Abstract
Barley (Hordeum vulgare L.) is well known for its relatively high salt tolerance among cereal crops. However, the genetic variation of cultivated barley becomes narrower due to continuous artificial selection and breeding processes. Compared with cultivated barley, wild barley contains wider genetic variation and abundant sources for abiotic stress tolerance, considering as an elite resource for mechanism study on salt tolerance. In this study, Tibetan wild barley accession XZ113 identified with high salt tolerance, was used to investigate ionic responses and to identify proteins involved in salt tolerance in roots and shoots at early stage of salt stress, during 48 h. Exposed to salinity, shoot growth is more sensitive than root growth. Conversely, K/Na ratio in the shoots was larger than that in the roots, and both were above 1.0. Steady-state K+ flux experiment showed XZ113 had a strong K+-retaining ability under salt stress, maybe contributing to its good performance of the absolute growth rate. Proteomic results suggested that monodehydroascorbate reductase and peroxidases related to reactive oxygen species scavenging in the roots and phosphoglycerate kinase, triosephosphate isomerase and sedoheptulose-1,7-bisphosphatase associated with photosynthesis and metabolisms in the shoots, played important roles in salt tolerance at early stage of salinity in wild barley.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Salt stress is one of major abiotic stresses restricting crop production and yield worldwide. Currently, around 20 % of irrigated land has been affected by salinity, and moreover the soil salinization is expanding rapidly due to unreasonable farming and unavoidable geological factors (Munns and Tester 2008; Rengasamy 2010). To fight against salt stress, development of crop cultivars with high salt tolerance is considered as a most effective method. Plant species differ widely in salt tolerance, and most cereal crops are glycophytes (Glenn et al. 1999; Horie et al. 2012). However, barley (Hordeum vulgare L.), ranking the fourth in terms of planting area in the world among cereal crops, is well known for its relatively high salt tolerance. Hence, barley is commonly planted in the saline soil where other cereal crops cannot grow well, and meanwhile it is frequently used in the studies on the mechanisms of salt stress tolerance in plants (Munns and Tester 2008; Munns et al. 2012). But the physiological and molecular mechanisms underlying salt tolerance in barley still remain largely elusive, especially osmotic and ionic stresses at early phases. On the other hand, the genetic background of cultivated barley becomes narrower with development of intensive planting, thus resulting in the loss of elite genes, including those related to salt stress tolerance.
Comparatively, wild barley (Hordeum spontaneum) contains a wider source of genetic variation for abiotic stress tolerance (Ellis et al. 2000; Nevo 2007). Wild barley relatives have stronger adaptive ability to various abiotic stresses including cold, drought and salt salinity than cultivated barley. A recent study confirmed that Tibet is one of the centers of domestication of cultivated barley (Dai et al. 2012). Meanwhile, it was found that nearly 70 % of 189 Tibetan wild barley accessions showed higher salt tolerance than a well-known tolerant cultivar, CM72 (Wu et al. 2011; Qiu et al. 2011). Obviously, Tibetan wild barley is highly valuable in breeding for the cultivars of barley as well as other crops with high salt stress tolerance, and it is quite imperative to understand the tolerant mechanisms.
High concentration of soluble sodium (Na) initially causes osmotic and ionic stresses to plants (Zhu 2002; Munns and Tester 2008). Osmotic shock starts immediately when plant roots are exposed to salt solution at a certain threshold level, approximately 40 mM NaCl for most plants (Munns and Tester 2008). Osmotic stress significantly reduces water uptake and tissue growth rates of plants, leading to reduced development and productivity in later phases (Horie et al. 2012; Han et al. 2015). Ionic stress is mainly caused by Na accumulation at a toxic concentration, which causes leaf senescence and cell death (Munns and Tester 2008). Therefore, the effective strategies of fighting against salt stress for plants are to keep low cytosolic Na+ concentration at the cellular level and low shoot Na+ concentrations at the whole plant level in response to salt stress at the early phases (Horie et al. 2012). Previous studies also revealed that high cytosolic K+/Na+ ratio, especially in shoots is crucial for salt tolerance for glycophyte plants (Gorham et al. 1990; Blumwald 2000; Chen et al. 2005; Shabala et al. 2010). For example, a salt-tolerant rice genotype, PL177 showed significantly less salt damage, higher K+/Na+ ratio in shoots and lower Na+ translocation from roots to shoots than the sensitive genotype, IR64 (Wang et al. 2016). It is also suggested that salt tolerant genotypes are capable of maintaining a higher xylem K+/Na+ ratio by HKT transporter localized in plasma membrane and sequestering Na+ in vacuole by tonoplast-localized Na+/H+ exchangers in barley (Shabala et al. 2010). In short, the current studies demonstrate the importance of salt-induced ionic changes in the development of salt stress tolerance in plants.
