Abstract
We have cloned and characterized the phytochrome C (PHYC) gene from Stellaria longipes. The PHYC gene is composed of a 110-bp 5′-untranslated leader sequence, a 3,342-bp coding region, and a 351-bp 3′-untranslated sequence. The Stellaria PHYC contains three long introns within the coding region at conserved locations as in most angiosperm PHY genes. DNA blot analysis indicates that the Stellaria genome contains a single copy of PHYC. Stellaria PHYC shares 60%, 58%, and 57% deduced amino acid identities with rice, Sorghum, and Arabidopsis PHYC, respectively. Phylogenetic analysis indicates that Stellaria PHYC is located in the dicot branch, but is divergent from Arabidopsis PHYC. The Stellaria PHYC is constitutively expressed in different plant organs, though the level of PHYC gene transcript in roots is slightly higher than in flowers, leaves, and stems. When 2-week old seedlings grown in the dark were exposed to constant white light, PHYC mRNA quickly accumulates within 1–12 h. When plants grown in darkness for 7 days were exposed to different red/far-red light (R/FR) ratios, the levels of PHYC mRNA at R/FR=0.7 are much lower than under R/FR=3.5. The levels of PHYC mRNA under short-day (SD) photoperiod are higher than under long-day (LD) photoperiod. Plants under SD conditions do not elongate, and are only about 1.7 cm tall at 19 days. In contrast, plants under LD conditions elongate with an average height of 21.2 cm at 19 days. The plants do not flower under SD conditions, but do so at 18–19 days under LD conditions. These results indicate that under SD conditions the high level of PHYC mRNA may inhibit stem elongation and flower initiation. In contrast, under LD conditions the high level of PHYC mRNA may promote stem elongation and flowering.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phytochrome genes have been identified in a broad range of species throughout the plant kingdom, from the algae and mosses to gymnosperms and angiosperms (Mathews and Sharrock 1997; Schneider-Poetsch et al. 1998; Clapham et al. 1999). In all plants examined, the phytochrome apoproteins are encoded by small, multigene families, and have been classified into four subfamilies: A, B/D, C/F, and E (Mathews et al. 1995; Mathews and Sharrock 1996, 1997; Pratt et al. 1997; Schneider-Poetsch et al. 1998; Schmidt and Schneider-Poetsch 2002). In Arabidopsis, the PHY family is composed of five members (PHYA, PHYB, PHYC, PHYD, and PHYE) and is classified into four subfamilies (A, B/D, C, E; Sharrock and Quail 1989; Clack et al. 1994). We recently identified the PHY family in Stellaria, which is composed of six members (four PHYAs, one PHYB, and one PHYC), but lacks the PHYE subfamily (Li and Chinnappa 2003). Although a large number of PHY DNA sequences have been reported in the databases, a significant fraction of these are either ESTs or partial sequences generated by PCR using primers to a conserved region of the sequences (Mathews et al. 1995; Mathews and Sharrock 1997; Pratt et al. 1997; Schneider-Poetsch et al. 1998). Relatively few full-length cDNA sequences have been reported. Fewer PHY gene structures have been determined experimentally by comparison of DNA sequences to corresponding cDNAs (Basu et al. 2000).
The expression of the five PHY in Arabidopsis and tomato, as well as PHYA and PHYB in potato, has been described as uniform in different organs, at different developmental stages, and under different light conditions (Heyer and Gatz 1992a, 1992b; Clack et al. 1994; Hauser et al. 1997, 1998). Data from Northern blot analyses indicate that PHYC is less abundant compared to PHYA and PHYB, and is relatively unaffected by light in Arabidopsis and rice (Sharrock and Quail 1989; Clack et al. 1994; Basu et al. 2000).
The physiological functions of individual phytochrome species have been recently revealed by analysis of PHY-deficient mutants and overexpression studies of individual PHY genes. The results from analysis of Arabidopsis phyA-, phyB-, phyD-, or phyE-deficient mutants have revealed that individual members of the PHY family have different but partially overlapping functions in controlling plant responses (Whitelam and Devlin 1997; Devlin et al. 1998; Quail 2002). For example, PHYA plays a major role throughout the plant life cycle in sensing prolonged far-red (FR) light and in mediating very-low-fluence responses (Casal et al. 1997). PHYB and PHYD have highly overlapping roles in sensing the red/far-red light (R/FR) ratio and in mediating shade avoidance responses (Aukerman et al. 1997; Smith and Whitelam 1997; Devlin et al. 1999). PHYE seems to play a role in the regulation of shade-avoidance response in a manner partially conditionally redundant to PHYB (Devlin et al. 1998). PHYE is also involved in the control of seed germination (Hennig et al. 2002), and in the control of cotyledon expansion under constant red light (Franklin et al. 2003b). Although PHYC-deficient mutants have only recently been isolated, data from overexpression studies of Arabidopsis PHYC in both Arabidopsis (Qin et al. 1997) and tobacco (Halliday et al. 1997) suggest a possible specific role for PHYC in the control of Arabidopsis primary-leaf cell expansion and tobacco cotyledon expansion. A recent study with an Arabidopsis PHYC-deficient mutant has revealed a functional role for the PHYC gene throughout Arabidopsis photomorphogenesis (Franklin et al. 2003a; Monte et al. 2003). The PHYC gene may perform a significant role in the modulation of other photoreceptors, and PHYA and PHYC act together to regulate mature leaf morphology (Franklin et al. 2003a). PHYC function in seedling de-etiolation in constant red light, however, may require the presence of PHYB (Monte et al. 2003). In addition, the spectral characteristics of PHYC were strictly different from other PHY genes and the PHYC apoprotein chromophore adduct has undergone a strong dark reversion (Eichenberg et al. 2000). While most members of PHY genes are well characterized, information about the physiological function and expression pattern of PHYC is limited.
