Abstract
The prairie ecotype of Stellaria longipes (Caryophyllaceae) is highly responsive to light and is a shade avoider. Using the prairie ecotype as a model, we investigated the expression of the PHYTOCHROME B (PHYB) gene in response to different ratios of red to far-red light (R/FR), photoperiods and ethephon, an ethylene producing compound. This was done to examine the potential role of the PHYB and its interaction with ethylene in determining the plasticity of stem elongation. The PHYB gene was constitutively expressed in flowers, leaves, stems, and roots. The relative transcript abundance of the PHYB gene increased significantly upon transfer of etiolated-seedlings to light. The level of the PHYB gene transcript under elevated R/FR (3.5) was significantly higher than that under lower R/FR (0.7) in de-etiolated-mature plants. Long day photoperiod (16 h light/day, LD) significantly promoted stem growth, whereas plants under short day photoperiod (8 h light/day, SD) showed minor elongation. Interestingly, under LD, but not under SD, the level of PHYB mRNA significantly correlated with stem elongation. The plants treated with high concentration of ethephon (10.0 mol m−3) showed a 55% decrease in PHYB transcript within the first 4 days after treatment, and the level of PHYB transcript was significantly lower than that in control plants and the plants treated with a lower concentration of ethephon (0.1 mol m−3). Taken together with our earlier studies related to stem elongation, our results suggest that the level of PHYB gene transcription on day 4 is critical for the initiation of stem elongation. After this stage, a relatively high level of the PHYB gene transcription might be important for maintaining stem elongation of the prairie plants of S. longipes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Elongation of plant stems is a fundamental growth process that is considerably enhanced under situations like shading by neighbours (Smith 2000; Pierik et al. 2003; Franklin and Whitelam 2005). The major light signal triggering an increase in stem elongation is a reduction in the ratio of red to far-red light (R/FR), which is reduced by the selective reflection of FR in the leaves of neighbouring plants (Ballare 1999; Aphalo et al. 1999). Plants sense and respond to changes in R/FR through phytochromes (Whitelam and Devlin 1997; Smith 2000). Phytochromes in all plants examined so far are encoded by a small divergent gene family (Quail 1994) which is classified into four subfamilies A, B/D, C/F, and E (Mathews and Sharrock 1997; Pratt et al. 1997). In Arabidopsis thaliana, for example, five apophytochrome-encoding genes PHYA to PHYE have been sequenced, characterized, and classified into A, B/D, C/F, and E subfamilies (Sharrock and Quail 1989; Clack et al. 1994).
The functions of individual phytochromes in plant growth and development have been recently revealed by analyzing phytochrome deficient mutants and overexpression lines of individual phytochrome genes. In a number of plant species, studies using phyB-deficient mutants have confirmed a major role of PHYB in mediating shade avoidance by sensing the low R/FR signal (Reed et al. 1993, 1994; Takano et al. 2005; Finlayson et al. 2007). Other phytochromes are likely to play a role in mediating stem elongation, but their roles are less clear. For example, studies using phyD deficient mutant and phyE knockout line revealed that both PHYD and PHYE played minor roles in modulating shade avoidance in Arabidopsis. PHYA plays a major role in perceiving the FR signal during early seedling growth (Tuinen et al. 1995; Takano et al. 2001, 2005; Weller et al. 1997; Franklin et al. 2007). It also plays an important role in responding to changes in R/FR ratio (Johnson et al. 1994; Li et al. 2009). PHYC may play a minor role in mediating shade avoidance in Arabidopsis (Monte et al. 2003; Takano et al. 2005) and Stellaria longipes (Li and Chinnappa 2004), but this has been disputed by Franklin et al. (2003). Given that the strongest evidence for phytochrome involvement in stem elongation was for PHYB, we chose to study this gene in Stellaria longipes following photoperiod and light quality manipulations.
Along with phytochromes and environmental signals, ethylene has also been shown to play a role in mediating shade avoidance response (Goeschl et al. 1967; Imaseki et al. 1971; Samimy 1978; Vangronsveld et al. 1988; Finlayson et al. 1998, 1999; Pierik et al. 2003, 2004; Kurepin et al. 2007). Low R/FR can increase ethylene production in light-grown Sorghum bicolor (Finlayson et al. 1998, 1999). The Sorghum null mutant of PHYB (phyB-1) exhibited a constitutive shade avoidance phenotype and a higher level of ethylene production (Finlayson et al. 1998). In addition, two studies in tobacco revealed that the shade avoidance response was reduced in transgenic ethylene-insensitive tobacco plants (Pierik et al. 2003) and the ethylene production was enhanced by low R/FR in wild type tobacco plants resulting in shade avoidance response (Pierik et al. 2004). They suggested that ethylene positively modulated phytochrome-mediated elongation responses. However, Kurepin et al. (2007) showed that the levels of endogenous ethylene in both sunflower hypocotyls and internodes were lower when the plants were grown under the lower ratio of the R/FR. Those authors suggested that the low ratio of the R/FR caused a reduction of ethylene, allowing maximum stem elongation in sunflower seedlings (Kurepin et al. 2007).