There are complex signaling and metabolic pathways in plants to cope with salt stress (Zhang et al. 2012a, b). Physiological studies indicated that increased energy metabolism such as glycolysis and tricarboxylic acid cycle (TCA) were necessary to alleviate osmotic and ionic effects caused by salt stress (Kosová et al. 2013a), but little is known on the relationship between signaling and metabolism at the whole plant level (Wang et al. 2016). So, it is imperative to determine the molecular responses to salt stresses in order to understand signaling and metabolic pathways associated with salt tolerance. Obviously, among these molecular components, proteins play direct roles in biological metabolisms and set up the tolerant system in response to salt stresses. Therefore, proteomics analysis is powerful and useful in studies on abiotic tolerance in order to identify the key genes associated closely with stress tolerance (Kosová et al. 2011, 2013b). Zhang et al. (2012a, b) summarized 2,171 salt-responsive proteins in 34 plant species by proteomic studies including those linked with photosynthesis, reactive oxygen species (ROS) scavenging, ion homeostasis, osmotic modulation, signaling transduction and protein synthesis. Those stress-induced proteins may be helpful for obtaining an entire knowledge of molecular networks and beneficial for mining salt-associated genes. However, few attempts have been conducted to investigate the ionic and proteomic responses of wild barley to salt stress, although it shows much higher salt tolerance relative to cultivated barley. Therefore, the current research was done in order to understand ionic responses to salt stress at early stage, and to identify proteins involved in ionic changes and salt tolerance using a Tibetan wild barley accession XZ113, which is high salt tolerant (Qiu et al. 2011).
Materials and methods
Plant materials and hydroponic culture
Seeds of Tibetan wild barley accession XZ113 were surface sterilized with 3 % H2O2 solution for 20 min and rinsed thoroughly with distilled water. Seed germination was conducted according to Wu et al. (2014) in a growth condition (22/18 °C, day/night; 14/10 h, light/dark). Seven-days-old seedlings were transplanted into 5 L black plastic containers containing 1/2 Hoagland solution. Aerated hydroponic solution was renewed every 5 days (pH 6.0). Seven days after transplanting, the treatment of 200 mM NaCl was initiated and the solution without NaCl addition was used as the control. For non-invasive MIFE (Microelectrode Ion Flux Estimation) experiments, seeds were placed vertically into floating filter papers in an aerated basic salt medium (BSM) containing 0.1 mM CaCl2 and 0.5 mM KCl, and kept in a growth chamber for 3 days without light until use for measurement.
Root K+ and H+ flux measurement
Root net ion fluxes were measured using MIFE technique (Chen et al. 2007). One hour prior to measurement, a root of 3-days-old barley seedling was immobilized to a slide and placed horizontally in a Perspex container with 10 ml BSM. The calibration and operation of K+ and H+ ion-selective microelectrodes were followed according to the protocol described by Shabala et al. (2010). The electrode tips were positioned 40 μm above the surface of mature root zone (~10 mm from the root tip). Steady-state net K+ and H+ ion fluxes were measured at each time point of 0, 2, 6 and 24 h after 80 mM NaCl was added into BSM solution according to Chen et al. (2007). Six biological replicates were measured in this experiment. The different significance among treatments was tested by SAS 9.1 software and the difference at p < 0.05 and p < 0.01 were considered as significant and highly significant.