In addition, it is known that phytochromes also play an important role in adaptive plasticity in natural light environments in plants (Weinig 2002; Alokam et al. 2002). For example, Stellaria longipes has been used as a model system to understand the genetic regulation of phenotypic plasticity in various ecotypes (Emery et al. 1994; Kathiresan et al. 1998; Alokam et al. 2002). During the past two decades, we have studied the growth responses of several ecotypes under various light conditions (Macdonald et al. 1984; Emery et al. 1994; Alokam et al. 2002). We have recently determined the phytochrome gene family in the S. longipes complex (Li and Chinnappa 2003). In this study, we cloned the PHYC gene from S. longipes and identified its structure by comparison with the cDNA sequence. We also report characterization of light regulation, and patterns of expression of the Stellaria PHYC gene using semiquantitative RT-PCR.
Materials and methods
Plant materials and growth conditions
Prairie genotypes of S. longipes were originally obtained from Kananaskis valley, Alberta, Canada. The plants were clonally propagated and maintained in 8 cm pots containing peat moss, sand and Terra green, a crushed baked clay growing medium (2:1:1), in the University of Calgary greenhouse. The plants used in this experiment were first transferred from the greenhouse into a short day cold (SDC) growth chamber (8 h/day photoperiod, 8°C day/4°C night) for a minimum of 90 days to simulate the winter cycle. (1) To study expression of PHYC in different organs, the plants were transferred from the SDC chamber to a long day warm (LDW) chamber (22°C day/18°C night, 16 h photoperiod) and maintained under LDW for 19 days until flowering. To obtain roots, the young ramets were planted in 8 cm pots containing sterile sand and maintained in a tray containing 1/2 liquid Murashige and Skoog basal medium under LDW for 14 days and then fresh roots were collected. (2) To study the effects of different R/FR ratios on the expression of PHYC, 2-week-old seedlings were used. Seeds were surface-sterilized for 10 min in 70% (v/v) alcohol and then for 10 min in 20% (v/v) bleach, rinsed at least five times with sterile water, and germinated in glass jars containing Murashige and Skoog basal agar medium (Sigma, St. Louis, Mo.) plus 2% (w/v) sucrose. The jars were kept in the dark at 4°C for 2 days, treated for 2 h with white light to induce germination, and then placed in complete darkness at 22°C for 14 days. After that, the jars were transferred to continuous white light at 22°C for 1–12 h. (3) To study the effects of different photoperiods on the expression of PHYC and stem elongation, the plants were transferred from SDC to a warm chamber (22°C day/18°C night) with 8 h/day (short day; SD) or 20 h/day (long day; LD) conditions. White light sources were provided by standard cool white fluorescent bulbs with a flux rate of 35 μmol s−1m−2. Red and far-red lighting was provided by a light emitting diode (LED) system (Snap-Lite Solid State Lighting System for Plant Growth, Quantum Devices, Barneveld, Wis.). Fluency rates and spectral distribution of the light sources were recorded by the cosine-corrected remote probe of a calibrated LI-1800 Spectroradiometer (LI-COR, Lincoln, Neb.). The probe was held horizontally at the level of the shoot apices. Since PHYC gene expression showed a diurnal rhythm in Arabidopsis (Toth et al. 2001), we assumed a similar trend in Stellaria. Hence, all samples were collected between 10:00 am and 11:00 am, unless otherwise indicated.
Isolation of the Stellaria PHYC gene
A 3,237-bp Stellaria PHYC cDNA fragment was first cloned using degenerate primer PCR and 3′rapid amplification of cDNA ends (RACE; Li and Chinnappa 2003). Since this clone was missing a portion of exon 1, the sequence of the 5′ end of the Stellaria PHYC was obtained using a 5′RACE kit (Invitrogen, Gaithersburg, Md.). Total RNA was isolated from young ramets using an RNeasy Plant Mini kit (Qiagen, Hilden, Germany). Two reverse Stellaria PHYC gene-specific primers, PhyCR1 and PhyCR3 (Table 1) near the 5′ end of the open reading frame (ORF) were designed based on the 3,237 bp PHYC cDNA sequence (Li and Chinnappa 2003). First-strand cDNAs were synthesized with primer PhyCR1 (Table 1). After synthesis of the first strand, the cDNAs were purified and tailed with dCTP, and then used directly in the 5′RACE PCR reaction. Primary PCR amplification reactions were achieved using gene-specific primer PhyCR1 and Abridged Anchor Primer (AAP, Invitrogen), and a secondary PCR was run using gene-specific primer PhyCR3 and Abridged Universal Amplification Primer (AUAP, Invitrogen) in a 50-μl reaction containing 1×PCR buffer (Invitrogen), 1.5 mM MgCl2, 200 μM dNTP, 10 pmol each primer, and 2.5 U DNA polymerase (Invitrogen). The PCR program was 3 min at 94°C, 5 min at 72°C [at this point 0.5 μl 5 U/μl Taq DNA polymerase (Invitrogen) was added to each reaction] for 1 cycle; 0.5 min at 94°C, 1 min at 55°C, 2 min at 72°C for 35 cycles, and 10 min at 72°C for 1 cycle. RACE fragments were gel-purified using a QIAquick gel extraction kit (Qiagen), and then cloned into the pGEM-T Easy vector (Promega, Madison, Wis.) for sequencing.
To obtain the genomic sequence of the Stellaria PHYC gene, Stellaria genomic DNA was used as a template and amplified using PCR with three pairs of PHYC-gene-specific primers (PhyCF7/PhyCR7, QmCF/QmCR, and PhyCF6/PhyCR6, Table 1), which were designed based on the Stellaria PHYC cDNA sequence. PCR conditions were identical to those described above. Three fragments (1,790-bp, 2,351-bp, and 2,036-bp) were gel-purified, cloned and sequenced as described above. A 6-kb fragment was obtained by overlapping the three fragments using the NCBI BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/).
DNA sequencing and data analysis
DNA samples for sequencing were prepared with QIAprep Miniprep Kit (Qiagen) and sequenced in both directions with T7 and SP6 primers using an ABI Prism BigDye Sequencing kit (Applied Biosystems, Foster City, Calif.) at the University of Calgary DNA Sequencing Center. Analysis of the DNA sequences was carried out using the Advanced BLASTX search program (Altschul et al. 1990). Multiple sequence alignments were performed using CLUSTAL X version 1.8 (Jeanmougin et al. 1998). Characterization of the nucleotide sequences and conceptual translation of nucleotide sequences were made using DNAStrider 1.1 (Christian Marck, Department de Biologie Institut de Recherche Fondamentale, CEA, France). Phylogenetic analyses were performed with all full-length plant PHYC and PHYF protein sequences available in the databases (Stellaria PHYC AF544029, Arabidopsis PHYC X17343, Oryza PHYC AF141942, and Solanum PHYF AF178568). CLUSTAL X (Jeanmougin et al. 1998) was used to align the amino acid sequences. A neighbor-joining tree was created with NJplot software (Perriere and Gouy 1996) using these aligned amino acid sequences. Arabidopsis PHYA was the outgroup in this study.
