Abstract
Drought and salinity stresses are adverse environmental factors that affect crop growth and yield. Proteomic analysis offers a new approach to identify a broad spectrum of genes that are expressed in living system. We applied this technique to investigate protein changes that were induced by salinity in barley genotypes (Hordeum vulgare L.), Afzal, as a salt-tolerant genotype and L-527, as a salt-sensitive genotype. The seeds of two genotypes were sown in pot under controlled condition of greenhouse, using a factorial experiment based on a randomized complete block design with three replications. Salt stress was imposed at seedling stage and leaves were collected from control and salt-stressed plant. The Na+ and K+ concentrations in leaves changed significantly in response to short-term stress. About 850 spots were reproducibly detected and analyzed on 2-DE gels. Of these, 117 proteins showed significant change under salinity condition in at least one of the genotypes. Mass spectrometry analysis using MALDI-TOF/TOF led to the identification some proteins involved in several salt responsive mechanisms which may increase plant adaptation to salt stress including higher constitutive expression level and upregulation of antioxidant, upregulation of protein involved in signal transduction, protein biosynthesis, ATP generation and photosynthesis. These findings may enhance our understanding of plant molecular response to salinity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil salinity is a prevalent abiotic stress that limits the productivity and geographical distribution of plants [1]. Increasing salinization of agricultural soils is one of the most challenging issues faced by modern agriculture. In excess of 30% of cultivated soils are affected by salinity [2, 3]. Much of this salinization is attributed to the infiltration and accumulation of NaCl [2], often resulting in soil Na+ concentration above 40 mM and growth suppression in most crop [3]. Natural phenomena and human activities such as irrigation can cause salts to accumulate salts in soil [1].The United Nation Environmental program estimated that 20% of agricultural land and 50% of cropland in the world is salt-stressed [4]. Salinity causes water deficit, ion toxicity and nutrient deficiency leading to molecular damage, growth and yield reduction even plant death [5].
Barley is one of the most salt-tolerant crop species among the glycophytes, with genotypes that can germinate in seawater (i.e., about 47 ds m−1). However, the literature is replete with conflicting reports as to whether a positive phenotypic or genetic correlation exists between its tolerance at the germination and seedling growth stages under stress and non-stress conditions, or whether salt tolerance at the germination and seedling growth stages is expressed at subsequent growth stages [4]. Screening for salt-tolerant barley germplasm is important to determine whether there is a genetic basis for selection and breeding purposes [6]. But the screening of large numbers of germplasm accession for small, repeatable and quantifiable differences is very difficult [7].
Proteomics is a powerful tool to analyze biochemical pathway and the complex response of plants to environmental stresses. Also this is a recent addition to the molecular tools used to analyze salinity affected plants. Patterson et al. examined Boron toxicity responsive proteins of two barley lines. The concentration of 5 mM of Boric acid was submitted at seedling stage. Proteins were extracted from root and leaves and analyzed by 2-DE. They identified several responsive proteins belonged to metabolic processes like photosynthetic carbon dioxide assimilation and photorespiration [8].
The salt responsive proteins in the roots of salt-tolerant rice variety pokkali and the salt-sensitive variety IR29 were studied. They identified several salt responsive proteins including an ABA-stress-responsive protein (ASR1), ascorbate peroxidase, and caffeoyl CoA O-methyltransferase. The last of these proteins showed similarity to one of CCOMTs present in the rice genome and plays an important role in suberin and lignin biosynthesis [9].
To further understand the mechanism of plant response to salinity, we employed a proteomic approach to profile the protein change of barley seedling leaves. To the best of our knowledge, this is the first report utilizing proteomics to discover Iranian barley seedling leaves response to salinity.
Materials and methods
Plant material and treatments
Seeds of salt-tolerant genotype, Afzal and salt-sensitive genotype, L.527 were obtained from the seeds and plant improvement institute of Iran. Uniformly sized and weighted seeds were selected, surface-sterilized with hypochlorite 1% and planted into pots under controlled condition of greenhouse by using a factorial experiment based on a randomized complete block design with three replications. Salinity was applied at seedling stage. 50 mM NaCl was added twice a day over 3 days to a final concentration of 300 mM, 24 h after final concentration of NaCl leaves were collected from control and salt-stressed plants. Fresh leaf samples were ground to a fine powder in liquid nitrogen and then were preserved in −80°C until use.
Ion measurements
The leaves were oven dried at 70°C for 48 h and ground after being weighed. The Na+ and K+ concentrations were measured in triplicate using flame photometrical after extraction with 500 mM HNO3[10].
