Abstract
Genomic restructuring was detected in newly synthesized tritordeum by molecular and cytogenetic tools. Genomic stability is expected for advanced tritordeum lines (HchHchAABB; 2n = 42) with multiple generations of self-fertilization. This study intends to confirm or decline this hypothesis by characterizing three advanced tritordeum lines and their parental species using cytogenetics, inter-simple sequence repeat (ISSR) and retrotransposon-based markers. Mitotic chromosomes of each tritordeum line were hybridized with six synthetic oligonucleotide probes using non-denaturing fluorescence in situ hybridization. Polymorphic hybridization patterns and structural rearrangements involving SSR regions were detected. The same chromosome spreads were re-hybridized with genomic DNA of Hordeum chilense Roem. et Schult. and the 45S ribosomal DNA (rDNA) sequence pTa71. These FISH experiments allowed for parental genome discrimination, identification of nucleolar chromosomes, and detection of structural rearrangements, mostly involving rDNA loci. The chromosomes bearing SSR hybridization signals and/or chromosomes involved in structural rearrangements were identified. ISSR, retrotransposon-microsatellite amplified polymorphism, inter-retrotransposon amplified polymorphism and inter-primer binding site markers evidenced genomic reshuffling in all tritordeum lines relative to their parents. Line HT28 was considered the most genetically stable. This work demonstrated that cytogenetic and molecular monitoring of tritordeum is needed, even after several self-fertilization generations, to guarantee the selection of the most stable lines for improvement and sustainable agriculture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Allopolyploids result from the merging of two or more divergent genomes within a single nucleus. Such genomic “shock” induces reorganization (McClintock 1978) at the DNA and/or chromosomal levels (Song et al. 1995; Wendel et al. 1995; Leitch and Bennett 1997; Ozkan et al. 2001; Cabo et al. 2014a); gene expression (Scheid et al. 1996; Comai et al. 2000); changes in methylation patterns (Shaked et al. 2001; Han et al. 2003); activation of retroelements (Zhao et al. 1998; Fedoroff 2000); and elimination of high- and/or low-copy DNA sequences (Feldman et al. 1997; Salina et al. 2000; Ozkan et al. 2001; Chen et al. 2007). Sequence elimination constitutes the most studied response to allopolyploidy in various species. The non-random elimination of coding and noncoding sequences of parental origin was the most common event in the first generations of newly formed allopolyploids (Ozkan et al. 2001, 2003; Chen and Chen 2007; Bento et al. 2008, 2010). Such a feature explains why allopolyploids have a genome size smaller than the expected sum of the sizes of their parental species (Ozkan et al. 2003).
Tritordeum (×Tritordeum Ascherson et Graebner) is a synthetic amphiploid derived from chromosome doubling of interspecific hybrids produced by crosses between wild barley (Hordeum chilense Roem. et Schult.) (HchHch; 2n = 2x = 14) and cultivated durum wheat (Triticum turgidum L. convar. durum (Desf.) MacKey (AABB; 2n = 4x = 28) (Martín and Sánchez-Monge Laguna 1982; Martín et al. 1996). Tritordeum could be an alternative crop, as it presents high contents of protein and carotenoids (Millán et al. 1988; Ballesteros et al. 2005; Atienza et al. 2007; Mellado-Ortega et al. 2015) as well as high adaptive potential to Mediterranean environments (Villegas et al. 2010). This cereal has been used as bridge material in breeding programs for the transference of useful agronomic traits from H. chilense to cultivated wheat (Martín 1988; Alvarez et al. 1992; Martín et al. 1999; Ballesteros et al. 2005).
Intergenomic translocations were reported previously in allopolyploids of the tribe Triticeae, namely in wheat–barley addition and translocation lines (Koba et al. 1997); tetraploid tritordeum (DDHchHch; 2n = 28) (Martín et al. 1998); derivatives of wheat–barley hybrids multiplied in vitro (Molnár-Láng et al. 2000); and F1 multigeneric hybrids (AABBRHch; 2n = 6x = 42) (Carvalho et al. 2009). Genomic in situ hybridization is an accurate technique for detection of intergenomic translocations, determination of breakpoints, and estimation of the amount of alien chromatin involved in structural rearrangements (Schwarzacher et al. 1989; Jiang et al. 1994).
Synthetic oligonucleotide probes for Triticeae species were developed based on sequences of cloned repetitive DNA sequences published in public databases (Tang et al. 2014a). These oligonucleotides can replace the role of the cloned repetitive probes, being less time-consuming and more cost-effective (Tang et al. 2014a). Furthermore, these synthetic oligonucleotides proved to be suitable for non-denaturing fluorescence in situ hybridization (ND-FISH) (Fu et al. 2015). This technique does not require chromosomal denaturing, preserving the chromosome morphology and enabling multiple reprobing experiments on the same chromosome spread (Cuadrado et al. 2009; Cuadrado and Jouve 2010; Cabo et al. 2014a). ND-FISH proved to be suitable for synthetic oligonucleotide probes, such as simple sequence repeats (SSRs), telomeric sequences (Cuadrado et al. 2009; Cuadrado and Jouve 2010; Cabo et al. 2014a; Pavia et al. 2014), and other synthetic oligonucleotide probes (Fu et al. 2015).