Identification of introns
Identification of introns was achieved by comparing the Stellaria PHYC genomic sequence with the cDNA sequence. The exact locations of the splice sites were found. These intron/exon designations were subsequently compared with those of Arabidopsis and rice PHYC genes.
Southern blot analysis
Genomic DNA was isolated from young ramets of S. longipes prairie ecotype, using the method of Doyle and Doyle (1987). Southern blot analysis was carried out with 10 μg/lane genomic DNA digested with DraI, EcoRV, or HindIII. Blots were hybridized with a 660 bp fragment of the Stellaria PHYC gene (Fig. 2b). The 660 bp fragment was digested with BamHI and EcoRI from p7B-PHYC-1 (Li and Chinnappa 2003), gel-purified (QIAquick gel extraction kit; Qiagen), and labeled with 32P-dCTP using the Random Primers DNA Labeling System (Invitrogen) according to the manufacturer’s instructions. The 32P-labeled probe was purified with a Nick Column (Amersham Pharmacia, Little Chalfont, Buckinghamshire, UK) following the manufacturer’s instructions. The blots were prehybridized at 42°C for 2–4 h in 50% deionized formamide, 5× SSC, 5× Denhardt’s buffer, 0.5% (w/v) SDS, then hybridized with 32P-labeled PHYC probe overnight with the same conditions and buffer as prehybridization, and washed twice in 2× SSC, 1% (w/v) SDS at room temperature for 10 min each, following by washing once in 0.5× SSC, 0.5% (w/v) SDS at 55°C for 30 min, and once in 0.1× SSC, 0.1% (w/v) SDS at 55°C for 10 min. The blots were exposed to XAR-5 film (Kodak X-Omat AR-5) using intensifying screens at −80°C for 3 weeks. The sizes of the hybridizing bands were estimated by comparing their mobility with λDNA/HindIII fragments (Invitrogen).
Semiquantitative RT-PCR
Total RNA (5 μg) treated with DNase I was annealed to an oligo (dT)-containing adapter primer (Invitrogen); first-strand cDNAs were then synthesized using Superscript II RT (Invitrogen) and used directly in RT-PCR. A pair of Stellaria PHYC gene-specific primers (QmCF and QmCR, Table 1), designed based on the Stellaria PHYC cDNA sequence, were used to amplify a 626-bp fragment of Stellaria PHYC. A 2,351-bp fragment of the genomic PHYC gene was amplified if total RNA was contaminated by genomic DNA. Both of these fragments can be distinguished from any contamination of genomic DNA by the fragment size. A pair of Stellaria 23S rRNA primers (23SF and 23SR, Table 1) were designed based on the Stellaria 23S rRNA gene cDNA sequence (GenBank accession number: AY251296), and used to amplify a 381-bp fragment as an internal standard. RT-PCR was carried out in a 50-μl reaction containing 1× PCR buffer (Invitrogen), 1.5 mM MgCl2, 200 μM dNTP, 10 pmol each primer (QmCF and QmCR), 3 pmol each primer (23SF and 23SR) [except in the organ-specific expression experiment (10 pmol each primer used)], and 2.5 U Taq DNA polymerase (Invitrogen). A master mix was made excluding DNA polymerase and template cDNAs. From the master mix, an aliquot was added to 0.5 ml thin wall tubes, followed by addition of 2 μl 10× diluted first-strand cDNAs. We selected one sample in which the PHYC gene was expected to be the most abundant and used it to determine the PCR cycle number that gave a linear range of gene amplification. A cycle number in the linear range (26 cycles) was chosen for subsequent experiments. The amounts of the 23S rRNA internal standard primers were determined in order to amplify both PHYC and 23S at a similar level. PCR cycling conditions were 3 min at 94°C, 5 min at 72°C for 1 cycle; 0.5 min at 94°C, 1 min at 56°C, 1.5 min at 72°C for 21–28 cycles; and 10 min at 72°C for 1 cycle.
RT-PCR products (5 μl each) were separated on 1.7% (w/v) agarose gels, visualized by ethidium bromide staining under UV light, and quantified by quantitative image analysis of PHYC and 23S rDNA bands using NIH Image 1.62 software (Research Services Branch of the National Institute of Mental Health, USA). PHYC gene transcript abundance levels were given as relative values normalized to 23S mRNA levels that were the ratio of the optical density of PHYC/23S gene bands. The results shown were from at least two individual experiments.
Results
Identification and structure of the Stellaria PHYC gene
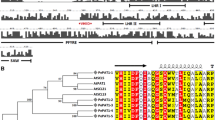
A 3,237-bp Stellaria PHYC cDNA was initially cloned using degenerate primer PCR and 3′RACE (Li and Chinnappa 2003). Since this clone was missing a portion of exon 1 (Fig. 1), the 5′ end of Stellaria PHYC sequence was obtained by 5′RACE. A 756-bp fragment, including PCR primers, was obtained by direct sequencing of the 5′RACE product. After overlapping the 756-bp fragment of the 5′end of PHYC cDNA with a 3,237-bp initial PHYC cDNA, a 3,803-bp full-length PHYC cDNA sequence was obtained, comprising a 110-bp 5′-untranslated leader sequence, a 3,342-bp coding region, and a 351-bp 3′-untranslated sequence (Fig. 1).