Growth and element concentration determination
Before sampling, roots were thoroughly rinsed with deionized water for several times. Three biological replicates of roots and shoots were sampled at 0, 2, 6, 24 and 48 h after salt treatment. Fresh weight (FW) of roots and shoots were measured using an electronic balance (Sartorius BSA124S, Germany), then dried at 80 °C for 72 h, and weighed. Absolute growth rate (GR) was calculated according to the formula: GR (mg/h) = (DWT − DW0)/T, here T is the treatment time. Tissue relative water content (RWC) was calculated by the formula: RWC (%) = (FW − DW)/DW × 100 %. For element extraction, dried samples were digested with 6 ml HNO3 and 200 μl 30 % H2O2 in boiling tubes using a microwave digestion instrument at 200 °C for 1.5 h (Multiwave 3000, Anton Paar GmbH, Australia). Then the contents of Na, K, Ca and Mg in the extracted solution were determined using an ICP–OES spectrometer (Optima 8000 series, PerkinElmer Inc, USA). The different significance was also tested by SAS 9.1 software, as mentioned above.
Protein extraction and purification
Fresh tissue of roots and shoots were sampled at 48 h after salt treatment, frozen immediately in liquid nitrogen, and then stored at −80 °C before use. Total proteins were extracted from 3 g fresh tissue and purified using ReadyPrep™ 2-D cleanup kit (BIO-RAD, Hercules, USA). The Bradford kit (BIO-RAD, Hercules, USA) was used to determine the concentration of proteins.
Two-dimensional electrophoresis
Two-dimensional electrophoresis was conducted according to the method described by Cheng et al. (2009). IEF was carried out on a 24 cm IPG strip (pH 4–7). A total of 460 μl rehydration buffer containing 200 μg (for gel image) or 1200 μg (for MS analysis) protein sample mixed with loading buffer solution (1 % DTT, 1 % IPG Buffer, 1× BPB and rehydration buffer) was loaded in the gels. The gel electrophoresis was set in a total voltage of 52 kVh with variable programs in Ettan IPGphor 3 system (GE Healthcare, Germany) according to Wu et al. (2014). After IEF, the strips were equilibrated in an equilibration buffer for 15 min and placed in the dark for another 15 min. The second dimension of SDS-PAGE was performed on top of 12.5 % running gel, with first voltage of 2 W for 45 min and second 17 W per gel for 5 h. Then the gels were strained with silver staining method and CBB G-250 staining method for further analysis.
2-D gel image analysis
For gel images analysis, 2D gels were scanned by a GS-800 2-DE scanner (BIO-RAD, Hercules, USA) and analyzed by Image Master™ 2D version 5.0 (Amersham Biosciences) according to automated matching and manual editing. Protein spots were analyzed according to Wu et al. (2014). Three biological and two technical replicates were set in the experiments.
MALDI-TOF/TOF MS analysis
Differentially expressed spots (p < 0.05) were excised from the 2-D gels, then washed clearly and digested in trypsin digestion solution according to Cheng et al. (2009). Analysis of peptide mass spectra were operated by an Autoflex speed™ MALDI-TOF–TOF mass spectrometer (Bruker Dalton, Germany) in the mode described by Wu et al. (2014). To analyze data and search for database of MS/MS, MASCOT program was conducted by BioTools (Bruker Dalton, Germany) along with NCBI in the following parameters: peptide mass tolerance of 50 ppm; fragment mass tolerance of ±0.5 Da; peptide charge state of +1; fixed modification of cysteine carbamidamethyl; variable modification of methionine (Met) oxidation. Only target peptides as displayed by Mascot probability analysis (p < 0.05) were accepted as candidate significant proteins defined by biological function.
Results
Growth performance of XZ113 in response to salt stress
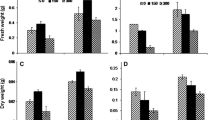
Tibetan wild barley accession XZ113 is identified as a salt-tolerant genotype in a previous study (Qiu et al. 2011). To understand physiological and molecular responses of XZ113 to salt stress, we firstly determined its growth performance in terms of absolute growth rate (GR) and relative water content (RWC) in a time-course of salt treatment (200 mM NaCl) during 48 h. Obviously, salt treatment caused a significant decrease of GR in roots and shoots, and the decreased extent was more obvious with the treatment time (Fig. 1a). During 24 h treatment, nearly eightfold decrease of GR was found for both roots and shoots, and then kept a low GR after 24 h (Fig. 1a). Compared with the roots, shoots showed larger inhibition of GR in each sampling time, indicating that shoot growth is more sensitive to salt stress than root growth. Meanwhile, salt stress only caused a slight water loss in roots, merely 2 % of RWC decrease after salt treatment. While shoot RWC showed less significant change during 48 h treatment (Fig. 1b). Surprisingly, root RWC increased at 6 h to reach a maximum value and then decreased over the treatment time (Fig. 1b).