The study of the physical distribution and abundance of SSRs has given insights into the organization, dynamics, and evolution of genomes (Schmidt et al. 1993; Depeiges et al. 1995; Cuadrado and Schwarzacher 1998). SSR-rich regions also have been useful for the understanding of genomic restructuring in allopolyploids, including their use as probes. Structural rearrangements on mitotic wheat-origin chromosomes of newly synthesized hexaploid tritordeum were detected after ND-FISH performed with the SSR (AG)10 probe (Cabo et al. 2014a).
Transposable elements are involved in genome evolution through sequence and size modifications (Charles et al. 2008). A significant rate of polyploidization-induced rearrangements have unraveled the involvement of retrotransposons (RTNs) and/or SSR sequences in newly formed allopolyploids (Han et al. 2003; Chen et al. 2007; Bento et al. 2008, 2010; Cabo et al. 2014b). This type of genomic restructuring has been evidenced by inter-retrotransposon amplified polymorphism (IRAP), retrotransposon-microsatellite amplified polymorphism (REMAP), inter-simple sequence repeats (ISSRs) and inter-primer binding site (iPBS) markers (Bento et al. 2008, 2010; Cabo et al. 2014b; Delgado et al., unpublished).
The genomic restructuring studies developed so far relied on newly formed allopolyploids and evidenced rapid genomic modifications in the first generations after polyploidization (Ozkan et al. 2001; Han et al. 2003; Chen et al. 2007; Bento et al. 2008, 2010; Cabo et al. 2014a, b, c). Such studies allowed us to question whether multiple generations of self-fertilization could reduce the high genomic instability found in lines of newly formed tritordeum. To answer this question, we decided to study three advanced lines of hexaploid tritordeum that were previously characterized by our group at the mitotic, meiotic, molecular, and morphological levels (Lima-Brito 1998; Carvalho 2004). Furthermore, these lines were used as male parents in widespread hybridization with triticale, durum, and bread wheat, and some of the F1 interspecific hybrids showed structural rearrangements and intergenomic translocations (Carvalho et al. 2008, 2009).
With this study, we aimed to evaluate the genomic stability of three highly inbred lines of hexaploid tritordeum (HT9, HT28, and HT31; HchHchAABB; 2n = 42) by comparison with H. chilense and durum wheat using cytogenetic techniques and four molecular marker systems.
Materials and methods
Plant material
Five plants of each inbred advanced line of tritordeum (HT9, HT28, and HT31; HchHchAABB; 2n = 6x = 42) were cytogenetically and molecularly characterized. These advanced lines have a complex genealogy (Rubiales et al. 1991; Chauhan and Singh 1997; Ballesteros et al. 2005). We should highlight that the production of lines HT9 and HT31 involved crosses with an octoploid tritordeum (HchHchAABBDD; 2n = 56), whereas line HT28 resulted from an intraspecific cross between two hexaploid tritordeum lines (HchHchAABB; 2n = 42) (Rubiales et al. 1991; Chauhan and Singh 1997; Ballesteros et al. 2005). The plants of the three HT lines used in this studied resulted, at least, from 11 generations of self-fertilization performed at UTAD.
For further comparison with the three advanced HT lines, five plants of H. chilense line H1 (HchHch; 2n = 2x = 14) and five plants of durum wheat line T846 (AABB; 2n = 4x = 28) (involved in the genealogy of the advanced lines) were also studied. From now on, these lines will be referred to as parental species.
Seeds of lines HT9, HT28 and HT31, as well as those of H1 and T846, were allowed to germinate on moistened filter paper in the dark at 25 °C. Root tips 1.5 cm in length were collected, treated in ice-cold water for 24 h, and fixed in a freshly prepared solution of absolute ethanol and glacial acetic acid, 3:1 (v/v). The fixed root tips were maintained at −20 °C until the preparation of mitotic chromosome spreads.
The plants were grown in greenhouse conditions for further harvesting of young leaves and genomic DNA isolation.
ND-FISH with synthetic oligonucleotide probes
The SSR sequences (AG)10, (CT)10, (AGA)5, and (TTC)5 were 5′-end labeled with biotin or fluorescein isothiocyanate and commercially purchased from StabVida (Oeiras, Portugal). These synthetic oligonucleotides were used as probes in ND-FISH experiments that were performed according to methods of Cuadrado and Jouve (2010). Pre-treatments of the mitotic chromosome spreads using the Schwarzacher and Heslop-Harrison (2000) protocol were also performed. The biotin-labeled SSR probes were detected with the antibody fluorescein-avidin (Vector Laboratories, Burlingame, CA, USA).
The SSR sequence (AAC)5 was labeled with biotin-16-dUTP (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany) with a Random Primed DNA Labeling kit (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany). This SSR probe required a different ND-FISH protocol, as described below. The pre-treated chromosome spreads were incubated with 30 μL of hybridization mixture containing the following: 1 pmol of SSR (AAC)5 probe; 1.5 μL of 100 % Denhardt’s solution (composition: 20 g L−1 of Ficoll; 20 g L−1 of Polyvinylpyrrolidone (PVP); and 20 g L−1 of bovine serum albumin (BSA); 1 % of sodium dodecyl sulfate (SDS); 7.5 μL of 20× salt sodium phosphate EDTA (SSPE) solution (composition: 3.6 M of NaCl; 200 mM of NaH2PO4; and 20 mM EDTA, pH 7.4); and a final concentration of 50 ng μL−1 of sheared unlabeled genomic DNA of Escherichia coli.