A comparison of the Stellaria, rice and Arabidopsis PHYC gene structures over the protein-coding region. Solid bars Exons, connecting lines introns and untranslated portions of exons, small hatched boxes chromophore attachment sites. The number of nucleotides in each intron, and untranslated region is indicated
In order to obtain PHYC genomic sequence, three pairs of PHYC gene-specific primers (PhyCF7/PhyCR7, QmCF/QmCR, and PhyCF6/PhyCR6, Table 1) were designed based on the Stellaria PHYC cDNA sequence. A 1,790-bp fragment was amplified with PhyCF7/PhyCR7, a 2,351-bp fragment was amplified with QmCF/QmCR, and a 2,036-bp fragment was amplified with PhyCF6/PhyCR6 by PCR using Stellaria genomic DNA as template. A 6-kb genomic DNA fragment was obtained by overlapping these three fragments. Exon/intron boundaries were established by comparing the genomic sequence with the cDNA sequence. The physical organization of the Stellaria PHYC gene is similar to that of most other angiosperm phytochromes, with four exons of the coding region interrupted by three introns; an exception is Arabidopsis PHYC (Cowl et al. 1994), which lacks the third intron toward the 3′end of the coding region (Fig. 1). The Stellaria PHYC had three long introns of 1,724-bp, 784-bp and 207-bp in length, respectively (Fig. 1).
Southern blot analysis
S. longipes genomic DNA was digested with three different restriction enzymes and subjected to Southern blot analysis (Fig. 2a). Using a coding region fragment of Stellaria PHYC as a probe (Fig. 2b), the homologous fragments showed strong hybridization signals. A single band was observed with DraI or EcoRV digestion, and two bands were observed for HindIII digestion. Since PHYC has two HindIII recognition sites within the region targeted by the probe (Fig. 2b), digestion with HindIII should result in three bands. One of them should be 224-bp (Fig. 2a). However, we did not observe a third band corresponding to the 3′ HindIII fragment, presumably because either the region of overlap between the probe and the 3′ HindIII fragment (117-bp) is too short, or two PHYC HindIII fragments possibly comigrated. Since hybridization was carried out under highly stringent conditions, it is likely that only PHYC fragments were observed. The hybridization patterns indicate that the PHYC gene is present as a single copy in the Stellaria genome (Fig. 2a).
Southern blot analysis and physical map of Stellaria PHYC gene. a Southern blot analysis. Total DNA (10 μg) from Stellaria longipes prairie ecotypes was digested with DraI, EcoRV, or HindIII. Hybridization with an internal probe including the chromophore binding site was carried out under highly stringent conditions. The positions of λDNA/HindIII markers (kb) are shown on the left. b Restriction map of the Stellaria PHYC gene
Stellaria PHYC protein
The polypeptide encoded by the Stellaria PHYC gene is predicted to be 1,114 amino acids in length (Fig. 3a). The nucleotide and amino acid sequence identities of the Stellaria PHYC compared with the individual members of the rice, Sorghum, Arabidopsis, and tomato PHY gene families have been analyzed by the alignment of complete Stellaria PHYC with other phytochromes (Table 2). The Stellaria PHYC shares 60%, 58% and 57% deduced amino acid identities with rice, Sorghum, and Arabidopsis PHYC, respectively. However, Stellaria PHYC and tomato PHYF share 62% identity (Table 2). The conserved residues are distributed throughout the polypeptide, but the region of highest conservation lies within the amino-terminal one-third of the protein (Fig. 3a). This portion of phytochrome has been suggested to be important for chromophore attachment to the apoprotein and for spectral integrity of the holo-phytochrome (Cherry et al. 1993; Clack et al. 1994). The chromophore-binding region is the same as in Arabidopsis and rice PHYC, which have a cysteine at position 318. In addition, the COOH-terminal domain of Stellaria PHYC has a copy of a sequence designated the histidine-kinase domain. The histidine residue of the canonical prokaryotic two-component sensor ‘H-box’ present in the majority of phytochromes (Quail 1997a, 1997b) is absent in Stellaria PHYC, PHYA1 and PHYA2 proteins (Li and Chinnappa 2003), as well as in rice and sorghum PHYC proteins (Fig. 3b; Basu et al. 2000).
Stellaria PHYC protein. a Alignment of the deduced amino acid sequences of the Stellaria, Arabidopsis and rice PHYC using CLUSTALX. Identical amino acids are highlighted in black, similar residues in gray. Dashes Deletions compared to the other genes, * predicted chromophore binding site (Cysteine). GenBank accession numbers: Stellaria PHYC (AF544029), Arabidopsis PHYC (X17343), rice PHYC (AF141942). b Multiple alignment of the histidine kinase-like domain (HKLD) of the Stellaria PHYC, PHYA1 (AF544027), PHYA2 (AY190013), Arabidopsis PHYC, rice PHYC, and Sorghum PHYC (U56731) proteins. * Histidine residue within HKLD domain present only in Arabidopsis PHYC and absent in all other PHY proteins
Phylogenetic analysis
Figure 4 presents the phylogenetic tree resulting from analysis of all full-length plant PHYC and PHYF amino acid sequences currently available in the databases. The tree indicates that all phytochromes examined are divided into two groups, monocots and dicots. Stellaria PHYC is located in the dicot branch as expected, but is relatively divergent from Arabidopsis PHYC.
A distance-based, neighbor-joining tree relating the complete Stellaria PHYC amino acid sequence to all known full-length plant phytochromes of PHYC/PHYF subfamily. Sequences were aligned with CLUSTALX 1.8 program. A neighbor-joining tree was constructed with NJplot. A bootstrap analysis (100 replicates) was performed with BOOTSTRAP. Arabidopsis PHYA was used as an outgroup. Sequences examined: Arabidopsis PHYA (X17341), PHYC (X17343), Oryza PHYC (AF141942), Solanum PHYF (AF178568), Sorghum PHYC (U56731), and Stellaria PHYC (AF544029)
PHYC gene expression
Stellaria PHYC gene expression has been determined in different organs, under different photoperiods, and at different R/FR ratios using semiquantitative RT-PCR. A 23S rRNA gene fragment was used as an internal control for RNA amount standardization. PHYC and 23S gene-specific primers (QmCF/QmCR, 23SF/23SR; Table 1) were designed based on the PHYC and 23S cDNA sequences. They were initially tested with PCR and RT-PCR. A 2,351-bp PHYC fragment was amplified by PCR and a 627-bp PHYC fragment was amplified by RT-PCR with primer pair QmCF/QmCR. A 381-bp 23S fragment was amplified with primer pair 23SF/23SR by RT-PCR (data not shown). Both fragments (627-bp PHYC and 381-bp 23S) were amplified at the same time with the two primer pairs (QmCF/QmCR and 23SF/23SR; data not shown). If total RNAs were contaminated with genomic DNA, a 2,351-bp PHYC fragment would also be amplified by RT-PCR, in addition to the 627-bp PHYC fragment. Therefore, the primers for PHYC are not only gene specific, but can also distinguish the PHYC cDNA fragment from any contamination of genomic DNA by fragment size.