Dynamic changes of element concentration in roots and shoots in response to salt stress
The effect of salt treatment on ionic change was significant, and differed between roots and shoots (Table 1). In roots, Na concentration showed significant increase at 2 h after treatment, followed by a slight decrease at 6 h, and then kept rising to 20.6 mg g−1 DW at 48 h, nearly twofold higher than that at 2 h. The change of Na concentration in roots was completely opposite to that of root RWC. In contrast, K concentration decreased from nearly 40 mg g−1 DW at 0–6 h to 21.39 mg g−1 DW at 48 h (Table 1). In shoots, ionic change was slower and less in response to salt stress in comparison with roots. Until 24 h after treatment, Na concentration showed significant increase relative to the control, and then continued increase, reaching to 14.2 mg g−1 DW at 48 h, nearly 1.5 fold higher than that at 24 h. While shoot K concentration remained less change during the whole treatment (Table 1). In terms of K/Na ratio, it showed the opposite change as Na concentration did in both roots and shoots. The value of K/Na ratio in roots and shoots was above 1.0 and was larger in shoots than in roots at 48 h after salt treatment (Table 1). In addition, in comparison with control, Ca and Mg concentrations decreased by 39.1 and 26.8 % in roots, and by 11.8 and 21.0 % in shoots, respectively, at 48 h after salt treatment (Table 1).
In order to reveal the effect of salt stress on K+-retaining ability in XZ113, steady-state K+ and H+ flux in the roots exposed to 80 mM NaCl was measured using MIFE technology within 24 h (Fig. 2). The result indicates that K+ efflux was significantly reduced under salt stress, falling into −20 nmol m−2 s−1 at 2 h and into −100 nmol m−2 s−1 at 6 h exposure. However, at 24 h exposure, K+ efflux was obviously recovered (Fig. 2a, b). Meanwhile, a fast H+ efflux was observed at 2 h exposure, being sixfold increase compared with that at starting time, and become twofold increase at 24 h (Fig. 2c, d). Apparently, H+ recovered more quickly and effluxed more than K+ in the roots of XZ113 when subjected to salt stress. Hence, XZ113 may maintain relatively high K concentration under salt stress, resulting in high K/Na ratio after salt stress, contributing to its salt tolerance.
Steady-state K+ (a) and H+ (c) fluxes measured by MIFE from the mature root zone of 3-day-old seedlings exposed to 80 mM NaCl for 0, 2, 6 and 24 h. The results in b, d are averaged over 10 min of K+ and H+ flux measurement, respectively. Six replications were performed at each time point. **Indicates highly significant between treatment and control at p < 0.01 by Tukey HSD
Identification of salt stress induced proteins in roots
A total of 8 differentially expressed protein spots were successfully identified in roots of XZ113 under salt stress (Table 2; Fig. 3a). Among these protein spots, 4 up-regulated protein spots were putative monodehydroascorbate reductase (AU1), putative class III peroxidase (AU2, AU4) and porin family protein (AU5), respectively. Meanwhile, the spot abundance of succinyl-CoA ligase beta-chain (AD1), predicted protein (AD3), hydroxyacylglutathione hydrolase (AD4) and dehydroascorbate reductase (AD5) were greatly reduced (Table 2). Here, 5 proteins mainly participated in ROS scavenging, were listed as AD4, AD5, AU1, AU2 and AU4. Among them, AU1 and AU4 showed protein abundance specifically under salt stress (Table 2; Fig. 4), indicating highly active ROS activity at early stage of salt stress.