The hybridization occurred overnight at 37 °C on a Thermo Scientific™ Hybaid OmniSlide Thermal Cycler System (Thermo Fisher Scientific, Inc., Rockford, IL, USA). The post-hybridization washes started with the incubation of chromosome spreads in 6× salt sodium citrate (SSC) buffer (prepared from the stock solution 20× SSC: 3 M of NaCl and 0.3 M of sodium citrate) for 30 min at room temperature (RT). During this period, the 6× SSC solution was renewed three times. The more stringent wash was performed in 6× SSC at 35 °C for 2 min. The annealing temperature of the oligonucleotide (AAC)5 resulted from the reduction by 5 °C of its melting temperature (Tm), which was calculated according to the equation of Wallace et al. (1981). Then, 100 μL of a solution of 5 % BSA prepared in 6× SSC was loaded over each chromosome spread, and an incubation of 10 min at RT was performed. The chromosome spreads were washed three times for 15 min in the detection buffer 6× T (composition: 6× SSC and 0.05 % Tween 20). The hybridization signals were detected with a diluted solution (1:100) of the antibody fluorescein-avidin (Vector Laboratories, Peterborough, UK) in 6× SSC. The chromosome spreads were incubated with 100 μL of the antibody solution at 37 °C for 1 h and then washed for 15 min in detection buffer 6× T (renewed three times). The slides were mounted with Vectashield Antifade mounting medium with DAPI (Vector Laboratories, Peterborough, UK). The images were captured on an Olympus BX51 fluorescence microscope (Olympus America, Inc., Hauppauge, NY, USA) after double or triple exposure with appropriated filters by an XC-50 charge-coupled device (CCD) digital camera (Olympus America, Inc., Hauppauge, NY, USA) using the software cellSens (Olympus Soft Imaging Solutions GmbH, Münster, Germany).
The synthetic Oligo-pTa535-1 and Oligo-pTa535-2 probes (Tang et al. 2014a) were labeled at their 5′-ends with fluorochrome 6-carboxytetramethylrhodamine (6-TAMRA) and commercially purchased from StabVida (Oeiras, Portugal). An amount of 6 pmol μL−1 of each probe was diluted in 2× SSC and 1× TE buffer (composition: 10 mM Tris and 1 mM EDTA, pH 8.0). To obtain all hybridization signals presented by Tang et al. (2014a), equal amounts of the Oligo-pTa535-1 and Oligo-pTa535-2 probes were included in the hybridization mixture. Throughout this study, the combined Oligo-pTa535-1 and Oligo-pTa535-2 probes will be referred to only as Oligo-pTa535.
The reprobing ND-FISH experiments were performed with the Oligo-pTa535 probe, and they followed the hybridization conditions recommended by Fu et al. (2015). The following steps were completed as described above for the SSR (AAC)5 probe, replacing the 6× SSC and 6× T buffers with 4× SSC and 4× T, respectively.
FISH experiments and reprobing
Genomic DNA of H. chilense and the 45S ribosomal DNA (rDNA) sequence, pTa71 (Gerlach and Bedbrook 1979), were labeled with biotin-16-dUTP (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany) and digoxigenin-11-dUTP (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany), respectively, using the Nick Translation Kit (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany). Unlabeled and fragmented genomic DNA of durum wheat was used as blocking DNA. The amount of blocking DNA exceeded the amount of genomic probe in 80 times. The FISH protocol was performed according to methods of Schwarzacher and Heslop-Harrison (2000).
Each chromosome spread was re-hybridized at least three times following the reprobing method of Heslop-Harrison et al. (1992).
Chromosome identification
The individual wheat- and H. chilense-origin chromosomes of tritordeum bearing SSR hybridization signals or those chromosomes involved in structural rearrangements were identified. For that purpose, their hybridization patterns were compared with ideograms published previously by other authors (Mukai et al. 1993; Cuadrado et al. 2000, 2008a; Komuro et al. 2013; Tang et al. 2014a; Fu et al. 2015). We also used our previously constructed ideogram for H. chilense-origin chromosomes of tritordeum based on the hybridization patterns of the synthetic oligonucleotide probe, Oligo-pTa535 (Delgado et al. 2016).
Genomic DNA extraction
Young leaves of plants from lines HT9, HT28, and HT31; H. chilense line H1; and durum wheat line T846 were collected and frozen immediately in liquid nitrogen. The leaves were maintained at −80 °C until genomic DNA isolation using the Cetyl trimethylammonium bromide (CTAB) method of Doyle and Doyle (1987).
The integrity of the genomic DNA samples was evaluated after electrophoresis on 0.8 % agarose gels stained with ethidium bromide. The DNA samples were quantified on a Nanodrop ND-1000® spectrophotometer (Thermo Scientific™, Thermo Fisher Scientific, Inc., Rockford, IL, USA) and diluted to 40 ng µL−1 for further amplification of molecular markers. The genomic DNA of H. chilense and durum wheat was also used for genomic probes and for blocking DNA production, respectively.