Figure 5 shows that PHYC is constitutively expressed in flowers, leaves, stems, and roots of mature Stellaria plants, but the level of PHYC mRNA in roots is slightly higher than in other organs. The relative transcript abundances of PHYC are 0.71 in roots, and 0.41–0.5 in flowers, leaves and stems (Fig. 5b).
Analysis of PHYC transcript levels in different organs. Stellaria plants grown in short-day cold (SDC) conditions for at least 90 days were transferred into a growth chamber with long-day warm (LDW) conditions (22°C day/18°C night, 16 h photoperiod) until reproductive stage, and then leaves (L), stems (S), and flowers (F) were collected. Roots (R) were obtained from the young remets planted in 8 cm pots containing sterile sand under LDW for 14 days. a RT-PCR analysis of PHYC expression; 5 μl RT-PCR product for each sample was separated on a 1.7% (w/v) agarose gel and visualized with ethidium bromide under UV light. b PHYC gene expression relative to 23S control. PHYC transcript abundance levels are given as the ratio of optical density of PHYC/23S bands. Error bars Standard error of the mean. The results are from at least two individual experiments
To characterize the light stability of Stellaria PHYC, 2-week-old seedlings that were grown in the dark, and grown in the dark but irradiated with 1 h, 6 h or 12 h of continuous white light before harvest, were analyzed by semiquantitative RT-PCR. The levels of PHYC gene transcription increased slightly when Stellaria seedlings grown in complete darkness for 14 days (0 h) were exposed to continuous white light for 1 h, 6 h, or 12 h (Fig. 6a, b). The relative transcript abundance reached 0.7 at 12 h, which is about 1.5-fold higher than the control (0 h).
PHYC gene expression patterns in seedlings. Seedlings were grown for 14 days in the dark (0 h), and then transferred to continuous white light for the times indicated (1 h, 6 h, 12 h). PHYC transcript levels were analyzed by semiquantitative RT-PCR. a RT-PCR analysis of PHYC expression; 5 μl RT-PCR product for each sample was separated on a 1.7% (w/v) agarose gel and visualized with ethidium bromide under UV light. b Graph of PHYC gene expression relative to 23S control. Levels are given as the ratio of optical density of PHYC/23S bands. Error bars Standard error of the mean. The results are from at least two individual experiments
Plants grown in SDC were transferred to darkness for 7 days and then transferred to LDW with R/FR=0.7 or 3.5. Dark-adapted plants showed the lowest level of PHYC mRNA (Fig. 7a–c). Transfer of the dark-adapted plants into R/FR=0.7 or 3.5 increased PHYC gene transcript levels within 1 h of exposure. Maximum accumulation of PHYC transcript occurred after 9 h of exposure to R/FR=0.7 (Fig. 7a, c), or 6 h of exposure to R/FR=3.5 (Fig. 7b, c). An interesting observation is that the level of PHYC gene transcript mRNA under low R/FR ratio (0.7) was much higher than under high R/FR ratio (3.5) (Fig. 7c).
PHYC transcript levels under varied red/far-red (R/FR) ratios. Stellaria plants grown under SDC conditions for at least 90 days and in the dark for 7 days (0 h), were transferred into two different chambers with R/FR=0.7 or =3.5 for 1–24 h. PHYC gene expression was analyzed by semiquantitative RT-PCR; 5 μl RT-PCR product for each sample was separated on a 1.7% (w/v) agarose gel, visualized with ethidium bromide staining under UV light, and analyzed with NIH Image analysis software (Research Services Branch of the National Institute of Mental Health, USA). a PHYC expression in R/FR=0.7. b PHYC expression in R/FR=3.5. c Graph of PHYC gene expression relative to 23S control. The number is given as a ratio of optical density of PHYC/23S bands. Error bars Standard error of the mean. The results are from at least two individual experiments
The plants grown in SDC were transferred to short-day (8 h/day) and long-day (20 h/day) photoperiods, and were used to determine the effect of different photoperiods on PHYC gene expression. Under SD conditions, PHYC transcript levels were generally higher, except at 11 days (Fig. 8a, c), while under the LD conditions they remained at levels similar to those in SDC-grown plants except at 3 days (Fig. 8b, c). Maximum accumulation of PHYC transcript occurred at 15 days exposure to SD or LD conditions, which reached 1.11 and 0.85, respectively (Fig. 8c). The results of stem elongation under the two different photoperiods are shown in Fig. 8d. Plants grown under SD conditions did not elongate; however, plants grown under LD conditions did elongate. Average heights reached about 1.7 cm and 21.2 cm tall at 19 days under SD and LD conditions, respectively (Fig. 8d). In addition, we observed that plants did not flower (at 60 days) under SD conditions. In contrast, plants flowered at 18–19 days under LD conditions.