To better understand the potential role of phytochrome B and its interaction with ethylene under the conditions that facilitate stem elongation, this study used the shade-avoiding prairie ecotype of Stellaria longipes as a model to investigate the expression of the PHYB gene in response to different photoperiods, R/FR manipulations, and ethephon applications.
Materials and methods
Plant materials and growth conditions
Plants of the prairie ecotype of Stellaria longipes were originally collected from the Chain Lakes (1,310 m) area in south-western Alberta, Canada. Clonal ramets were raised in 4 cm pots containing peat moss, sands, and Terra green growing medium (2:1:1) and maintained in a growth chamber (Conviron, Winnipeg, Canada) under short-day cold condition (SDC, 8 h photoperiod, 8°C day/4°C night, R/FR = 1.4) for at least 90 days to simulate the winter cycle. Photosynthetically active radiation (PAR) was maintained at μmol photons m−2 s−1. These environmental conditions allow the growth of new shoots and elicit maximum growth and flowering (Macdonald et al. 1984).
Experimental design
To study the expression of the PHYB gene in different organs, plants were transferred from the SDC chamber to a summer-simulated long-day warm (LDW) chamber (16 h photoperiod, 22°C day/18°C night, R/FR = 1.4, PAR = 120 μmol photons m−2 s−1) and maintained under this condition until flowering, at which leaves, stems, and flowers were collected. To obtain roots, the young ramets were planted in 8 cm pots containing sterile sands and maintained in a tray containing 1/2 strength liquid Murashige and Skoog basal medium under LDW for 14 days, at which the fresh roots were collected.
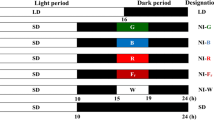
To study the effect of different R/FRs on the expression of the PHYB gene, it was examined in two-week-old seedlings grown in the dark for 14 days and in plants grown under SDC for at least 90 days. To obtain seedlings, seeds were surface-sterilized by soaking in 70% (v/v) alcohol for 10 min and then in 20% (v/v) bleach for 10 min followed by at least five rinses with sterile water. These seeds were subsequently germinated on Murashige and Skoog basal agar medium (Sigma, St. Louis) with 2% (w/v) sucrose in glass jars. The jars were kept in the dark at 4°C for 2 days, treated with white light for 2 h to induce germination, and then placed in the dark at 22°C. After 14 days in the dark, the etiolated-seedlings were transferred to a LDW growth chamber, kept there for 1–12 h, and sampled at specific time points. To obtain etiolated-mature plants, four pots of plants maintained under SDC for at least 90 days were transferred to darkness at 22°C and kept under this condition for 7 days. The etiolated-mature plants were then transferred to the growth chambers with different R/FRs (0.7 or 3.5) and LDW conditions for 1–24 h and sampled at specific time points.
To study the effects of different photoperiods on the expression of the PHYB gene and stem elongation, plants were transferred from the SDC chamber to a warm chamber (22°C day/18°C night) with either 8 h light/day (short day; SD) or 16 h light/day (long day; LD) and kept there until flowering. Samples were collected at specific time points.
To study the effect of exogenous ethephon on the expression of the PHYB gene, four pots of the plants maintained under SDC for at least 90 days were selected for each treatment. Before the plants were treated with ethephon, young shoots were collected and used as the sample for day 0. Based on previous studies (Emery et al. 1994), two concentrations of ethephon (0.1 and 10.0 mol m−3) were applied to each group of plants, respectively. The control plants were sprayed with distilled water (pH 3.0) to match the acidic pH of the ethephon solution. After spray, the pots were covered with a plastic tray to maintain consistent humidity. After 24 h, they were uncovered and transferred to a LDW chamber, and kept under this condition until flowering. Samples were collected at specific time points.