Statistical analysis
Statistical analysis was done using SPSS version 13 and Melanie version 7.0 for spot statistic analysis.
Protein extraction and 2-DE
Protein was extracted from the leaves according TCA/acetone method [11]. Triplicate tissue samples were ground in liquid nitrogen with mortal and pestle. Approximately 1 g of ground leave was suspended in 10 ml extraction buffer consisting of 10%w/v TCA in acetone with 0.07%w/v DDT at −20°C for 2 h, followed by centrifugation for 15 min at 3500×g. The pellets were washed with ice cold acetone containing 0.07% DDT, incubated at −20°C for 1 h and centrifuged again at 4°C. This step was repeated for twice then pellets were lyophilized. The sample powder then was solubilized in lysis buffer (9 M urea, 2%w/v CHAPS, 0.8%w/v pharmalyte, pH 3–10, 1%w/v DDT), and then protein concentration was determined by the Bradford assay with BSA as the standard.
The IEF was performed at 20°C with using Multiphore II and dry strip kit (Amersham pharmacy Biotech). For analytical and preparative gels, the 18 cm IPG strip (pH 4–7) were rehydrated overnight with 350 μl of rehydration buffer (8 M urea, 0.5% CHAPS, 20 mM DDT, 0.5%v/v IPG buffers) in a reswelling tray (Amersham Pharmacia, Uppsala, Sweden) at room temperature. For analytical and preparative gels, 120 μg and 1.2 mg of protein were loaded, respectively. The running program was as follows 500 V 2 h, followed by 1000 V for 2 h, and finally 3000 V for 14 h. The focused IPG strips were equilibrated twice for 15 min in 10 ml of equilibration solution. The first Equilibration solution contained 50 mM Tris–HCL buffer, pH 8.8, 6 M urea, 30% (v/v) glycerol, 2% (w/v) SDS and 1% (w/v) DDT with a few grains of bromophenol blue. The second equilibration was performed in solution modified by the replacement of DDT by 2.5%w/v iodoacetamide. The second dimension was developed by SDS-PAGE in vertical slab of acrylamide (12.5% monomer, with 2.6% cross linker) using a PROTEAN II MULTI Cell (Bio-Rad, Hercules, CA, USA). Analytical gels were stained with silver nitrate and preparative gels were stained with colloidal Coomassie Brilliant Blue G-250 [12].
Image and data analysis
GS-800 densitometer (Bio-Rad) at a resolution of 600 dots per square inch (dpi) was used for scanning the silver-stained gels. The scanned gels were saved as TIF images for subsequent analysis. Spot quantification was carried out using the Melanie 7 software (GeneBio, Geneva, Switzerland). The molecular masses of proteins on gels were determined by electrophoresis of standard protein markers (Amersham Biosciences) and pI of the proteins determined by the migration of the protein spots on 18 cm IPG (pH 4–7, linear) strips. The percent volume of each spot was estimated and analyzed to determine protein abundance. Statistical analysis of the relative abundance of each matched protein spot among the control and treated leaves of tolerant and sensitive genotype was accomplished using a Two-tailed T-test. Only quantitative differences with a P-value of at least <0.05 were considered.
Protein identification and database search
Protein spots were excised from preparative CBB- or silver-stained gels and digested with trypsin [13]. The digestion was performed over 4 h at 37°C using a minimum of 10 ml of digest solution (5 ng/ml trypsin in 10 mM ammonium bicarbonate). Tryptic peptides were eluted using 10 μl of a mixture of 50% ACN/water with 1% TFA.
MALDI-MS was used to acquire PMFs and MS/MS spectra from tryptic fragments. The supernatant (5 ml) was placed on a thin layer of CHCA covering the area of the anchor. The sample was shrunk, dried and concentrated onto an MTP-384-AnchorChip™ sample plate (Bruker Daltonik, Bremen, Germany) with a diameter of the anchors of 800 μm [14]. In order to eliminate salts and other water soluble contaminants, the preparation was washed using 10 ml of 10 mM diammonium phosphate and 0.1% TFA and recrystallized with 0.5 μl of acetone/ethanol/0.1% TFA (3:6:1). The PMF and MS/MS were performed using Ultraflex II TOF/TOF (Bruker Daltonik). The MS/MS was carried out by means of the LIFT-TOF/TOF mode of the UltraflexII. The samples were measured, post processed, and identified in a fully automated fashion using the software AutoXecute™, Flex-Analysis™, and Proteinscape. Mass spectral data were then submitted for NCBInr database searching program MASCOT version 2.1.04 (http://www.matrixscience.com) for protein identification.