PHYC gene expression and stem elongation under two different photoperiods. Stellaria plants grown under SDC conditions for at least 90 days (0 days) were transferred into two different growth chambers with 8 h/day (SD) or 16-h/day (LD) conditions for the indicated times. PHYC gene expression was analyzed by semiquantitative RT-PCR; 5 μl RT-PCR product for each sample was separated on a 1.7% (w/v) agarose gel, visualized with ethidium bromide under UV light, and analyzed with NIH Image analysis software. a PHYC gene expression under SD conditions. b PHYC gene expression under LD conditions. c Graph of PHYC gene expression relative to 23S control. Levels are given as the ratio of optical density of PHYC/23S bands. Error bars Standard error of the mean. The results are from at least two individual experiments. d Stem elongation under SD and LD conditions. Error bars Standard error of the mean. The stem height results are from six individual plants
Discussion
PHY gene structure
All plant PHY genes thus far reported contain three introns located at relatively conserved regions within their coding regions, with the exception of the Arabidopsis PHYC which lacks the third intron (Cowl et al. 1994). Since rice, Sorghum, and other lower-plant PHYC genes have three introns, Basu et al. (2000) suggested that the absence of the Arabidopsis third intron is the result of a relatively recent evolutionary event after monocot/dicot divergence. However, our data indicates that Stellaria PHYC (dicot) also contains three introns at all conserved locations within the coding region, which is similar to rice PHYC (Fig. 1) and Sorghum PHYC (data not shown). Therefore, it is not clear as to when the Arabidopsis PHYC would have lost its third intron. In addition, the intron sizes between different PHY genes are significantly different. The first introns in the Arabidopsis and rice PHYC genes are only 137-bp and 104-bp in length, respectively. In contrast, the first intron of Stellaria PHYC is 1,724-bp in length (Fig. 1). The function of this long intron in Stellaria PHYC is currently unknown. A BLAST search using the 110-bp 5′-UTR of the Stellaria PHYC showed no match to the 5′-UTR of the Arabidopsis, rice, or Sorghum PHYC genes.
Stellaria PHYC protein
The deduced amino acid sequence of rice PHYC shares significantly higher identity with Sorghum PHYC (86%) than with Arabidopsis PHYC (59%) (Basu et al. 2000). Since both Stellaria and Arabidopsis are dicots, the Stellaria PHYC should share a much higher identity with Arabidopsis PHYC than with rice or Sorghum PHYC. In fact, Stellaria PHYC shares lower identity (57%) with Arabidopsis PHYC than with Sorghum (58%), or rice PHYC (60%) (Table 2). This result indicates that the Stellaria PHYC has diverged more from Arabidopsis PHYC than from rice or Sorghum PHYC.
Comparison of the Stellaria PHYC amino acid sequence with other phytochromes indicates that it is similar to rice, Sorghum, and Arabidopsis PHYC protein. The Stellaria PHYC has a number of sequence features typical of conventional plant phytochromes (Quail 1997a, 1997b), including a C-terminal histidine-kinase-related domain (Fig. 3b). Although the histidine in the ‘H-box’ is present in the majority of eukaryotic phytochromes (Quail 1997b), e.g., rice and Sorghum PHYC, Stellaria PHYC does not have this histidine in the ‘H-box’. Stellaria PHYA1 and PHYA2 (Li and Chinnappa 2003) also lack this histidine, in contrast to rice and Arabidopsis PHYA, which do contain this histidine residue (Fig. 3b). Thus, based on this analysis, the absence of the histidine in the ‘H-box’ is a feature not only of PHYC members (Basu et al. 2000), but also of some PHYA members.
PHYC gene phylogeny
The phytochromes in all plants examined are encoded by small, multigene families that have been classified into four subfamilies: A, B/D, C/F, and E (Mathews et al. 1995; Mathews and Sharrock 1996, 1997; Pratt et al. 1997). In the Arabidopsis PHY gene family, three diversification events have occurred: the PHYA/C-PHYB/D/E duplication near the origin of seed plants, and the PHYA-PHYC and PHYB/D-PHYE duplications near the origin of flowering plants (Mathews and Sharrock 1997). The Stellaria PHY gene family is composed of six members (four PHYAs, one PHYB, and one PHYC) that have been classified into three subfamilies: PHYA, PHYB, and PHYC (Li and Chinnappa 2003). Analysis of PHYC gene phylogeny indicates that the Stellaria PHYC branch is located at the middle of the rice/Sorghum PHYC branch (monocots) and Arabidopsis PHYC/tomato PHYF branch (dicots), though Stellaria PHYC belongs to the dicot group (Fig. 4). This is similar to the results we observed in the phylogenic analysis of Stellaria PHYA and PHYB (Li and Chinnappa 2003).
Stellaria PHYC expression
The Stellaria PHYC gene is expressed in different organs, but the level of PHYC mRNA in roots is slightly higher than in flowers, leaves and stems (Fig. 5a, b). These results are different from those reported for PHYC in Arabidopsis (Sharrock and Quail 1989; Clack et al. 1994). Based on analysis of Northern blots, the Arabidopsis PHYC mRNA has been found to be fairly uniformly distributed in roots, leaves, stems, and flowers of mature plants (Clack et al. 1994). This difference may be due to the different materials and methodologies used. In addition, mRNA levels increased slightly after seedlings were exposed to continuous white light for 1–12 h, reaching a relative level of 0.69 at 12 h (Fig. 6a, b), which was about 141% of that in the dark (data not shown).
Plants have evolved an endogenous circadian clock to allow the synchronization of internal events with daily changes in the external environment. For a circadian clock to function correctly, however, it must have not only a 24-h period but should also be entrained in the correct relationship to the local day/night cycle. Light signals at dawn and dusk are most important in entraining circadian clocks, though temperature cycles also contribute (Lumsden and Millar 1998). The entraining light signals are transduced by light input pathways, which are involved in at least two classes of photoreceptors, phytochromes and cryptochromes. However, the rhythmic expression of many genes becomes arrhythmic upon transfer to constant darkness, for example, adopting a constant level within two or three cycles. This could occur due to the effect of light on the clock via the input pathways, since many rhythmic processes are regulated by light, or because light affects the coupling between the clock and some target proteins (Hall et al. 2001). In tomato, following a light-to-dark transition, all five PHY transcripts exhibit modest increases during the subsequent 48 h. However, following a dark-to-light transition, PHYA and PHYB2 transcripts exhibit a marked decline. PHYF exhibits a negligible response to the transition. PHYB1 and PHYE undergo a transient increase during the first few hours, followed by a decline and, at least in the case of PHYB, a second transient increase (Hauser et al. 1998). In order to study the effect of different R/FR ratios on PHYC gene expression in Stellaria, we used 2-week-old seedlings grown in the dark as the initial material to minimize the effect of the rhythmic expression of the PHYC, followed by transfer to R/FR=0.7 or 3.5 for 1–24 h (see Fig. 7a–c). The results indicate that patterns of Stellaria PHYC gene expression are different under various R/FR ratios. The level of PHYC mRNA under R/FR=0.7 is much higher than under R/FR=3.5. The peak period was 9 h for R/FR=0.7 and 6 h for R/FR=3.5 (Fig. 7a–c).