In this study, white light was provided by standard cool white fluorescent bulbs with a flux rate of 120 μmol photons m−2 s−1. Red and far-red lights were provided by a Light Emitting Diode (LED) System (Snap-Lite Solid State Lighting System for Plant Growth, Quantum Devices Inc, Barneveld, WI, USA). Fluence rates and spectral distribution of the light sources were recorded by the cosine-corrected remote probe of calibrated LI-1800 Spectroradiometer (LI-COR, Lincoln, NE, USA) held horizontally at the level of the shoot apices. PAR and photon irradiance were measured in the range of 400–800 nm. R/FR was calculated as the ratio of fluence rates over wavelength intervals of 650–675 and 720–745 nm. Since the PHYB gene expression showed a diurnal rhythm in Arabidopsis (Toth et al. 2001), we assumed that a similar trend would occur in Stellaria. Hence, all samples were collected between 10:00 am and 11:00 am, unless otherwise indicated.
Growth measurements
On day 0, 20 ramets were randomly selected from four pots of the prairie ecotype maintained under SDC conditions for at least 90 days for each experiment. Ramets in the pots were then transferred to a warm chamber with either SD or LD conditions. The stem height of the individual ramets was measured from the bottom of the first internode to the shoot tip every 2 days for 20 days. The experiments were replicated three times. The stem heights presented in this study were means ± SE of three replicates with a total n = 60.
RNA Extraction and semi-quantitative RT-PCR
Total RNA was extracted from each sample using RNeasy Plant Mini kit (QIAgen, Hilden, Germany) following the manufacturer’s instructions. A total of 100 mg frozen tissues were used and finally about 30 μg of total RNA in about 50 μl of RNase-free water were obtained. Prior to reverse transcription, RNA samples were treated with RNase-free DNase I following the manufacturer’s instructions (Invitrogen, Gaithersburg, MD). Total RNA (4 μg) was reverse transcribed using primer AP (Table 1) and Superscript II RT (Invitrogen) following the manufacturer’s instructions. The reaction containing the first-strand cDNAs was directly used in the PCR reactions.
A pair of the PHYB gene specific primers (QmBF and QmBR, Table 1) was designed based on the Stellaria PHYB cDNA sequence (GenBank accession no. AF544028; Li and Chinnappa 2003). A pair of Stellaria 23S rRNA primers (23SF and 23SR, Table 1) was designed based on the Stellaria 23S rRNA sequence (GenBank accession no. AY251296; Li and Chinnappa 2004). RT-PCR was carried out in a 50 μl reaction containing 1× PCR buffer (Invitrogen), 1.5 mM MgCl2, 200 μM dNTP, 10 pmol of each PHYB primer, 3 pmol of each 23S primer (Table 1), and 2.5 units of Platinum Taq DNA polymerase (Invitrogen). Each reaction also contained 2 μl of 10× diluted first-strand cDNAs. PCR cycling conditions were 94°C 3 min for an initial cycle, followed by 94°C 30 s, 56°C 30 s, 72°C 40 s for 20–24 cycles; and a 72°C 5 min as a final extension cycle. A 23S rRNA gene was used as an internal control for standardization of RNA loading. A 381-bp amplicon of 23S rRNA gene (Fig. 1a, lane 1) was amplified with the primers 23SF/23SR (Table 1) and two amplicons of PHYB and 23S rRNA (Fig. 1a, lane 2) were amplified with the primers QmBF/QmBR and 23SF/23SR (Table 1) in the same reaction. The seedlings grown under light for a week were used to determine the PCR cycle number that gave a linear range of gene amplification. The amount of each PCR product was approximately doubled in each successive cycle from 21 to 24 (Fig. 1b). A 23 cycle, which gave a linear range for both PHYB and 23S gene amplifications, was chosen for the subsequent experiments.
Primer specific test and determination of PCR cycle numbers. a Primer specific test. A 381-bp 23S rRNA (lane 1) was amplified by RT-PCR with a pair of primers 23SF/23SR, whereas a 381-bp 23S and a 615-bp PHYB (lane 2) were amplified by RT-PCR with two pairs of primers 23SF/23SR and QmBF/QmBR (Table 1). The 1-kb plus DNA marker (Invitrogen) was loaded in lane M. b Determination of PCR cycle numbers. To obtain a linear range for both PHYB and 23S amplifications, seedlings grown under light for a week were used to determine the PCR cycle number which gave a linear range of gene amplification
Three major factors were considered in this experiment when using RT-PCR as a quantitative method. First, it is important to ensure that only cDNA is used as a template. Prior to reverse transcription, all RNA samples were treated with RNase-free DNase I to eliminate genomic DNA contamination. Meanwhile, the positions of the PHYB primers spanned an intron to ensure that the genomic DNA was not amplified. Second, it is also important to account for the difference in the efficiency of amplification. For this matter, an internal control was used to eliminate a potential source of error. Third, PCR can be used as a quantitative method only during exponential amplification. Therefore, PCR products were determined during the exponential amplification phase.