Result and discussion
Ion concentrations
Na+ and K+ concentration of leaves in tolerant and sensitive genotypes were markedly affected by salinity (Table 1). Genotypic differences in ion accumulation rate were found between genotypes. Na+ concentration in the leaf of genotypes was significantly increased in response to salinity and Afzal as a tolerant genotype exhibited good control of Na+ accumulation compared to L-527 on a Na+ concentration basis. The Na+ concentration of leaf increased in tolerant and sensitive genotype up to 0.6 and 1.3 (mg/g dry weight) respectively. However, the control of Na+ accumulation was better in tolerant genotype and on a Na+ concentration basis, salt sensitive genotype differed 2-fold in leaf accumulation of Na+ from tolerant genotype.
There was a significant difference between genotypes in K+ concentration and Afzal remarkably showed the higher K+ concentration than L-527. The K+ concentration was significantly increased in response to salinity in tolerant genotype but it was decreased in sensitive genotype (Table 1).
It has been shown that low sodium concentration does not have considerable inhibitory effects on the metabolism and enzyme activity. Instead it may affect the apical meristem through a signaling process [15]. Higher Na+ concentration in roots restricted K+ adsorption. Na+ adsorption competes with K+ and decreases K+ adsorption in the root [16]. Moreover, the increase of K+ concentration was observed in some plant leaves under salinity [17]. Although whole plant and individual tissue K+ decreased with increasing salinity but it plays important roles in salt tolerance [18]. However, the mechanism which be caused to maintain either retention of K+ or preventing Na+ from accumulating in leaves, is a key feature for salt tolerance.
Identification of salt responsive proteins
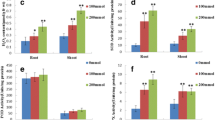
We applied 2-DE to analyze the leaf proteomes of the two barley genotypes in response to short-term salt stress. Salt-responsive proteins were excised from preparative gels and examined by MALDI-TOF/TOF. A spot that consistently showed an equal position in the different gels was considered to be the same protein. About 850 spots were reproducibly detected in three replications of each genotype, that is, Afzal, salt-tolerant (Fig. 1), and L.527. Out of these, 117 proteins showed significant change under salinity condition in at least one of the genotypes (Fig. 1 and Supporting Information Table 2). Mass spectrometry analysis using MALDI-TOF/TOF led to the identification of 22 proteins (Supporting Information Table 2).
2-D PAGE protein pattern. Protein (120 μg) was loaded on 18 cm IPG strip with linear gradient (pH 4-7) and SDS-PAGE was performed with 12.5% gel. Proteins were visualized by silver staining. Arrow represents salt-responsive spots of which have been identified by MS (Table 2)
Total number of salt-responsive proteins were higher in sensitive genotype but the number of up-regulated proteins (P < 0.05) in tolerant genotype, Afzal, were more than the sensitive genotype, L.527 (Fig. 2). Most of the proteins up-regulated in salt tolerant genotype have shown change in sensitive genotype. On the contrary, a small number of proteins up-regulated in the sensitive genotype either showed no change or down-regulated in the tolerant counterpart (Fig. 3).
Function of salt-responsive proteins
The proteins function in fundamental processes, including stress defense, metabolism, protein synthesis and photosynthesis.
Proteins involved in photosynthesis
Present concepts of the photosynthetic carbon reduction cycle require the synthesis of three ATP molecules per CO2 molecule reduced to the level of carbohydrate.
It is interesting that preparations which have the highest capacity for CO2 fixation are notoriously poor in catalyzing photophosphorylation [19].
Phosphoribulokinase (PRK), and sedoheptulose-1,7-bisphosphatase (SBPase) have been considered as key enzymes in Benson–Calvin cycle, because their regulation by dark/light transitions is responsible for the regulation of the cycle. Phosphoribulokinase seems to be a regulatory enzyme within the photosynthetic carbon reduction cycle. It provides the immediate CO2 acceptor; therefore, the control of its activity by a photo-reduced reducing agent is a logical site of regulation. Since it is activated very rapidly, the kinase cannot, by itself, account for the much longer lag periods encountered in the kinetics of CO2 fixation is isolated chloroplasts [20]. Sedoheptulose-1,7-bisphosphatase catalyses the dephosphorylation of Sed(1,7)P2, forming sedoheptulose 7-phosphate (Sed7P) and inorganic phosphate, and this is an essentially irreversible reaction which commits intermediates to the regenerative part of the photosynthetic carbon-reduction cycle. The activity of sedoheptulose-1,7-bisphosphatase is increased significantly in light by reduced thioredoxin [21].