Amplification of molecular markers
For REMAP, IRAP, ISSR and iPBS amplifications, we used 14 combinations of long terminal repeat (LTR), SSR and/or PBS primers (Table 2) previously designed and reported by other authors (Teo et al. 2005; Kalendar et al. 1999, 2008; Wegscheider et al. 2009). The sequences of all primers are presented in the Online Resource 1. For the amplification and visualization of the IRAP, REMAP, ISSR and iPBS markers we used the conditions described by Cabo et al. (2014b).
Each band was considered a marker. Each PCR reaction was repeated twice and only reproducible bands were considered for presence (1) or absence (0) analysis. Since the amplification conditions were the same for ISSR, IRAP and REMAP markers, in each REMAP matrix, bands with similar molecular weights to ISSRs and/or IRAPs produced with the same SSR or LTR primers respectively, were discarded to ensure the analysis of effective REMAPs as suggested by Kalendar et al. (1999).
Results
Physical location of SSRs and parental genome discrimination
The ND-FISH experiments were performed with the SSR probes (AG)10, (CT)10, (AGA)5, (TTC)5, and (AAC)5 in mitotic chromosome spreads of plants of tritordeum lines HT9, HT28, and HT31; H. chilense line H1; and durum wheat line T846.
Table 1 presents the number of chromosomes per metaphase cell that showed hybridization of the probes SSR (AG)10, (CT)10, (AAC)5, (AGA)5, and (TTC)5 in each plant material.
Probes (AG)10 and (CT)10 showed a pericentromeric or subtelomeric location in durum wheat and in the three advanced HT lines (Fig. 1a, c–e; S1). After reprobing the same metaphase cells with H. chilense genomic DNA and pTa71, we confirmed that both SSR (AG)10 and SSR (CT)10 hybridized on wheat-origin chromosomes of tritordeum (Fig. 1b, f). These SSR probes did not hybridize on chromosomes of H. chilense line H1.
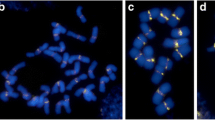
Mitotic metaphase cells of the hexaploid tritordeum after ND-FISH performed with SSR probes (a, c, d, e, g, i, k) and the reprobing of some of them with genomic DNA of H. chilense and pTa71 (b, f, h, j). a (AG)10 in one cell of plant HT28-5 showing six hybridization signals; c (AG)10 in one cell of plant HT9-15 showing 10 hybridization signals, with two of them located in the same chromosome (arrow); d (CT)10 in one cell of plant HT31-19 presenting six hybridization signals and a detached satellite region (yellow arrow) of one 6B wheat-origin chromosome (yellow); e (CT)10 in one cell of plant HT9-15 showing a total of 10 hybridization signals; g (AGA)5 in one cell of plant HT31-7 showing hybridization on 36 chromosomes; i (TTC)5 in one cell of plant HT28-5 showing hybridization on 36 chromosomes; and k (TTC)5 in one cell of plant HT31-7 presenting hybridization on 36 chromosomes. The nucleolar chromosomes were identified by pTa71 (b, f, h, j). Some chromosomes bearing SSR (AG)10 and SSR (CT)10 hybridizations were identified based on previously published ideograms (Cuadrado et al. 2000, 2008a; Cabo et al. 2014a). Scale bars 20 μm. (Color figure online)
The number of chromosomes showing hybridization signals of SSR (AG)10 and (CT)10 probes differed among HT lines and also among cells of the same plant (Table 1; Fig. 1a, c–e). Lines HT28 and HT31 showed six hybridization signals per metaphase cell when hybridized with the SSR (AG)10 probe (Table 1; Fig. 1a, d). Based on the previous ideograms of Cabo et al. (2014a) for SSR (AG)10 and Cuadrado et al. (2008a) for SSR (AG)12, we assigned the hybridization of (AG)10 in tritordeum to the wheat-origin chromosome pairs 5A, 3B, and 6B (Fig. 1a, b). Plants of line HT9 exhibited metaphase cells with 10 hybridization signals of SSR (AG)10 distributed by 10 chromosomes (Table 1). In fact, one metaphase cell of plant HT9-15 showed 10 hybridization signals of SSR (AG)10 on nine chromosomes, being two of the signals detected in a single chromosome (Table 1; Fig. 1c). After reprobing with additional repetitive DNA sequences, we identified the wheat-origin chromosomes of plants from line HT9 bearing the SSR (AG)10 hybridization signals as 5A, 3B, 4B, 5B, and 6B (Online Resource 3). The chromosome of the metaphase cell of plant HT9-15 with two SSR (AG)10 hybridization signals (one per arm) (Fig. 1c, arrow) was identified as being the derivative 5A chromosome (Fig. 3h).