RT-PCR products (5 μl) were separated on 1.7% (w/v) agarose gels and visualized with ethidium bromide under UV light. Individual PHYB and 23S rRNA bands separated on the same gel were quantified using the Quantitative Image Analysis of the NIH Image 1.62 software (Research Services Branch of the National Institute of Mental Health, USA). The relative amount of the PHYB mRNA in each sample was expressed relative to normalized 23S mRNA levels i.e. the ratio of optical density of PHYB to 23S. The results presented here were the average of three individual experiments.
Statistical analysis
One way ANOVA was performed to determine statistical significance of the differences of the level of the PHYB gene expression between different organs and conditions at a 95% confidence interval, followed by a Bonferroni test to determine the differences among group means (Microcal Origin 7.5, Origin Lab Corporation, Northampton, MA). The correlation between the relative transcript abundance of the PHYB gene and the increment of stem elongation was also assessed using Origin 7.5. P value of ≤0.05 was considered to be significant.
Results
Expression of the PHYB gene in different organs
Expression of the PHYB gene in different organs was analyzed using semi-quantitative RT-PCR. The PHYB gene was constitutively expressed in flowers, leaves, stems, and roots of the plants (Fig. 2). The range of the relative transcript abundance of the PHYB gene in these organs was 0.73–0.84. The relative transcript abundance of the PHYB gene in different organs was not significantly different except that the level in flowers was higher than in leaves (Fig. 2).
Expression of the PHYB gene in different organs. Plants grown under R/FR = 1.4, 8 h photoperiod, 8°C day/4°C night (SDC) for at least 90 days were transferred into a growth chamber with R/FR = 1.4, 16 h photoperiod, 22°C day/18°C night (LDW) and maintained under the same conditions until flowering. Leaves, stems, and flowers were collected. Roots were obtained from young ramets planted in 8 cm pots containing sterile sands under LDW for 14 days. The level of the PHYB transcript was analyzed by semi-quantitative RT-PCR. The transcript abundance was given as a ratio of optical density of the PHYB band to 23S band. Values are means ± SE of independent experiments (n = 3). Means with the same letters are not statistically significantly different at the 95% confidence interval
Expression of the PHYB gene in etiolated-seedlings and -mature plants
To determine if the PHYB gene expression was regulated by de-etiolation, the etiolated-seedlings were transferred to light. The relative transcript abundance of PHYB gene increased significantly by 57% at h 6 after transfer of the etiolated-seedlings from the dark to light. After 6 h, the level decreased until the end point of sampling (12 h), but the change was not statistically significant (Fig. 3a).
Expression of the PHYB gene in etiolated-seedlings and etiolated-mature plants. The level of the PHYB transcript was analyzed using semi-quantitative RT-PCR. The transcript abundance was given as a ratio of optical density of the PHYB band to 23S band. Values are means ± SE of independent experiments (n = 3). a Expression of the PHYB gene in etiolated-seedlings. Seedlings grown in the dark at 22°C for 14 days (0 h) were transferred into a growth chamber with R/FR = 1.4 and LDW for 1, 6, and 12 h. Means with the same letters between the different light exposures are not statistically significantly different at the 95% confidence interval. b Expression of the PHYB gene in the etiolated-mature plants. Plants grown under R/FR = 1.4, 8°C day/4°C night, 8 h photoperiod (SDC) for at least 90 days were transferred into the dark at 22°C for 7 days (0 h), followed by transferring into a growth chamber with R/FR = 3.5 or 0.7 at 22°C for 1, 6, 12, and 24 h. Means with the same letters at each time point are not statistically significantly different at the 95% confidence interval
To investigate the effects of different R/FRs on the expression of the PHYB gene, plants maintained under SDC for at least 90 days were transferred to the dark at 22°C and kept at this condition for 7 days. The etiolated-mature plants were then transferred to different R/FRs (3.5 and 0.7) for 1–24 h. The relative transcript abundance of the PHYB gene under R/FR 3.5 was significantly higher than that under R/FR 0.7 at all time points tested (Fig. 3b). Within treatments, there was an increasing trend to a peak at h 6 upon exposure to R/FR 3.5, but the relative transcript abundance from h 0 to h 24 was not statistically significantly different (Fig. 3b). The level of the PHYB transcript under R/FR 0.7 decreased significantly by 15% within 1 h upon transfer of the etiolated-mature plants to light. After 1 h, the level increased slightly and maintained until the end of the experiment (Fig. 3b).