Plastocyanin (PC) is a copper-containing protein of around 12 KDa in size. Photosystem I subunit VII serves as a electron carriers between the Cyt b6f complex and PSI in higher plants. Plastocyanin is located in the lumen of the thylakoid membrane system in chloroplasts and is essential for linear as well as cyclic electron transport [22, 23]. The plastocyanin gene is expressed in photosynthetic tissue in a developmentally regulated manner and the expression is stimulated by light [24]. It has been concluded that a PC-dependent flux control adjusts the electron transport rate in such a way that PSI is protected against over reduction under stress conditions where carbon assimilation is limited as it occurs under sink limitation or sugar depression of photosynthesis [23].
Spot 314 and 216 were identified as Phosphoribulokinase and sedoheptulose 1,7-bisphosphatase, respectively. Both of them were up-regulated in tolerant genotype and down-regulated in the sensitive genotype. Spot 44 and 278 were identified as plastocyanin and photosystem I subunit VII proteins. Our results showed differential expression of these proteins in tolerant and sensitive genotype. Down-regulation of these proteins suggests reduce of carbon reduction in the stroma of chloroplast is due the stomata closure in sensitive genotype [25].
Spot 165 was identified as Oxygen-evolving enhancer protein 2(OEE2), chloroplastic which is up regulated in tolerant and down-regulated in sensitive genotype. This polypeptide has been implicated in photosynthetic oxygen evolution, and it is associated with the photosystem II complex, the site of oxygen evolution exist in all higher plants and algae. Increased expression of OEE2 under salt stress has been reported in Mangrove and it is known that OEE2 and OEE3 can easily be removed from the PSII complex in the presence of NaCl [26]. Increased expression levels of OEE2 might be needed to repair protein damage caused by NaCl. Murota et al. reported a role for OEE2 in salt adaptation in photoautotrophically cultured green tobacco cells. They observed strong association of OEE2 with thylakoid membranes in NaCl adapted cells and dissociation in non-adapted cells. This is consistent with a role for OEE2 in protecting barley from salt stress [27].
ROS detoxification key enzymes
There are many potential sources of ROS in plants. Some are involved in normal metabolism such as photosynthesis and respiration. Reactive oxygen species (ROS), such as the superoxide anion and hydrogen peroxide, are generated by the photosystems because photo excited electrons are often generated in excess of requirements for CO2 fixation and used for reducing molecular oxygen, even under normal environmental conditions. Moreover, ROS generation is increased in chloroplasts if plants are subjected to stresses, such as drought, high salinity and chilling [28, 29]. The enhanced amount of ROS can be viewed as a threat for cell, and as secondary messengers involved in the stress-response signal transduction pathway.
Therefore, plant cells require two different mechanisms which will enable the detoxification of excess ROS and fine modulation of ROS for signaling purpose.
SOD acts as the first line of defense converting a superoxide to the less toxic hydrogen peroxide molecules. The detoxification of H2O2 is accomplished with ascorbate peroxidase, glutathione peroxidase, thioredoxin peroxidase, catalase, and 2-Cys peroxiredoxin [30]. Thioredoxin is a member of a family of small proteins that appear to be ubiquitous, Thioredoxin functions in scavenging, but perhaps more importantly, the protein regulates a number of fundamental seed processes [31, 32].
Thioredoxin is also involved in a range of biochemical processes. These include the mobilization of protein and starch in germinating cereal seeds, self-incompatibility and cellular protection against oxidative stress, particularly during seed desiccation and germination.
Thioredoxin reduce a variety of target proteins that contain disulfide bonds, including storage proteins, such as glutenins and gliadins in wheat, hordein and glutelin in barley and proteins related to oxidative stress such as peroxiredoxin [33]. Spot 382 and 283 identified as super oxide dismutase and thioredoxine peroxidase. These enzymes showed decisive differential expression in the tolerant and sensitive genotypes.