Since SSR (CT)10 is complementary to sequence (AG)10 and presented hybridization in chromosomes with similar shape and size, we also assigned the location of SSR (CT)10 to the chromosome pairs 5A, 3B, and 5B in plants of lines HT28 and HT31 (Fig. 1d) and to chromosome pairs 5A, 3B, 4B, 5B, and 6B in plants of line HT9 (Fig. 1e). Only one metaphase cell of plant HT31-20 showed five hybridization signals of SSR (CT)10 instead of the six signals scored in their remaining cells and in other plants of the same line (Table 1; Online Resource 2). Furthermore, all metaphase cells of the HT31 plants presented a heteromorphic 6B wheat-origin chromosome pair due to the deletion of the satellite region of one 6B chromosome that was noticed by the observation of its rDNA locus located at the telomere (Fig. 1h). In some metaphase cells, a detached satellite was observed (Fig. 1d, yellow arrow). The reprobing of the metaphase cell presented in Fig. 1e with H. chilense genomic DNA and pTa71 allowed the detection of a homozygous pericentric inversion of the rDNA locus in the H. chilense-origin chromosome pair 6Hch, located on the 6HchL arm of the derivative chromosomes (Fig. 1f, orange arrows).
SSR probes (AAC)5, (AGA)5, and (TTC)5 showed highly intense and interspersed hybridization signals on most of the chromosomes of both H. chilense and durum wheat parental species and tritordeum. No polymorphic patterns of (AAC)5, (AGA)5, nor (TTC)5 were detected within and among HT lines (Table 1; Fig. 1g, i, k). The number of chromosomes with hybridization of the SSRs (AAC)5, (AGA)5, and (TTC)5 in tritordeum constitutes the sum of those observed in the parental species (Table 1).
The SSR (AAC)5 hybridized on 12 chromosomes of line H1 (all H. chilense chromosomes except for the pair 5Hch) and on 20 chromosomes of durum wheat line T846, resulting in 32 chromosomes labeled with this sequence in each tritordeum line (Table 1). Both SSRs (AGA)5 and (TTC)5 did not hybridize on nucleolar chromosome pairs 5Hch and 6Hch of H. chilense line H1, nor in any of the HT line; however, these SSRs hybridized on 26 wheat chromosomes, resulting in 36 chromosomes labeled with these sequences in each HT line (Table 1; Fig. 1g, i, k).
The chromosome spreads of all HT plants were re-hybridized with H. chilense genomic DNA and pTa71 simultaneously, allowing for parental genome discrimination and unequivocal identification of the nucleolar chromosomes of wheat and H. chilense origin. This procedure was also useful for the identification of the parental origin of chromosomes bearing SSR hybridization signals (Fig. 1b, f, h, j).
As expected, a total number of eight rDNA loci per metaphase cell of tritordeum, corresponding to chromosome pairs 1B and 6B of wheat origin and to pairs 5Hch and 6Hch of H. chilense origin, was observed (Fig. 1b, f, h, j).
Detection and identification of structural rearrangements
Structural rearrangements involving SSR-rich regions (Fig. 1c; S1) and rDNA loci (Fig. 2) were detected. In fact, polymorphisms in number and location of hybridization signals of the probes SSR (AG)10 and (CT)10 were observed among the three HT lines, relative to the wheat parent.
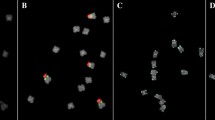
Mitotic metaphase cells of tritordeum plants HT9-15 (a–d) and HT31-20 (e) after reprobing FISH with genomic DNA of H. chilense (green) and pTa71 (red), simultaneously, as probes, showing structural rearrangements (arrows). Wheat chromosomes were counterstained with DAPI (blue). d Corresponds to the reprobing of Fig. 1c. Scale bars 20 μm. (Color figure online)
Reprobing FISH experiments using H. chilense genomic DNA and pTa71 as probes also enabled the identification of spontaneous intergenomic translocations and other structural rearrangements (Fig. 2). Most of the rearrangements involved one rDNA locus of wheat or H. chilense origin (Fig. 2a–e) and were detected in two HT plants: (1) HT9-15 (2n = 40/2n = 41/2n = 42) and (2) HT31-20 (2n = 42) (Fig. 2). Fifty metaphase cells were observed in each chromosome spread of plants HT9-15 and HT31-20. Most of the metaphase cells (92 %) of plant HT9-15 showed 42 chromosomes with the expected genomic constitution, namely 14 H. chilense-origin chromosomes and 28 wheat-origin chromosomes (Fig. 2). Nonetheless, aneuploidy (2n = 40 and 2n = 41) was observed in 8 % of the observed metaphase cells of this plant as well as the presence of structurally rearranged chromosomes (Fig. 2). Therefore, plant HT9-15 was considered a mosaic individual.
Figure 2e represents the unique metaphase with structural rearrangements detected in plant HT31-20, which corresponds to 2 % of the observed metaphase cells.
All nucleolar chromosomes of wheat and H. chilense origin were identified with the pTa71 probe (Fig. 2) using the ideogram of Mukai et al. (1993). The identification of most of the wheat-origin chromosomes involved in structural rearrangements of plants HT9-15 and HT31-20 (Fig. 2) was possible after reprobing the chromosome spreads with Oligo-pTa535 (Online Resource 3). Those hybridization patterns were compared with the ideograms reported by Komuro et al. (2013) and Tang et al. (2014a) for wheat chromosomes. The identification of structurally rearranged chromosomes of wheat origin was also complemented with the (AAC)5 sequence and by using the ideograms published by Cuadrado et al. (2000, 2008a). For the identification of structurally rearranged chromosomes involving H. chilense chromatin in the present and other studies, it was necessary to construct an ideogram for the hybridization patterns of Oligo-pTa535 in H. chilense-origin chromosomes of tritordeum (Delgado et al. 2016).