Interaction between photoperiod and the PHYB transcript in regulating stem elongation
The plants of the prairie ecotype kept under SDC for at least 90 days were transferred to a warm temperature (22°C) with either SD or LD photoperiod. These plants showed a significant difference in stem elongation (Fig. 4a). LD photoperiod significantly promoted stem elongation and the plants reached about 21 cm high by day 20 (Fig. 4a). By contrast, the plants grown under SD photoperiod showed only a slight increase in plant height which reached about 2 cm by day 20 (Fig. 4a). In addition, the plants grown under LD started flowering on day 18, whereas no plants grown under SD flowered before 60 days (Fig. 4b).
Cumulative stem elongation and the PHYB gene expression during growth period. a The cumulative stem elongation of the prairie ecotype of Stellaria longipes. Plants grown under 8 h photoperiod and 8°C day/4°C night (SDC) for at least 90 days were transferred into a growth chamber with 22°C day/18°C night and either 8 h photoperiod (short-day, SD) or 16 h photoperiod (long-day, LD) and maintained under the same conditions until flowering. The stem height of the individual ramets was measured from the bottom of the first internode to the shoot tip every 2 days for 20 days. Values are mean ± SE of three replicates with a total n = 60. b Ramets of the prairie genotype of Stellaria longipes grown under SD and LD after 20 days. c Daily growth increments of plants. d Expression of the PHYB gene during the growth period. The plants were grown under the same conditions as described in a The level of PHYB gene transcript was determined by semi-quantitative RT-PCR. The transcript abundance was given as a ratio of optical density of the PHYB band to 23S band. Values are means ± SE of independent experiments (n = 3). Means with the same letters at different time points under the same growth conditions were not statistically significantly different at the 95% confidence interval. e and f The correlation between the relative transcript abundance of the PHYB gene and the increment of stem elongation. The correlation was assessed using Origin 7.5 (Origin Lab Corporation, Northampton, MA). P value of ≤0.05 was considered to be significant
The daily growth increments over 20 days in LD and SD were shown in Fig. 4c. Stem growth under LD was slow in the first 4 days, but began to accelerate and peaked at day 12. After that, it decelerated. However, the stem elongated very slowly over 20 days in SD. For both LD and SD conditions, the levels of the PHYB mRNA at the same time points were determined using semi-quantitative RT-PCR. Relative transcript abundance of the PHYB gene under SD was significantly higher than that under LD, but it reached a similar level at day 20 (Fig. 4d). The relative transcript abundance of the PHYB gene under SD remained stable until day 8, but dropped dramatically from day 8 to day 12 at which the abundance reached the lowest level. After day 12 until day 20, the abundance increased slightly (Fig. 4d). In contrast, the relative transcript abundance of the PHYB gene under LD decreased significantly as soon as the plants were transferred to light. The abundance reached the lowest level on day 12, which was consistent with that observed under SD. After day 12, the level kept significantly increasing until day 20 (Fig. 4d). Furthermore, the correlation between the relative transcript abundance of the PHYB gene and the increment of stem elongation was assessed using Origin 7.5 (Fig. 4e, f). Interestingly, the level of the PHYB mRNA significantly correlated with the increment of stem elongation under LD, with r 2 = 0.88, P = 0.01 (Fig. 4e). Under SD, however, there was no significant correlation between the level of the PHYB mRNA and the increment of stem elongation (Fig. 4f).
Effect of exogenous ethephon on the expression of the PHYB gene
Application of exogenous ethephon was shown to significantly increase the evolution of ambient ethylene in the prairie plants. It was found that stem elongation of the prairie plants of S. longipes was stimulated by low ethephon concentration (0.1 mol m−3) and inhibited by high ethephon concentration (10.0 mol m−3) (Emery et al. 1994). In this study, the correlation between the PHYB gene expression and exogenous ethephon treatment and the role of the PHYB gene in stem elongation were investigated using the same prairie ecotype (Fig. 5). The transcript level of the PHYB gene in plants treated with the higher concentration of ethephon decreased significantly on day 4 and remained relatively stable until day 16. It then declined significantly by day 20. Transcript levels remained stable in control plants and plants treated with lower concentration of ethephon throughout the experiment (Fig. 5).