Two-dimensional gels of differentially expressed protein spots isolated from roots and shoots of XZ113 under 200 mM NaCl treatment or control for 2 day. a 2-D gels of root proteome; and b 2-D gels of shoot proteome. Arrows indicate the location of differentially expressed protein spots. Numbers in gels showed different abundance of proteins in roots or shoots after salinity. As D2 and U3 were not identified in MS analysis, they were not discussed in this study
‘Spot view’ of the abundance of differentially expressed proteins in roots of XZ113 exposed to salt stress for 2 day. Green circles with black arrows indicate spots showing decrease (listed as D) and white arrows indicate spots showing increase (listed as U). Protein spot ID corresponds to spot number in Table 2 and Fig. 3a. (Color figure online)
Identification of salt stress induced proteins in shoots
In shoots, there were 8 differentially expressed proteins identified under salt stress when compared with its control (Table 3; Fig. 3b). Three protein spots showing reduction under salt stress were characterized as chloroplastic ATP synthase (BD1, BD2, BD3), belonging to two types of subunit groups, alpha and gamma. On the other hand, 5 significantly increased proteins were identified as chloroplastic ATP synthase alpha subunit (BU1, BU4), phosphoglycerate kinase (BU2), triosephosphate isomerase (BU3), Sedoheptulose-1,7-bisphosphatase (SBPase, BU5) (Table 3; Fig. 5). Here, 5 ATP synthase subunits (BD1, BD2, BD3, BU1 and BU4) were identified, indicating important roles of ATP synthases in response to salt stress in shoots. In addition, SBPase (BU5) showed the maximum change (7.29 folds) as a key enzyme in Calvin cycle of plant photosynthesis, while BU2 and BU3 are two key enzymes in the pathway of glycolysis, indicating the enhanced activity of metabolisms were occurred in shoots of XZ113 under salt stress.
‘Spot view’ of the abundance of differentially expressed proteins in shoots of XZ113 exposed to salt stress for 2 day. Green circles with black arrows indicate spots showing decrease (listed as D) and white arrows indicate spots showing increase (listed as U). Protein spot ID corresponds to spot number in Table 3 and Fig. 3b. (Color figure online)
Discussion
Salt tolerance in plants involves complex components of signaling and metabolic regulation networks, and also toughly depends on species, organ, dose and time of salt exposure (Deinlein et al. 2014; Ellouzi et al. 2014). Up to date, most studies on salt resistance are concentrated on a long-term or high salt level, so as to reveal the diverse mechanisms mainly linked to metabolic and ionic adjustment, but few studies were done to understand osmotic and ionic regulation at early stage of salt stress (Munns and Tester 2008; Deinlein et al. 2014). Our previous studies demonstrated that Tibetan wild barley had high salt tolerance (Qiu et al. 2011; Wu et al. 2011, 2013a). In this study, we used a wild barley accession with high salt tolerance, XZ113 as material, to reveal the ionic and proteomic responses to salt stress at early exposure stage. It is interesting to note that the plants could survive well under 200 mM NaCl, although growth was significantly inhibited relative to control, (Fig. 1a), indicating its extraordinary salt tolerance at seedling stage, and confirming the previous finding (Qiu et al. 2011).
Ionic concentrations were significantly changed under salt stress, especially for Na and K. On the whole, the response to salt stress is characterized by increased Na concentration and decreased K concentration in plant tissues. Maintaining of a high cytosolic K/Na ratio is normally considered as a key feature of salt tolerance in plants (Maathuis and Amtmann 1999; Tester and Davenport 2003; Shabala and Cuin 2008). Moreover, a K/Na ratio of around 1.0 is regarded as a minimum value for keeping ion homeostasis (Maathuis and Amtmann 1999). For example, a salt-sensitive accession of Tibetan wild barley, XZ169 showed K/Na ratios in roots and shoots less than 1.0 even under 150 mM NaCl at germination and seedling stages (Wu et al. 2013b). In the current study, K/Na ratio in roots and shoots of XZ113 was 3.48 and 1.04 at 48 h salt exposure (Table 1), respectively, indicating the capacity of maintaining an optimal K/Na ratio for conferring its salt tolerance. Otherwise, most glycophytes prefer to prevent K loss from cells rather than Na+ exclusion (Shabala 2000; Chen et al. 2007; Zhang et al. 2012a). Chen et al. (2005) estimated salt tolerance by K+ flux measurement and suggested the parameter of K+ efflux is a reliable and heritable indicator of salt tolerance in barley. In this study, we found XZ113 had a strong ability of maintaining K+, i.e. keeping a low level of efflux under salt stress and even being able to recover slow efflux in the late stage of salt exposure (Fig. 2a, b).