Glutathione S-Transferase contributes in ROS scavenging. The enzyme limits oxidative damage by removing ROS formed in stress and by detoxifying xenobiotics under normal conditions. The endogenous products of oxidative damage, for example, membrane lipid peroxides and products of oxidative DNA degradation are highly cytotoxic [33]. Spot 136 was identified as this enzyme. Our result showed the increase of this enzyme in tolerant genotype compared to the sensitive genotype. In cotton ovules cultured in vitro, tolerance of salt stress is associated with higher constitutive levels of ascorbate peroxidase and glutathione reductase and a greater capacity to up-regulate superoxide dismutase [34]. Over expression of an Arabidopsis peroxisomal ascorbate peroxidase gene in tobacco increased protection against oxidative stress [35]. It was shown that in the absence of stress, ascorbate peroxidase is 4.4-fold more abundant in salt-tolerant rice genotype Pokkali roots than in salt sensitive genotype IR29 roots [36]. They suggested that the greater salt tolerance of Pokkali compared with IR29 may be due to a higher constitutive level of antioxidant capacity. In salt-tolerant rice genotype, IR651, SOD and thioredoxin do not show significantly change compared to the control under 75 mM NaCl stress at young panicle [37].
Glutathione S-transferase, thioredoxin and 1-cys-peroxiredoxin have increased in tolerant genotype, Khazar, under drought stress but they had opposite change in drought-sensitive genotypes, Arvand and Afghani [33].
In 1996, cDNA sequences referred to as plant peroxiredoxins (Prx), i.e., a 1-Cys Prx and a 2-Cys Prx, were reported from barley [38]. Peroxiredoxins are ubiquitous antioxidant enzymes found in all organisms, with the single exception, to our knowledge, of Borrelia burgdorferi (and other Borrelia species). The broad distribution of Prxs and the high levels of expression suggest that they are both an ancient and important enzyme family [39].
Prx has several functions in plant. (A) They interact with proteins such as glutaredoxins, thioredoxins, and cyclophilins as reductants, but also non-dithioldisulphide exchange proteins. (B) By work with transgenic plants, their activity was shown to affect metabolic integrity, (C) protect DNA from damage in vitro and in vivo, and (D) pathogen defense [38].
Chloroplast-localized 2-cys peroxiredoxins assume the principal role of scavenging hydrogen peroxide [40]. Spot 119 identified as 2-cys peroxiredoxin increased significantly in response to salinity stress in tolerant genotype, but in sensitive genotype had opposite change. It is possible that the higher constitutive level of antioxidants expression confer the greater oxidative stress tolerance of Afzal salt-tolerant genotype compared to the L.527 sensitive genotype.
ATP generating proteins
Triosephosphate isomerase (spot 152) and FBP aldolase (spot211) have differential expression and up-regulated in salt-tolerant genotype. ATP is required for many biosynthetic pathways in plant cells, and during external stress maintenance of energy requirements may increase considerably. The up-regulation of these enzymes has been observed under salt, drought, and cold conditions [36]. Because of the central role of these enzymes in carbon metabolism and energy transduction, their increased abundance under stress presumably reflects altered patterns of carbon flux in response to reduced photosynthesis and increased need for osmotic adjustment in panicle. The up-regulation of Triosephosphate isomerase and FBP aldolase were observed in rice leaves and panicles under salinity and drought conditions [36, 37].
The salt-responsive protein spot 211 was identified as chloroplasts type FBP aldolase, an enzyme of the glycolytic pathway, which reversibly catalyzes the conversion of FBP to glyceraldehyde-3-phosphate and dihydroxyacetone phosphate. Enhanced expression of FBP aldolase in salt-treated rice was previously reported [41] also the up-regulation of this enzyme in mangrove plant under salinity [42]. In this study, the expression of FBP aldolase has up-regulate significantly in tolerant and sensitive genotype. These results and earlier reports of enhanced expression of aldolases under stress conditions demonstrate that expression of these proteins may play a role in acclimation of barley seedling to anaerobic condition created by oxidative stress. Enhanced expression of FBP aldolase would increase the flow of carbon through the Calvin cycle and lead to an increase in sucrose and amino acid production via increases in carbon flux through glycolysis. These traits would also lead to osmolite production and contribute to stress tolerance. It was reported that the activity of photosynthesis-related enzymes including FBP aldolase was inhibited by salt stress in tobacco; however, genetically engineered tobacco with the ability to synthesize glycinebetaine showed increased salt tolerance and reduced inhibition of the activity of these enzymes, suggesting that glycinebetaine protects the enzymes against salt stress [43]. These results suggest that FBP aldolase plays an important role in salt tolerance mechanisms.