Effect of different concentrations of ethephon on the expression of the PHYB gene. The prairie plants maintained in winter-simulated short-day cold (SDC) conditions for at least 90 days were selected for each treatment. The samples were collected before ethephon treatment (day 0) and on d 4, 8, 12, 16, and 20 after ethephon treatment. The transcript abundance of the PHYB gene was determined using semi-quantitative RT-PCR. The abundance was given as a ratio of optical density of the PHYB band to 23S band. Values are means ± SE of independent experiments (n = 3). Means with the same letters at each time point of each treatment are not statistically significantly different at the 95% confidence interval
Discussion
Transcript profiles in different organs and de-etiolated plants
In this study, the PHYB gene was expressed constitutively in different organs in the prairie plants of Stellaria. The expression levels among different organs were not significantly different except that the level in leaves was lower than in flowers (Fig. 2). These results are consistent with previous observations in Arabidopsis (Clack et al. 1994) and potato (Heyer and Gatz 1992). However, the PHYB gene showed tissue-specific expression in tobacco (Adam et al. 1996). In addition, Hauser et al. (1997) determined the absolute level of the PHYB gene transcript in tomato and confirmed that it was substantial in organ-specific expression. The considerable variation among these reports might be explained by the different plant species tested.
Following a dark-to-light transition of the prairie plants, the PHYB transcripts exhibited a transient increase during the first 6 h (Fig. 3a). This was in agreement with tomato PHYB1, but it was different from tomato PHYB2 which exhibited a sharp decline in abundance during the first 10 h after the plants were transferred from dark to light (Hauser et al. 1998). Phylogenetic analyses revealed that Stellaria PHYB shared a higher nucleotide identity with tomato PHYB1 than with tomato PHYB2, which are 72.1 and 70.7%, respectively (data not shown). Taken all together, we speculate that Stellaria PHYB gene might be an ortholog of tomato PHYB1.
Expression of the PHYB gene under different R/FRs
The major light signal triggering shade avoidance response is the ratio of the R/FR (Ballare 1999; Aphalo et al. 1999). PHYB, C, and D are responsible for perception and transduction of low R/FR signal and initiation of shade avoidance (Franklin and Whitelam 2005). For the prairie ecotype, the initiation of shade avoidance must be rapid. This is essential for survival of the plants in the prairie habitat (Chinnappa et al. 2005). In this study, our results demonstrated that the expression of the PHYB gene in de-etiolated plants was correlated with the variation of the R/FR ratio (Fig. 3b), whereby lowering R/FR was coincident with a down regulation of the PHYB gene transcription in de-etiolated plants.
Stem elongation and expression of the PHYB gene
The plants grown under SD showed very limited stem elongation. However, the plants grown under LD attained an average height of 21 cm by day 20 (Fig. 4a, b). The level of the PHYB gene expression under SD was significantly higher than that under LD (Fig. 4d). Moreover, the transcript abundance of the PHYB gene was significantly correlated with the increment of stem elongation under LD, whereas no correlation was observed under SD (Fig. 4e, f). We therefore suggest that higher level of the PHYB mRNA may inhibit stem elongation.
Interaction of ethephon, ethylene, and PHYB in mediating stem elongation
Ethylene is involved in many plant growth and development responses and is generally known as an inhibitor of vegetative growth (Abeles et al. 1992; Kurepin et al. 2007). However, several studies have demonstrated a positive correlation between growth and the level of endogenous ethylene (Raskin and Kende 1984; Jackson 1985; Rijnders et al. 1997; Finlayson et al. 1998, 2007; Pierik et al. 2003). Low R/FR enhanced ethylene production in wild-type tobacco plants resulting in shade avoidance responses, whereas in transgenic ethylene-insensitive tobacco plants, it reduced shade avoidance responses to neighbours (Pierik et al. 2003, 2004). The results obtained from sorghum phyB-1 mutant also demonstrated that the ethylene production in phyB-1 mutant was 10 times more than in wild-type (Finlayson et al. 1998). These results indicated that ethylene was involved in phytochrome-mediated shade avoidance responses (Pierik et al. 2004) and that both PHYB and ethylene are required for this response (Finlayson et al. 1998).