The pattern of PHYC gene expression under 8 h/day photoperiods is significantly different from 20 h/day (Fig. 8a–c). Under 8 h photoperiods, PHYC transcript levels were generally higher. However, under 20 h photoperiods PHYC transcript levels remained at levels similar to those in SDC-grown plants. The apparent inconsistencies in this data set are the PHYC transcript levels under 8 h photoperiods at 11 days and under 20 h photoperiods at 3 days (Fig. 8a–c). These results were from three replicated experiments. In each individual experiment, we observed similar results. In addition, we also determined the expression pattern of the PHYC gene under 16 h photoperiods. The patterns under 16 h photoperiods were identical to those under 20 h photoperiods (data not shown). The phenotype under 16 h photoperiods was identical to that under 20 h photoperiods, but was significantly different from that under 8 h photoperiods. However, although these observations were confirmed experimentally, it is not understood why the levels of PHYC mRNA were lower at 3 days under SD conditions or at 11 days under LD conditions. We also determined PHYC mRNA levels at 1 day, and 2 days after the plants were transferred from SDC to SD or LD conditions. Under LD conditions, the levels of PHYC mRNA decreased significantly at 1 days or 2 days. This result also confirms that the lowest level of PHYC mRNA occurred at 3 days. In contrast, under SD conditions, the levels of PHYC increased significantly at 1 day and 2 days (data not shown). Thus, we propose that the levels of PHYC mRNA between the 1st and 3rd day may play an important role in the control of stem elongation in S. longipes. During the period between 1 day and 3 days, the low or high levels of PHYC mRNAs may induce different physiological and developmental pathways.
Plants grown under an 8 h/day photoperiod did not elongate at all and were only about 1.7 cm tall at 19 days. In contrast, plants grown under a 20 h/day photoperiod elongated and had an average height of 21.2 cm at 19 days. In addition, plants did not flower under SD conditions, while plants flowered at 18–19 days under LD conditions. These results indicate that SD photoperiods may initially stimulate PHYC gene expression, and the subsequently higher levels of PHYC mRNA may inhibit stem elongation and flower initiation. If this hypothesis is correct, the question remains as to why there is a reduction of PHYC mRNA from 7 days to 19 days under a 20 h photoperiod, where elongation is induced. Recently, Monte et al. (2003) reported that PHYC is involved in photomorphogenesis throughout the life cycle of the plant, with a photosensory specificity similar to that of PHYB/D/E and with a complex pattern of differential crosstalk between PHYA and PHYB in the photoregulation of multiple developmental processes. In the PHYA background, for example, the phyc mutation led to late flowering under LD conditions compared with the wild type and phya mutants. The data suggested that PHYC has a positive effect on flowering under LD conditions in a manner redundant with PHYA. In contrast, under SD conditions, the phyc mutant exhibited early flowering compared with wild type, suggesting that PHYC has an inhibitory role in flowering induction. Thus, PHYC is able to promote flowering under LD conditions, and inhibit initiation of flowering under SD conditions (Monte et al. 2003). Hence, we can assume that Stellaria plants did not flower under SD conditions due to the high levels of PHYC mRNA, which inhibits flower initiation under SD conditions. Under LD conditions, Stellaria plants that flower normally are likely to flower due to the high levels of PHYC during the initiation of flowering (~15 days) since PHYC is able to promote flowering under LD conditions. Hence, it is possible that PHYC may have functions in the control of stem elongation and in the control of floral initiation in S. longipes similar to those found in Arabidopsis studies (Franklin et al. 2003a; Monte et al. 2003).
Temperature is one of the most important factors regulating plant development (Blazquez et al. 2003; Halliday and Whitelam 2003; Halliday et al. 2003). Many plants, for example, have adopted a reproductive strategy that requires long periods of cold (1–10°C) to promote flowering. Halliday et al. (2003) demonstrated that ambient temperature is a significant modulator of photoreceptor action in the control of flowering. A modest reduction in growth temperature, from 22°C to 16°C, completely abolished the phyB mutant early-flowering phenotype frequently observed at higher temperature. Thus, small changes in ambient temperature can have a large impact on photoreceptor action (Halliday and Whitelam 2003). In addition, Blazquez et al. (2003) reported that the expression of some genes involved in the control of flowering is higher at 23°C than at 16°C, while others are lower at 23°C than at 16°C. In this study, we also observed that the levels of PHYC transcript increased markedly when the plants were transferred from SDC (8-h light/16-h dark, at 4°C; 0 days) to SDW (8-h light/16-h dark, at 22°C; 3 days) (Fig. 8c). The data suggests that PHYC gene expression is upregulated by temperature in S. longipes. The differences observed in the PHYC gene expression studies, however, are probably not due to the effects of temperature since the temperatures were identical, whereas R/FR ratios were different. Therefore, any differences in the levels of PHYC mRNA between R/FR=0.7 and 3.5 (Fig. 7) or between 8 h/day and 20 h/day photoperiod (Fig. 8) were likely due to quality of light and photoperiod rather than temperature.