Meanwhile, some salt stress induced proteins were noted in both roots and shoots by 2-DE (Tables 2, 3). In roots, four induced proteins showed extremely expressed changes in comparison with controls, referred to AD5, AU1, AU4 and AU5, respectively (Table 2; Fig. 4). Most of them are involved in ROS scavenging. Wu et al. (2014) identified that monodehydroascorbate reductase (AU1) and class III peroxidase (AU2 and AU4) showed nearly 2 and 0.2 fold changes of protein level at 48 h after 200 mM salinity in CM72, a well-known salt tolerant cultivar. Here, the above two proteins performed more fold increase of protein level in roots of XZ113 than that in the previous study, after salt treatment. Obviously, these proteins may be attributed to dramatic oxidative activity caused by salt stress and rapid development of antioxidant defenses system in the plant tissues of XZ113. Additionally, AD1 is an important component of TCA, and A D3 may be helpful for plasma membrane stability. AU5 belongs to porin family, which is well known as transmembrane proteins and has the function in cell protection as anion channels (Geiger et al. 1999; Wen et al. 2011). Besides, aquaporins are members of porin family, which transport water and low-molecular weight neutral compounds (Tyerman et al. 2002; Maurel et al. 2008; Horie et al. 2012), enhancing salt tolerance in Arabidopsis (Guo et al. 2006). It may be assumed that ATP synthases act as a pioneer in response to salt stress in shoots of XZ113, and five proteins identified in this study belong to two types of subunits (Table 3; Fig. 5). Here, four subunit α displayed the same molecular mass (55.32 kDa) with different electric charges, identified as BD1, BD2, BU1 and BU4 (Table 3; Fig. 5). These subunits belong to CF1 complex (Kohzuma et al. 2013). Wu et al. (2014) indicated that H+-ATPases participated in development of ion homeostasis in salt-tolerant barley genotypes. Interestingly, fast H+ efflux activity was observed in the roots of XZ113 (Fig. 2c, d), which may help keeping balance voltage and pH in a proper range. Kosová et al. (2013b) suggested that many CF1 subunits in both halophytes and glycophytes involved in salt tolerance under salt stress. In addition, C3 photosynthesis cycle enzyme, SBPase (Listed as BU5) increased by 7 folds in comparison with control under salt stress (Table 3), hence leading to improved photosynthesis (Rosenthal et al. 2011). Similar result was also found in cultivated barleys, a salt-tolerant genotype Afzal had more up-regulation of SBPase than a sensitive genotype L-527 (Rasoulnia et al. 2011). Therefore the higher SBPase activity should be helpful for enhancing carbon assimilation in leaves of XZ113 under salt stress. Moreover, phosphoglycerate kinase (BU2) and triosephosphate isomerase (BU3) involved in glycolysis pathway were significantly up-regulated after salt stress, indicating that the metabolism level of glycolysis was enhanced by salt stress in shoots of XZ113 as consistent as other findings (Rasoulnia et al. 2011; Wu et al. 2014).
In conclusion, XZ113 showed relatively less growth inhibition in early stage of salt stress exposure, as reflected by the positive GR and relatively high water content. Its high salt stress tolerance may be attributed to the capacity in maintenance of optimal K/Na ratio and high K concentration in plant tissues. Meanwhile, proteomic analysis showed that the wild barley could set up rapidly ROS scavenging enzyme system in roots when meeting an early and transient oxidative burst caused by salt stress. In shoots, ATP synthases were pioneers for tolerant development at early stress exposure stage, and they belong to different subunits, with SBPase contributing to relatively stable photosynthesis and assimilation under salt stress. Further studies are still needed to reveal the exact roles of those proteins whether acting as essential components of salt tolerance in wild barley.