The effect of exogenous ethephon treatment on ethylene production and stem elongation in the prairie plants of Stellaria has been well investigated by our laboratory (Emery et al. 1994). Application of exogenous ethephon was shown to significantly increase the evolution of ambient ethylene in the prairie plants (Emery et al. 1994). Stem elongation in the prairie ecotype was stimulated by low ethephon concentration (0.1 mol m−3), but inhibited by high ethephon concentration (10.0 mol m−3). In this study, when the prairie plants were treated with higher concentration of ethephon (10.0 mol m−3), the level of the PHYB gene transcript was significantly lower than control plants and plants treated with lower concentration of ethephon (0.1 mol m−3) on day 4 (Fig. 5). It appears that day 4 might be a critical time point for initiation of stem elongation process. Our previous studies showed that stem elongation of the prairie ecotype commenced on day 4 after the plants were transferred from SDC to LDW (Alokam et al. 2002). Histological analyses showed that the number of cells in the second internode was relatively constant within the first 4 days after the plants were exposed to LDW and then it began to increase on day 5 (Choung 1998). Taken together, these lines of evidences suggest that a steady state level of the PHYB gene transcription on day 4 might be critical for initiation of stem elongation process in the prairie plant. After day 4, when plants pass through initiation of stem elongation process, a steady state level of the PHYB gene expression might then be required for maintaining stem elongation.
References
Abeles FB, Morgan PW, Saltveit ME Jr (1992) Ethylene in plant biology, 2nd edn. Academic Press, New York
Adam E, Kozma-Bognar L, Kolar C, Schafer E, Nagy F (1996) The tissue-specific expression of a tobacco phytochrome B gene. Plant Physiol 110:1081–1088
Alokam S, Chinnappa CC, Reid DM (2002) Red/far-red light mediated stem elongation and anthocyanin accumulation in Stellaria longipes: differential response of alpine and prairie ecotypes. Can J Bot 80:72–81
Aphalo PJ, Ballare CL, Scopel AL (1999) Plant-plant signaling, the shade avoidance response and competition. J Exp Bot 50:1629–1634
Ballare CL (1999) Keeping up with the neighbours: phytochrome sensing and other signaling mechanisms. Trends Plant Sci 4:97–102
Chinnappa CC, Donald GM, Sasidharan R, Emery RJN (2005) The biology of Stellaria longipes (Caryophyllaceae). Can J Bot 11:1367–1383
Choung SDX (1998) Stem elongation plasticity in Stellaria longipes: anatomical and biochemical studies. MSc thesis, University of Calgary, Calgary, Alta, Canada
Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25:413–427
Emery RJN, Reid DM, Chinnappa CC (1994) Phenotypic plasticity of stem elongation in two ecotypes of Stellaria longipes: the role of ethylene and response to wind. Plant Cell Environ 17:691–700
Finlayson SA, Lee IJ, Morgan PW (1998) Phytochrome B and the regulation of circadian ethylene production in sorghum. Plant Physiol 116:17–25
Finlayson SA, Lee IJ, Mullet JE, Morgan PW (1999) The mechanism of rhythmic ethylene production in sorghum. The role of phytochrome B and simulated shading. Plant Physiol 119:1083–1089
Finlayson SA, Hays DB, Morgan PW (2007) phyB-1 sorghum maintains responsiveness to simulated shade, irradiance and red light : far-red light. Plant Cell Environ 30:952–962
Franklin KA, Whitelam GC (2005) Phytochromes and shade avoidance responses in plants. Ann Bot 96:169–175
Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC (2003) Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131:1340–1346
Franklin KA, Allen T, Whitelam GC (2007) Phytochrome A is an irradiance-dependent red light sensor. Plant J 50:108–117
Goeschl JD, Pratt HK, Bonner BA (1967) An effect of light on the production of ethylene and the growth of the plumular portion of etiolated pea seedlings. Plant Physiol 42:1077–1080
Hauser BA, Pratt LH, Cordonnier-Pratt MM (1997) Absolute quantification of five phytochrome transcripts in seedlings and mature plants of tomato (Solanum lycopersicum L.). Planta 201:379–387
Hauser BA, Cordonnier-Pratt MM, Pratt LH (1998) Temporal and photoregulated expression of five tomato phytochrome genes. Plant J 14:431–439
Heyer A, Gatz C (1992) Isolation and characterization of a cDNA clone coding for potato type B phytochrome. Plant Mol Biol 20:589–600
Imaseki H, Pjon C-H, Furya M (1971) Phytochrome action in oryza sativa L. Plant Physiol 48:241–244