Figure 3 presents the derivative chromosomes identified by arrows in Fig. 2 and their identification after comparative analysis of their hybridization patterns. Figure S2 shows the same metaphase cells presented in Fig. 2 after reprobing with SSR (AAC)5 and/or Oligo-pTa535.
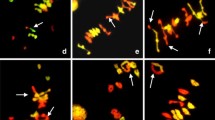
Hybridization patterns of the probes of genomic DNA of H. chilense, pTa71, four SSRs, and Oligo-pTa535 observed in 10 derivative (structurally rearranged) chromosomes of plants HT9-15 (a–h) and HT31-20 (i, j). The comparison with previously published ideograms allowed for their identification as follows: a der(6Hch/4A); b der(1A/sat6Hch); c der(6Hchinv/sat6B); d der(6B), with tandem duplication of the rDNA locus and deletion of the satellite region; e der(4HchLinv/sat6B); f der(4HchS/6B); g der(6Hch), with pericentric inversion of the rDNA locus; h der(5A/6HchL), also presenting an SSR (AG)10 hybridization signal on the 5AL arm, probably due to a duplication followed by a pericentric inversion; i der(4HchLinv.6BL); and j the reciprocal der(4HchSinv.6BS), with deletion of the 6B satellite region
Figure 2a shows a metaphase cell of plant HT9-15 with 2n = 40, showing 13 normal H. chilense-origin chromosomes (including the chromosome pair 5Hch and one chromosome 6Hch), 26 normal wheat-origin chromosomes, and one chromosome with a spontaneous intergenomic translocation (white arrow). This structurally rearranged chromosome was identified as der(6Hch/4A) (Figs. 2a, 3a).
The same plant HT9-15 contained a metaphase cell with 2n = 42 in which the following was observed: 13 normal H. chilense-origin chromosomes (including the chromosome pair 5Hch and one chromosome 6Hch); 25 normal wheat-origin chromosomes; and three rearranged chromosomes (Fig. 2b). The first rearranged chromosome (white arrow) exhibited a spontaneous intergenomic translocation of a wheat-origin chromosome with a terminal region of H. chilense in its long arm (Fig. 2b). The comparison of all hybridization patterns allowed us to conclude that this rearranged chromosome corresponds to der(1A/sat6Hch) (Fig. 3b). The second rearranged chromosome (Fig. 2b, green arrow) was derived from an inversion of the 6Hch chromosome and recombination with the satellite region of the 6B chromosome, resulting in der(6Hchinv/sat6B) (Fig. 3c). The third structurally rearranged chromosome also detected in the same metaphase cell consisted of one 6B wheat-origin chromosome with two adjacent rDNA loci (tandem duplication of the 6B rDNA locus) at the telomere, due to the deletion of the satellite region (Fig. 2b, yellow arrow; Fig. 3d).
A third metaphase cell of plant HT9-15 with 40 chromosomes is shown in Fig. 2c, presenting 12 normal H. chilense-origin chromosomes, 25 normal wheat-origin chromosomes, and three rearranged chromosomes. The rearranged chromosomes were identified as the following: der(4HchLinv/sat6B) (Fig. 2c, yellow arrow; Fig. 3e); der(4HchS/6B) (Fig. 2c, green arrow; Fig. 3f); and der(6Hch), with pericentric inversion of the rDNA locus (Fig. 2c, white arrow; Fig. 3g). The chromosomes der(4HchLinv/sat6B) and der(4HchS/6B) (Figs. 2c, 3e) resulted from the inversion of chromosome 4Hch positioning the rDNA locus of chromosome 6B on its long arm. One 1B wheat-origin chromosome is missing, but the four nucleolar chromosomes of H. chilense origin, one 1B, and one 6B wheat-origin chromosomes are present (Fig. 2c). Thus, we can conclude that the rDNA locus involved in the derivative chromosomes der(4HchLinv/sat6B) and der(4HchS/6B) (Figs. 2c, 3e, f) probably resulted from the rupture of the 6B rDNA locus.
Figure 2d corresponds to a fourth metaphase cell with 2n = 41 of plant HT9-15 that showed 12 normal H. chilense-origin chromosomes (including the chromosome pair 5Hch and one chromosome 6Hch), 25 normal wheat-origin chromosomes (including the nucleolar chromosome pairs 1B and 6B), and two rearranged chromosomes. This cell was previously probed with the SSR sequence (AG)10 (Fig. 1c). The two rearranged chromosomes were identified as being der(6Hch/4A) (Fig. 2d, white arrow), which was also detected in another cell of the same plant (Figs. 2a, 3a). The second rearranged chromosome (Fig. 2d, yellow arrow) that previously showed two (AG)10 hybridization signals (Fig. 1c) was identified as being der(5A/6HchL) (Fig. 3h). This derivative 5A chromosome presents one SSR (AG)10 hybridization signal on each arm, instead of one on its short arm, as verified for the normal 5A chromosome (Fig. 1a). This structural rearrangement probably resulted from a duplication of the SSR (AG)10 region followed by a pericentric inversion. In addition, the der(5A) chromosome also recombined the terminal region of its long arm with part of the 6HchL, since in the same cell, the der(6Hch/4A) chromosome was also detected (Figs. 2d, 3a).