Abbreviations
- LDW :
-
Long-day warm
- PHY :
-
Phytochrome gene
- R/FR :
-
Red/far-red light ratio
- SDC :
-
Short-day cold
References
Alokam S, Chinnappa CC, Reid DM (2002) Red/far-red light mediated stem elongation and anthocyanin accumulation in Stellaria longipes: differential response of alpine and prairie ecotypes. Can J Bot 80:72–81
Altschul F, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aukerman M, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino R, Sharrock RA (1997) A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9:1317–1326
Basu D, Dehesh K, Schneider-Poetsch HJ, Harrington SE, McCouch SR, Quail PH (2000) Rice PHYC gene: structure, expression, map position and evolution. Plant Mol Biol 44:27–42
Blazquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33:168–171
Casal JJ, Sanchez RA, Yanovsky MJ (1997) The function of phytochrome A. Plant Cell Environ 20:813–819
Cherry JR, Hondred D, Walker JM, Keller JM, Hershey HP, Vierstra RD (1993) Carboxy-terminal deletion analysis of oat phytochrome A reveals the presence of separate domains required for structure and biological activity. Plant Cell 5:565–575
Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25:413–427
Clapham DH, Kolukisaoglu Ü, Larsson CT, Qamaruddin M, Ekberg I, Wiegmann-Eirund C, Schneider-Poetsch HAW, von Arnold S (1999) Phytochrome types in Picea and Pinus. Expression patterns of PHYA-related types. Plant Mol Biol 40:669–678
Cowl JS, Hartley N, Xie DX, Whitelam GC, Murphy GP, Harberd NP (1994) The PHYC gene of Arabidopsis, absence of the third intron found in PHYA and PHYB. Plant Physiol 106:813–814
Devlin PF, Patel SR, Whitelam GC (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10:1479–1487
Devlin PF, Robson PRH, Patel SR, Goosey L, Sharrock RA, Whitelam GC (1999) Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol 119:909–915
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull 19:11–15
Eichenberg K, Baurle I, Paulo N, Sharrock RA, Rudiger W, Schafer E (2000) Arabidopsis phytochromes C and E have different spectral characteristics from those of phytochromes A and B. FEBS Lett 470:107–112
Emery RJN, Reid DM, Chinnappa CC (1994) Phenotypic plasticity of stem elongation in two ecotypes of Stellaria longipes: the role of ethylene and response to wind. Plant Cell Environ 17:691–700
Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC (2003a) Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 15:1981–1989
Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC (2003b) Phytochrome B, D and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131:1340–1346
Hall A, Kozma-Bognar L, Toth R, Nagy F, Millar AJ (2001) Conditional circadian regulation of phytochrome A gene expression. Plant Physiol 127:1808–1818
Halliday KJ, Whitelam GC (2003) Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiol 131:1913–1920
Halliday KJ, Thomas B, Whitelam GC (1997) Expression of heterologous phytochromes A, B, or C in transgenic tobacco plants alters vegetative development and flowering time. Plant J 12:1079–1090
Halliday KJ, Salter MG, Thingnaes E, Whitelam GC (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33:875–885
Hennig L, Stoddart WM, Dieterle M, Whitelam GC, Schafer E (2002) Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol 128:194–200
Hauser BA, Pratt LH, Cordonnier-Pratt MM (1997) Absolute quantification of five phytochrome transcripts in seedlings and mature plants of tomato (Solanum lycopersicum L.). Planta 201:379–387
Hauser BA, Cordonnier-Pratt MM, Pratt LH (1998) Temporal and photoregulated expression of five tomato phytochrome genes. Plant J 14:431–439
Heyer A, Gatz C (1992a) Isolation and characterization of a cDNA-clone coding for potato type A phytochrome. Plant Mol Biol 18:535–544
Heyer A, Gatz C (1992b) Isolation and characterization of a cDNA-clone coding for potato type B phytochrome. Plant Mol Biol 20:589–600
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405
Kathiresan A, Nagarathna KC, Moloney MM, Reid DM, Chinnappa CC (1998) Differential regulation of 1-aminocyclopropane-1-carboxylate synthase gene family and its role in phenotypic plasticity in Stellaria longipes. Plant Mol Biol 36:265–274
Li WZ, Chinnappa CC (2003) The phytochrome gene family in the Stellaria longipes complex. Int J Plant Sci 164:657–673
Lumsden PJ, Millar AJ (1998) Biological rhythms and photoperiodism in plants. BIOS Scientific, Oxford
Macdonald SE, Chinnappa CC, Reid DM (1984) Studies on Stellaria longipes complex: phenotypic plasticity. I. Response of stem elongation to temperature and photoperiod. Can J Bot 62:414–419
Mathews S, Sharrock RA (1996) The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol Biol Evol 13:1141–1150
Mathews S, Sharrock RA (1997) Phytochrome gene diversity. Plant Cell Environ 20:666–671
Mathews S, Lavin M, Sharrock RA (1995) Evolution of the phytochrome gene family and its utility for phylogenetic analyses of angiosperms. Ann Mo Bot Gard 82:296–321
Monte E, Alonso JM, Ecker JR, ZhangY, Li X, Young J, Austin-Phillips S, Quail PH (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling Pathways. Plant Cell 15:1962–1980
Perriere G, Gouy M (1996) WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369
Pratt LH, Cordonnier-Pratt MM, Kelmenson PM, Lazarova GI, Kubota T, Alba RM (1997) The phytochrome gene family in tomato (Solanum lycopersicum L.). Plant Cell Environ 20:672–677
Qin M, Kuhn R, Quail PH (1997) Overexpressed phytochrome C has similar photosensory specificity to phytochrome B but a distinctive capacity to enhance primary leaf expansion. Plant J 12:1163–1172
Quail PH (1997a) An emerging molecular map of the phytochromes. Plant Cell Environ 20:657–665
Quail PH (1997b) The phytochromes: a biochemical mechanism of signaling in site? BioEssays 19:571–579
Quail PH (2002) Phytochrome photosensory signaling networks. Nat Rev Mol Cell Biol 3:85–93
Schmidt M, Schneider-Poetsch HA (2002) The evolution of gymnosperms redrawn by phytochrome genes: the Gnetatae appear at the base of gymnosperms. J Mol Evol 54:715–724
Schneider-Poetsch HAW, Kolukisaoglu Ü, Clapham DH, Hughes J, Lamparter T (1998) Non-angiosperm phytochromes and the evolution of vascular plants. Physiol Plant 102:612–622
Sharrock RA, Quail PH (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3:1745–1757
Smith H, Whitelam GC (1997) The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant Cell Environ 20:840–844
Toth R, Kevei EE, Hall A, Millar AJ, Nagy F, Kozma-Bognar L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127:1607–1616
Weinig C (2002) Phytochrome photoreceptors mediate plasticity to light quality in flowers of the Brassicaceae. Am J Bot 89:230–235
Whitelam GC, Devlin PF (1997) Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ 20:752–758
Acknowledgement
This work is supported by an operating grant from the Natural Sciences and Engineering Research Council of Canada (A-7222).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, WZ., Chinnappa, C.C. Isolation and characterization of PHYC gene from Stellaria longipes: differential expression regulated by different red/far-red light ratios and photoperiods. Planta 220, 318–330 (2004). https://doi.org/10.1007/s00425-004-1337-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-004-1337-z