Jackson MB (1985) Ethylene and responses of plants to soil waterlogging and submergence. Annu Rev Plant Physiol 36:145–174
Johnson E, Bradley M, Harberd NP, Whitelam GC (1994) Photoresponses of light-grown phyA mutants of Arabidopsis. Plant Physiol 105:141–149
Kurepin LV, Walton LJ, Reid DM (2007) Interaction of red to far red light ratio and ethylene in regulating stem elongation of Helianthus annuus. Plant Growth Regul 51:53–61
Li W, Chinnappa CC (2003) The phytochrome gene family in the Stellaria longipes complex. Int J Plant Sci 164:657–673
Li W, Chinnappa CC (2004) Isolation and characterization of PHYC gene from Stellaria longipes: differential expression regulated by different red/far-red light ratios and photoperiods. Planta 220:318–330
Li W, Song ZH, Sasidharan R, Chinnappa CC (2009) Light and shade signals regulate four phytochrome A genes in Stellaria longipes. Int J Plant Sci 170:164–173
Macdonald SE, Chinnappa CC, Reid DM (1984) Studies on the Stellaria longipes complex: phenotypic plasticity. I. Response of stem elongation to temperature and photoperiod. Can J Bot 62:414–419
Mathews S, Sharrock RA (1997) Phytochrome gene diversity. Plant Cell Environ 20:666–671
Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, Young J, Austin-Phillips S, Quail PH (2003) Isolation and characterization of phyC mutants in Arabidopsis reveal complex crosstalk between phytochrome signaling pathways. Plant Cell 15:1962–1980
Pierik R, Visser EJW, de Kroon H, Voesenek LACJ (2003) Ethylene is required in tobacco to successfully complete with proximate neighbours. Plant Cell Environ 26:1229–1234
Pierik R, Cuppens MLC, Voesenek LACJ, Visser EJW (2004) Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol 136:2928–2936
Pratt LH, Cordonnier-Pratt MM, Kelmenson PM, Lazarova GI, Kubota T, Alba RM (1997) The phytochrome gene family in tomato (Solanum lycopersicum L.). Plant Cell Environ 20:672–677
Quail PH (1994) Phytochrome genes and their expression. In: Kendrick RE, Kronenberg GHM (eds) Photomorphogenesis in plants, 2nd edn. Kluwer Academic Publisher, Amsterdam, pp 71–104
Raskin I, Kende H (1984) The role of gibberellin in the growth response of submerged deep water rice. Plant Physiol 76:947–950
Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far red light photoreceptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5:147–157
Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104:1139–1149
Rijnders JGHM, Yang YY, Kamiya Y, Takahashi N, Barendse GWM, Blom CWPM, Voesenek LACJ (1997) Ethylene enhances gibberellin levels and petiole sensitivity in flooding-tolerant Rumex palustris but not in flooding-intolerant R. acetosa. Planta 2003:20–25
Samimy C (1978) Effect of light on ethylene production and hypocotyls growth of soybean seedlings. Plant Physiol 61:772–774
Sharrock RA, Quail PH (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3:1745–1757
Smith H (2000) Phytochromes and light signal perception by plants–an emerging synthesis. Nature 407:585–591
Takano M, Kanegae H, Shinomura T, Miyao A, Hirochika H, Furuya M (2001) Isolation and characterization of rice phytochrome A mutants. Plant Cell 13:521–534
Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, Shinomura T (2005) Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17:3311–3325
Toth R, Kevei EE, Hall A, Millar AJ, Nagy F, Kozma-Bognar L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127:1607–1616
Tuinen AV, Kerckhoffs LHJ, Nagatani A, Kendrick RE, Koornneef M (1995) Far-red light-insensitive, phytochrome A-deficient mutants of tomato. Mol Gen Genet 246:133–141
Vangronsveld J, Clijsters H, Van Poucke M (1988) Phytochrome-controlled ethylene biosynthesis of intact etiolated bean seedlings. Planta 174:19–24
Weller JL, Murfet IC, Reid JB (1997) Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol 114:1225–1236
Whitelam GC, Devlin PF (1997) Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ 20:752–758
Acknowledgments
We thank Dr. Dominic Rosso for valuable comments on this manuscript. This work is supported by a Discovery grant from the Natural Sciences and Engineering Research Council of Canada.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, W., Song, Z., Neil Emery, R.J. et al. Effects of day length, light quality and ethylene on PHYTOCHROME B expression during stem elongation in Stellaria longipes . Plant Growth Regul 63, 291–300 (2011). https://doi.org/10.1007/s10725-010-9529-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-010-9529-y