Abbreviations
- ATP:
-
Adenosine triphosphate
- BPB:
-
Bromophenol blue
- DTT:
-
Dithiothreitol
- DW:
-
Dried weight
- FW:
-
Fresh weight
- GR:
-
Growth rate
- ICP–OES:
-
Inductively coupled plasma–optical emission spectrometer
- IEF:
-
Isoelectric focusing electrophoresis
- IPG:
-
Immobilized pH gradient
- MALDI-TOF/TOF MS:
-
Matrix-assisted laser desorption/ionization time of flight mass
- MIFE:
-
Microelectrode ion flux estimation
- PM:
-
Plasma membrane
- PIPs:
-
Plasma membrane intrinsic proteins
- ROS:
-
Reactive oxygen species
- RWC:
-
Relative water content
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SBPase:
-
Sedoheptulose-1,7-bisphosphatase
- 2-DE:
-
Two-dimensional gel electrophoresis
- TCA:
-
Tricarboxylic acid cycle
- VDAC:
-
Voltage-dependent anion channel
References
Blumwald E (2000) Sodium transport and salt tolerance in plants. Curr Opin Cell Biol 12(4):431–434
Chen ZH, Newman I, Zhou MX, Mendham N, Zhang GP, Shabala S (2005) Screening plants for salt tolerance by measuring K+ flux: a case study for barley. Plant Cell Environ 28(10):1230–1246
Chen ZH, Pottosin II, Cuin TA, Fuglsang AT, Tester M, Jha D, Zepeda-Jazo I, Zhou MX, Palmgren MG, Newman IA, Shabala S (2007) Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol 145(4):1714–1725
Cheng YW, Qi YC, Zhu Q, Chen X, Wang N, Zhao X, Chen HY, Cui XJ, Xu LL, Zhang W (2009) New changes in the plasma-membrane-associated proteome of rice roots under salt stress. Proteomics 9(11):3100–3114
Dai F, Nevo E, Wu DZ, Comadran J, Zhou MX, Qiu L, Chen ZH, Beiles A, Chen GX, Zhang GP (2012) Tibet is one of the centers of domestication of cultivated barley. Proc Natl Acad Sci USA 109:16969–16973
Deinlein U, Stephan AB, Horie T, Luo W, Xu GH, Schroeder JI (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19(6):371–379
Ellis RP, Forster BP, Robinson D, Handley LL, Gordon DC, Russell JR, Powell W (2000) Wild barley: a source of genes for crop improvement in the 21st century? J Exp Bot 51(342):9–17
Ellouzi H, Ben Hamed K, Hernandez I, Cela J, Muller M, Magne C, Abdelly C, Munne-Bosch S (2014) A comparative study of the early osmotic, ionic, redox and hormonal signaling response in leaves and roots of two halophytes and a glycophyte to salinity. Planta 240(6):1299–1317
Geiger TR, Keith CS, Muszynski MG, Newton KJ (1999) Sequences of three maize cDNAs encoding mitochondrial voltage-dependent anion channel (VDAC) proteins (accession nos. AF178950, AF178951, and AF178952) (PGR 99-156). Plant Physiol 121(2):686
Glenn EP, Brown JJ, Blumwald E (1999) Salt tolerance and crop potential of halophytes. Crit Rev Plant Sci 18(2):27–255
Gorham J, Wyn Jones RG, Bristol A (1990) Partial characterization of the trait for enhanced K+–Na+ discrimination in the D genome of wheat. Planta 180:590–597
Guo L, Wang ZY, Cui WE, Chen J, Liu MH, Chen ZL, Qu LJ, Gu HY (2006) Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res 16(3):277–286
Han Y, Yin SY, Huang L (2015) Towards plant salinity tolerance-implications from ion transporters and biochemical regulation. Plant Growth Regul 76(1):13–23
Horie T, Karahara I, Katsuhara M (2012) Salinity tolerance mechanisms in glycophytes: an overview with the central focus on rice plants. Rice 5:11
Kohzuma K, Dal Bosco C, Meurer J, Kramer DM (2013) Light- and metabolism-related regulation of the chloroplast ATP synthase has distinct mechanisms and functions. J Biol Chem 288(18):13156–13163
Kosová K, Vítámvás P, Prášil IT, Renaut J (2011) Plant proteome changes under abiotic stress—contribution of proteomics studies to understanding plant stress response. J Proteomics 74:1301–1322
Kosová K, Vítámvás P, Urban MO, Prášil IT (2013a) Plant proteome responses to salinity stress—comparison of glycophytes and halophytes. Funct Plant Biol 40:775–786
Kosová K, Prášil IT, Vítámvás P (2013b) Protein contribution to plant salinity response and tolerance acquisition. Int J Mol Sci 14:6757–6789
Maathuis FJM, Amtmann A (1999) K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann Bot 84(2):123–133