The HT31-20 plant did not show mosaicism. The unique metaphase cell of plant HT31-20 (2n = 42) that showed spontaneous intergenomic translocations presented 13 normal H. chilense-origin chromosomes (including the chromosome pairs 5Hch and 6Hch), 27 normal wheat-origin chromosomes (including the chromosome pair 1B and one chromosome 6B), and two chromosomes with spontaneous intergenomic translocations (Fig. 2e). The rearranged chromosomes were identified as der(4HchLinv.6BL) (Fig. 2e, yellow arrow; Fig. 3i; Online Resource 3) and its reciprocal, der(6HchSinv.6BS) (Fig. 2e, white arrow; Fig. 3j; Online Resource 3). The remaining metaphase cells of plant HT31-20 and those observed in other plants of line HT31 could not be considered completely normal, since all of them showed the heteromorphic 6B chromosome pair with deletion of the satellite region in one 6B chromosome (Fig. 1d, h).
Most of the structural rearrangements detected in the plants HT9-15 and HT31-20 involved the nucleolar chromosomes 6B and 6Hch (Fig. 3). In these two HT plants, a total of 11 derivative chromosomes were detected corresponding to 10 different structural rearrangements, since the intergenomic translocation der(6Hch/4A) was present in two metaphase cells of plant HT9-15 (Figs. 2a, d, 3a). Among the 10 rearrangements, seven involved one rDNA locus of wheat or H. chilense origin (Fig. 3). The number of derivative chromosomes ranged from 1 to 3 per metaphase cell of plant HT9-15 (Fig. 2a–d), and a unique metaphase cell of plant HT31-20 showed two structural rearrangements (Fig. 2e).
Detection of molecular rearrangements
Since the cytogenetic approaches revealed genomic restructuring in two of 15 tritordeum plants relative to wheat and H. chilense species, the same plants were analyzed with REMAP, IRAP, and ISSR techniques in order to confirm the occurrence of molecular rearrangements.
First, REMAP, IRAP, ISSR and iPBS markers were amplified in all plants (15 HT plants, five H. chilense plants, and five durum wheat plants). Since no polymorphic molecular patterns were detected among plants of line HT28, H. chilense line H1 or wheat line T846 (see Online Resource 4), we proceeded with the molecular analyses using one plant representative of each one of these three lines. Plant HT9-4 represented the normal plants of this line. In the case of plants from line HT31, none could be considered normal, since all presented the deletion of the satellite region of one chromosome 6B. However, plant HT31-7 showed similar patterns with other two plants of this line. Thus, in all molecular analyses we included the “normal” plants HT9-4, HT28-5, and HT31-7 and those that showed structural rearrangements and/or polymorphic molecular patterns, HT9-15, HT31-19, and HT31-20.
Table 2 presents the global results of REMAP, IRAP, ISSR and iPBS markers amplified in the six HT plants, in the parental lines H. chilense (H1), and in durum wheat (T846).
ISSR and REMAP showed the highest values of total percentage of polymorphism, 93.75 and 80 %, respectively (Table 2).
The total number of polymorphic (P) bands in Table 2 includes the following: (1) those shared by individuals of lines HT9, HT28, and HT31 and one of the parental species (line H1 or line T846); (2) those common among HTs and both parents (conserved bands); and (3) those bands that were only amplified in one or both parental species (lost parental bands).
The total number of markers with wheat origin common to the HT individuals was higher (14 REMAP + 14 IRAP + 16 ISSR = 44) than that of H. chilense origin and common to the HT individuals (12 REMAP + 18 IRAP + 5 ISSR = 35). However, the number of IRAP markers of H. chilense origin inherited by the HT individuals was slightly higher (Table 2). Eighteen markers (4 REMAP + 10 IRAP + 4 ISSR = 18) amplified in the HT individuals were considered conserved bands (common to both parental species) (Table 2).
ISSR makers demonstrated that most of the bands were shared among HT individuals and wheat, and that the number of lost bands was equal between the two parental species (Table 2).
The lost parental bands were considered rearranged bands or molecular rearrangements, and were detected by the four molecular marker systems in plants of all lines (Table 2). In total, 22 REMAP, 12 IRAP, 20 ISSR, and 4 iPBS markers of parental origin were absent from the HT individuals (Table 2). The number of specific markers amplified in line H1 (11 REMAP + 4 IRAP + 9 ISSR = 24) was higher than that exclusively detected in line T846 (8 REMAP + 2 IRAP + 9 ISSR = 19) (Table 2).
No novel REMAP, IRAP, or ISSR bands (exclusively amplified in HT individuals and absent from the parental species) were detected.
Plants HT9-15 and HT31-20, which previously showed spontaneous intergenomic translocations and other structural rearrangements, presented molecular profiles similar to the remaining HT plants.