Magnesium chelatase 40-kDa subunit
Chloroplast development and the development of the entire plant are entirely dependent on chlorophyll biosynthesis. Magnesium chelatase, the first committed enzyme in the chlorophyll pathway, can therefore be expected to regulate not only chlorophyll biosynthesis but also the development of the plant in a larger context. Magnesium chelatase is a complex enzyme consisting of six 40-kDa subunits, six 70-kDa subunits [44], and an unknown number of 150-kDa subunits. In barley (Hordeum vulgare) the 40-, 70-, and 150-kDa subunits are called XanH, XanG, and XanF, respectively, as they are encoded by the genes Xantha-h, -g, and -f [45]. Magnesium chelatase catalyze the first unique step of the chlorophyll biosynthetic pathway. These findings suggest that chlorophyll biosynthetic intermediates could act as signaling molecules from plastid-to-nucleus and regulates the transcription of nuclear localized photosynthetic genes. Initially, the 150 kD, a magnesium chelatase subunit was, suggested to have a specific role in the plastid-to-nucleus signaling pathway [46].In this study spot 220 identified as magnesium chelatase and was up regulated in both genotype.
Cysteine synthase
Cysteine (Cys) synthesis in plants is a fundamental process for protein biosynthesis and all anabolic pathways that require reduced sulfur. Bacteria and fungi are able to reduce sulfate by assimilatory sulfate reduction to sulfide and to integrate it into Cys in the cytosol. In contrast, mammals need to take up reduced sulfur as Met and can synthesize Cys via trans-sulfurylation in the cytosol. Protein biosynthesis in the mitochondria in animals and fungi receives Cys, therefore, from the cytosol. In plants, the assimilatory sulfate reduction pathway is localized to plastids, whereas Cys synthesis is found in the cytosol, plastids, and mitochondria [47]. Enhanced expression of this enzyme was observed in rice seedling under oxidative stress of H2O2 [48]. Spot 200 was identified as cysteine synthase and up regulated in both genotypes. Cysteine synthase is responsible for the final step in cystein biosynthesis, the key limiting step in producing glutathione, which is involved in resistance to adverse stresses [49]. Regarding this result and earlier reports, it seems cysteine synthase has an important role in protein biosynthesis.
Concluding remarks
To the best of our knowledge, this is the first proteome analysis barley seedling in response to salinity stress. 2-DE coupled with MS led to the identification of several mechanisms and proteins which may be involved in plant adaptation to salt stress. In addition to providing new information on the response to salinity stress, the present study offers opportunities to pursue the breeding of barley with enhanced salt tolerance. Specific areas for further study include:
-
1.
The analysis of other salt sensitive and other salt-tolerant genotypes. Proteomic analysis of the other genotypes will show whether the protein responses observed here have broad adaptive significance.
-
2.
The analysis proteomes of recombinant inbred lines or doubled haploid lines derived from tolerant and sensitive lines. This analysis will verify the association of these proteins with salt tolerance.
-
3.
We are interested in Analyzing protein response of other tissues such root and reproductive stage of plant development because the structure of root differ from leaf and reproductive stage are sensitive to the salinity stress.
References
Wiebe BH, Eilers RG, Eilers WD, Brierly JA (2007) Application of a risk indicator for assessing trends in dry land salinization risk on the Canadian Prairies. Can J Soil Sci 87(2):213–224
Zhu Jk (2001) Plant salt tolerance. Trends Plant Sci 6(2):66–71. doi:10.1016/S1360-1385(00)01838-0
Munns RA (2005) Gene and salt tolerance: bringing them together. New Phytol 167(3):645–663. doi:10.1111/j.1469-8137.2005.01487.x
Jaradat AA, shahid M, Al Maskari AY (2004) Genetic diversity in the batini barley landrace from Oman. Crop Sci 44(3):304–315
Zeng L, Shannon MC (2000) Salinity effect on seedling growth and yield component of rice. Crop Sci 40(4):996–1003
Alonso SI, Guma IR, Clausen AM (1999) Variability for salt tolerance during germination in Lolium multiflorum Lam natural in the Pampean grassland. Genet Res Crop Evol 46(1):87–94. doi:10.1023/A:1008638325484