Maurel C, Verdoucq L, Luu DT, Santoni V (2008) Plant aquaporins: membrane channels with multiple integrated functions. Annu Rev Plant Biol 59:595–624
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Ann Rev Plant Biol 59:651–681
Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D, Gilliham M (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotech 30:360–364
Nevo E (2007) Evolution of wild wheat and barley and crop improvement: studies at the institute of evolution. Isr J Plant Sci 55(3–4):251–262
Qiu L, Wu DZ, Ali S, Cai SG (2011) Evaluation of salinity tolerance and analysis of allelic function of HvHKT1 and HvHKT2 in Tibetan wild barley. Theor Appl Genet 122:695–703
Rasoulnia A, Bihamta MR, Peyghambari SA, Alizadeh H, Rahnama A (2011) Proteomic response of barley leaves to salinity. Mol Biol Rep 38:5055–5063
Rengasamy P (2010) Soil processes affecting crop production in salt-affected soils. Funct Plant Biol 37:613–620
Rosenthal DM, Locke AM, Khozaei M, Raines CA, Long SP, Ort DR (2011) Over-expressing the C3 photosynthesis cycle enzyme sedoheptulose-1-7 bisphosphatase improves photosynthetic carbon gain and yield under fully open air CO2 fumigation (FACE). BMC Plant Biol 11:123
Shabala S (2000) Ionic and osmotic components of salt stress specifically modulate net ion fluxes from bean leaf mesophyll. Plant, Cell Environ 23(8):825–837
Shabala S, Cuin TA (2008) Potassium transport and plant salt tolerance. Physiol Plantarum 133(4):651–669
Shabala S, Shabala S, Cuin TA, Pang JY, Percey W, Chen ZH, Conn S, Eing C, Wegner LH (2010) Xylem ionic relations and salinity tolerance in barley. Plant J 61:839–853
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91(5):503–527
Tyerman SD, Niemietz CM, Bramley H (2002) Plant aquaporins: multifunctional water and solute channels with expanding roles. Plant Cell Environ 25:173–194
Wang WS, Zhao XQ, Li M, Huang LY, Xu JL, Zhang F, Cui YR, Fu BY, Li ZK (2016) Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. J Exp Bot 67(1):405–419
Wen GQ, Cai L, Liu Z, Li DK, Luo Q, Li XF, Wan JM, Yang Y (2011) Arabidopsis thaliana VDAC2 involvement in salt stress response pathway. Afr J Biotechnol 10(55):11588–11593
Wu DZ, Qiu L, Xu LL, Ye LZ et al (2011) Genetic variation of HvCBF genes and their association with salinity tolerance in Tibetan annual wild barley. PLoS ONE 6:e22938
Wu DZ, Cai SG, Chen MX, Ye LZ, Chen ZH, Zhang HT, Dai F, Wu FB, Zhang GP (2013a) Tissue metabolic responses to salt stress in wild and cultivated barley. PLoS One 8:55431
Wu DZ, Shen QF, Cai SG, Chen ZH, Dai F, Zhang GP (2013b) Ionomic responses and correlations between elements and metabolites under salt stress in wild and cultivated barley. Plant Cell Physiol 54(12):1976–1988
Wu DZ, Shen QF, Qiu L, Han Y, Ye LZ, Jabben Z, Shu QY, Zhang GP (2014) Identification of proteins associated with ion homeostasis and salt tolerance in barley. Proteomics 14:1381–1392
Zhang B, Li PF, Fan FC (2012a) Ionic relations and proline accumulation in shoots of two Chinese Iris germplasms during NaCl stress and subsequent relief. Plant Growth Regul 68(1):49–56
Zhang H, Han B, Wang T, Chen S, Li HY, Zhang YH, Dai SJ (2012b) Mechanisms of plant salt response: insights from proteomics. J Proteome Res 11:49–67
Zhu JK (2002) Salt and drought stress signal transduction in plants. Ann Rev Plant Biol 53:247–273
Acknowledgments
We are grateful to Prof. Dongfa Sun (Huazhong Agricultral University, China) for providing seeds of Tibetan wild barley accession. This research was supported by Natural Science Foundation of China (31330055, 31301246), China Agriculture Research System (CARS-05) and the Fundamental Research Funds for the Central Universities (2016QNA6013).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shen, Q., Fu, L., Qiu, L. et al. Time-course of ionic responses and proteomic analysis of a Tibetan wild barley at early stage under salt stress. Plant Growth Regul 81, 11–21 (2017). https://doi.org/10.1007/s10725-016-0180-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0180-0