE, Phillips PC (2005) Network thinking in ecology and evolution. Trends Ecol Evol 20(6):345–353
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28(14):1823–1829
Qian P-Y, Wang Y, Lee OO, Lau SC, Yang J, Lafi FF, Al-Suwailem A, Wong TY (2010) Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J 5(3):507–518
Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13(1):341–353
Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ (2011) Removing noise from pyrosequenced amplicons. BMC Bioinforma 12(1):38–55
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62(2):142–160
Reeder J, Knight R (2010) Rapid denoising of pyrosequencing amplicon data: exploiting the rank-abundance distribution. Nat Methods 7(9):668–669
Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, Hegarty R, Nusbaum C, Jaffe DB (2013) Characterizing and measuring bias in sequence data. Genome Biol 14(5):R51
Ruan Q, Dutta D, Schwalbach MS, Steele JA, Fuhrman JA, Sun F (2006) Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 22(20):2532–2538
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7(1):531–536
Soergel DA, Dey N, Knight R, Brenner SE (2012) Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J 6(7):1440–1444
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313
Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, Chow C-ET, Sachdeva R, Jones AC, Schwalbach MS (2011) Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5(9):1414–1425
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Vanwonterghem I, Jensen PD, Ho DP, Batstone DJ, Tyson GW (2014) Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr Opin Biotechnol 27:55–64
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267
Wright ES, Yilmaz LS, Noguera DR (2012) DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78(3):717–725
Xia Y, Cai L, Zhang T, Fang HH (2012) Effects of substrate loading and co-substrates on thermophilic anaerobic conversion of microcrystalline cellulose and microbial communities revealed using high-throughput sequencing. Int J Hydrog Energy 37(18):13652–13659
Xu Z, Malmer D, Langille MG, Way SF, Knight R (2014) Which is more important for classifying microbial communities: who’s there or what they can do&quest. ISME J 8:2357–2359
Yang Y, Jiang XT, Zhang T (2014) Evaluation of a hybrid approach using UBLAST and BLASTX for metagenomic sequences annotation of specific functional genes. PLoS One 9(10):e110947
Ye L, Shao MF, Zhang T, Tong AHY, Lok S (2011) Analysis of the bacterial community in a laboratory-scale nitrification reactor and a wastewater treatment plant by 454-pyrosequencing. Water Res 45(15):4390–4398
Yu K, Zhang T (2013) Construction of customized sub-databases from NCBI-nr database for rapid annotation of huge metagenomic datasets using a combined BLAST and MEGAN approach. PLoS One 8(4):e59831
Zhang T, Shao M-F, Ye L (2012) 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6(6):1137–1147
Acknowledgments
The authors wish to thank the Hong Kong General Research Fund (7195/06E, 7197/08E, 7202/09E, 7198/10E, 7201/11E, 7190/12E, and 172099/14E) for the financial support of this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ju, F., Zhang, T. 16S rRNA gene high-throughput sequencing data mining of microbial diversity and interactions. Appl Microbiol Biotechnol 99, 4119–4129 (2015). https://doi.org/10.1007/s00253-015-6536-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6536-y