Foolad MR, Chen FQ, Lin GY (1998) RFLP mapping of QTLs conferring salt tolerance during germination in an interspecific cross of tomato. Theor Appl Genet 97(7):1133–1144. doi:10.1007/s001220051002
Patterson J, Ford K, Cassin A, Natera S, Basic A (2007) Increased abundant of proteins involved in phytosiderophore production in Boron-tolerant barley. Plant Physiol 144:1612–1631. doi:10.1104/pp.107.096388
Salekdeh GhH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J (2002) A proteomics approach to analyzing drought- and salt-responsiveness in rice. Field Crop Res 76(2–3):199–219
Chapman HD, Pratt PF (1961) Methods of analysis for soils, plants and waters. Div Agric Sci Univ Calif Berkeley, California
Damerval C, de Vienne D, Zivy M, Thiellement H (1986) Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat seedlings proteins. Electrophoresis 7(1):52–54. doi:10.1002/elps.115007010
Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8(2):93–99. doi:10.1002/elps.1150080203
Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68(5):850–858. doi:10.1021/ac950914h
Gobom J, Schuerenberg M, Mueller M, Theiss D (2001) Alpha-cyano-4-hydroxycinnamic acid affinity sample preparation—a protocol for MALDI-MS peptide analysis in proteomics. Anal Chem 73(3):434–438. doi:10.1021/ac001241s
Munns R, Rawson HM (1999) Effect of salinity on salt accumulation and reproductive development in the apical meristem of wheat and barley. Aust J Plant Physiol 26(5):459–464. doi:10.1071/PP99049
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25(2):239–250. doi:10.1046/j.0016-8025.2001.00808.x
Garcia-sanchez F, Jifon JL, Carvajal M, Syvertsen JP (2002) Gas exchange, chlorophyll and nutrient contents in relation to Na+ and Cl− accumulation in ‘Sunburst’ mandarin grafted on different rootstocks. Plant Sci 162:705–712
Schachtman DP, Liu WH (1999) Molecular pieces to the puzzle of interaction between potassium and sodium uptake in plants. Trends Plant Sci 4(7):281–287
Avron M, Gibbs M (1974) Properties of phosphoribulokinase of whole chloroplasts. Plant Physiol 53(2):136–139. doi:10.1104/pp.53.2.136
Graciet E, Lereton S, Gontero B (2004) Emergence of new regulatory mechanisms in the Benson–Calvin pathway via protein- protein interactions: a glyceraldehyde-3-phosphatedehydrogenase/CP12/phosphoribulokinase complex. J Exp Bot 55(400):1245–1254. doi:10.1093/jxb/erh107
Christine AR, Julie CL, Nicola MW, Susan P, Tristan AD (1992) cDNA and gene sequences of wheat chloroplast sedoheptulose-1,7-bisphosphatase reveal homology with fructose-l,6-bisphosphatases. Eur J Biochem 205(3):1053–1059. doi:10.1111/j.1432-1033.1992.tb16873.x
Fischer N, Hippler M (1998) The PsaC subunit of photosystem I provides an essential lysine residue for fast electron transfer to ferredoxin. EMBO J 17(4):849–858. doi:10.1093/emboj/17.4.849
Schutze K, Steiner S, Pfannschmidt T (2008) Photosynthetic redox regulation of the plastocyanin promoter in tobacco. Physiol Plantarum 133(3):557–565. doi:10.1111/j.1399-3054.2008.01118.x
Nielsen PS, Causing K (1993) In vitro binding of nuclear proteins to the barley plastocyanin gene promoter region. Eur J Biochem 217(1):97–104. doi:10.1111/j.1432-1033.1993.tb18223.x
Chaves MM, Flaxe J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103(4):551–560. doi:10.1093/aob/mcn125
Sugihara K, Hanagata N, Dubinsky Z, Baba S, Karube I (2000) Molecular characterization of cDNA encoding oxygen evolving enhancer protein 1 increased by salt treatment in the mangrove Bruguiera gymnorrhiza. Plant Cell Physiol 41(11):1279–1285. doi:10.1093/pcp/pcd061
Murota KI, Ohshita Y, Watanabe A, Aso S, Sato F, Yamada Y (1994) Changes related to salt tolerance in thylakoid membranes of photoautotrophically cultured green tobacco cells. Plant Cell Physiol 35(1):107–113
Peskan-Berghöfer T, Neuwirth J, Kusnetsov V, Oelmüller R (2005) Suppression of heterotrimeric G-protein β-subunit affects anther shape, pollen development and inflorescence architecture in tobacco. Planta 220(5):737–746. doi:10.1007/s00425-004-1393-4
Bolwell GP, Wojtaszek P (1999) Role of active oxygen species and NO in plant defense responses. Curr Opin Plant Biol 2(4):287–294
Mehlhorn H, Lelandais M, Korth HG, Foyer CH (1996) Ascorbate is the natural substrate for plant peroxidases. FEBS Lett 378(3):203–206. doi:10.1016/0014-5793(95)01448-9
Schurmann P, Jacquot JP (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51:371–400. doi:10.1146/annurev.arplant.51.1.371
Yano H, Kuroda S, Buchanan BB (2002) Disulfide proteome in the analysis of protein function and structure. Proteomics 2(9):1090–1096. doi:10.1002/1615-9861(200209)2:9<1090:AID-PROT1090>3.0.CO;2-1
Hajheydari M, Eivazi A, Buchman BB, Wong JH, Majidi I, Salekdeh GH (2007) Proteomics uncovers a role for redox in drought tolerance in wheat. J Proteome Res 6(4):1451–1460. doi:10.1021/pr060570j
Rajguru SN, Banks SW, Gossett DR, Lucas MC, Millhollon EP (1999) Antioxidant response to salt stress during fiber development in cotton ovules. J Cotton Sci 3:11–18
Salekdeh GH, Siopongco J, Wada LJ, Ghareyazi B, Bennett JP (2000) Proteomic analysis of rice during drought stress and recovery. Proteomics 2:1131–1145
Wang J, Zhang H, Allen RD (1999) Overexpression of an Arabidopsis peroxisomal ascorbate peroxidase gene in tobacco increases protection against oxidative stress. Plant Cell Physiol 40(7):725–732
Dadashi Dooki A, Mayer-Posner FJ, Askari H, Ziaee AA, Salekdeh GH (2006) Proteomic response of rice young panicles to salinity. Proteomics 6(24):6498–6507. doi:10.1002/pmic.200600367
Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, Mariana S, Miranda ND, Barier M, Finkemeier I (2006) The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot 57(8):1697–1709. doi:10.1093/jxb/erj160
Hall A, Karplus AP, Poole BL (2009) Typical 2-Cys peroxiredoxins—structures, mechanisms and functions. FEBS J 276(9):2469–2477. doi:10.1111/j.1742-4658.2009.06985.x
Kitajima S (2008) Hydrogen peroxide-mediated inactivation of two chloroplastic peroxidases, ascorbate peroxidase and 2-Cys peroxiredoxin. Photochem Photobiol 84(6):1404–1409. doi:10.1111/j.1751-1097.2008.00452.x
Abbasi FM, Komatsu S (2004) A proteomic approach to analyze salt-responsive proteins in rice leaf sheath. Proteomics 4(7):2072–2081. doi:10.1002/pmic.200300741
Tada Y, Kashimura T (2009) Proteomic analysis of salt-responsive proteins in the mangrove plant, Bruguiera gymnorhiza. Plant Cell Physiol 50(3):439–446. doi:10.1093/pcp/pcp002
Yang X, Liang Z, Wen X, Lu C (2008) Genetic engineering of the biosynthesis of glycinebetaine leads to increased tolerance of photosynthesis to salt stress in transgenic tobacco plants. Plant Mol Biol 66(1–2):73–86. doi:10.1007/s11103-007-9253-9
Elmlund H, Lundqvist J, Al-Karadaghi S, Hansson M, Hebert H, Lindahl M (2008) A new cryo-EM single-particle ab initio reconstruction method visualizes secondary structure elements in an ATP-fueled AAA+ motor. J Mol Biol 375(4):934–947. doi:10.1016/j.jmb.2007.11.028
Von Wettstein D, Henningsen KW, Boynton C, Kannangara G, Nielsen OF (1971) The genetic control of chloroplast development in barley. North-Holland Publishing Company, Amsterdam
Gadjieva R, Axelson E, Olsson U (2005) Analysis of gun phenotype in barley magnesium chelatase and Mg-protoporphyrin IX monomethyl ester cyclase mutants. Plant Physiol Biochem 43(10–11):901–908. doi:10.1016/j.plaphy.2005.08.003
Haas FH, Heeg C, Queiroz R, Bauer A, Witz M, Hell R (2008) Mitochondrial serine acetyl transferase functions as a pacemaker of cysteine synthesis in plant cells. Plant Physiol 148(2):1055–1067. doi:10.1104/pp.108.125237
Wan XY, Liu JY (2008) Comparative proteomics analysis reveals an intimate protein network provoked by hydrogen peroxide stress in rice seedling leaves. Mol Cell Proteomics 7(8):1469–1488. doi:10.1074/mcp.M700488-MCP200
May MJ, Vernoux T, Leaver C, Van Montagu M, Inze D (1998) Glutathione homeostasis in plants: implications for environmental sensing and plant development. J Exp Bot 49(321):649–667
Acknowledgment
We are grateful to Professor Claire Gay, York university of UK, facility for performing the MS analysis and associated bioinformatics.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rasoulnia, A., Bihamta, M.R., Peyghambari, S.A. et al. Proteomic response of barley leaves to salinity. Mol Biol Rep 38, 5055–5063 (2011). https://doi.org/10.1007/s11033-010-0651-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0651-8