Discussion
Structural rearrangements detected by SSRs, 45S rDNA, and H. chilense probes
The occurrence of rapid genetic and epigenetic alterations has been widely described in newly synthesized allopolyploids of the tribe Triticeae (Liu et al. 1998a; Ozkan et al. 2001, 2003; Shaked et al. 2001; Madlung et al. 2005; Lukens et al. 2006; Ma and Gustafson 2006; Bento et al. 2008, 2010; Tang et al. 2012; Cabo et al. 2014a, b, c).
Tritordeum has been used as bridge material in breeding programs, providing the transference of interesting agronomic traits from H. chilense to cultivated wheat, and the production of alloplasmic bread wheat lines (Martín et al. 1998, 1999, 2008). This fertile amphiploid resulted from the high compatibility at the genomic and cytoplasmic levels between H. chilense and Triticum spp., enabling the production of tritordeum plants with different ploidy levels by chromosome doubling of F1 hybrids between H. chilense and tetraploid or hexaploid wheat (Martín et al. 1999). Recently, structural and/or molecular rearrangements were detected in two newly synthesized lines of hexaploid tritordeum (Cabo et al. 2014a, b, c), triticale, and wheat–rye addition lines (Bento et al. 2008, 2010) by comparison with their parental species. The advanced tritordeum lines studied here showed a higher number of structural rearrangements and a lower number of rearranged bands than those found previously in newly formed tritordeum (Cabo et al. 2014a, b).
The SSR probes (AAC)5, (AGA)5, and (TTC)5 revealed additive hybridization patterns in the three HT lines, showing a number of chromosomes equal to the sum of those that presented SSR hybridization in both parental species (Table 1).
According to Cuadrado et al. (2008b), the SSRs (AAC)5 and (AAG)5 (complementary to TTG and TTC, respectively) are the two most common motifs in barley and wheat genomes. The same could be extended to the SSR (AGA)5 and its complementary sequence (TCT). Overall, the trinucleotide repeats are the most abundant class of SSRs in the expressed sequence tags of all cereal species, ranging from 54 to 78 %, whereas the dinucleotide SSR repeats only represent 17.1–40.4 % (Varshney et al. 2002). This dominance of the trinucleotide SSR motifs over the other classes of SSRs, particularly the dimeric SSRs, may be explained on the basis of the suppression of the non-trimeric SSRs in coding regions to avoid the risk of frameshift mutations (Metzgar et al. 2000).
The SSR (AAC)5, (AGA)5, and (TTC)5 probes were not able to reveal genomic reshuffling in tritordeum relative to their parents. Although Carvalho et al. (2013) detected variable hybridization patterns of the SSR (AAC)5 among the nucleolar chromosomes of different Old Portuguese bread wheat cultivars, no polymorphic patterns were detected among the three HT lines. At least two ideograms of wheat chromosomes (genomes A, B, and D) are available for the SSR probe (AAC)5 (Cuadrado et al. 2000, 2008a). This is not the case for H. chilense chromosomes. In addition, since most of the tritordeum chromosomes were hybridized by the SSRs (AGA)5 and (TTC)5, the construction of ideograms for these probes would be extremely useful for the identification of individual wheat and H. chilense chromosomes.
The SSRs with trinucleotide motifs showed similar hybridization patterns in tritordeum chromosomes but differed from those SSRs with dinucleotide motifs. The distribution patterns of SSRs with trinucleotide motifs were considered similar between wheat and barley chromosomes and are preferentially associated with heterochromatin (Cuadrado et al. 2000; Cuadrado and Jouve 2007). The hybridization signals of the SSRs (AG)10 and (CT)10 were located at the pericentromeric or subtelomeric regions, presenting one or few sites per wheat-origin chromosome (Table 1). Contrastingly, the trinucleotide SSR probes showed interspersed signals in most of the wheat- and H. chilense-origin chromosomes of tritordeum. Several studies indicated that the distribution of SSRs on chromosomes is not random (Beckmann and Weber 1992; Lagercrantz et al. 1993; Tautz and Schlötterer 1994; Cardle et al. 2000; Toth et al. 2000; Katti et al. 2001; Subramanian et al. 2003), probably due to the role of SSRs in genome structure, function, and evolution (Cuadrado et al. 2008b). Cuadrado and Schwarzacher (1998) also explained the similar SSR hybridization patterns between wheat and rye genomes independently of the SSR repetition motif, as related to their ancestral origin in the tribe Triticeae. When we compared the present SSR (AG)10 hybridization patterns of wheat-origin chromosomes with that described by Cabo et al. (2014a) and with the wheat ideogram of SSR (AG)12 published by Cuadrado et al. (2008a), we also found high similarities. These ideograms were also used for the identification of wheat-origin chromosomes that showed hybridization of SSR (CT)10 (Fig. 1d, e; Online Resource 2). The SSRs (AG)10 and (CT)10 correspond to complementary sequences and, consequently, their chromosomal locations are the same in durum wheat and wheat-origin chromosomes of tritordeum, only differing in number among the three studied advanced lines (Table 1; Fig. 1a, d, e). The reprobing of the same cells with additional repetitive DNA sequences and the comparison of their hybridization patterns with other available ideograms of Mukai et al. (1993), Cuadrado et al. (2000, 2008a), Komuro et al. (2013), Tang et al. (2014a), and Fu et al. (2015) allowed the unequivocal identification of the wheat-origin chromosomes of tritordeum bearing SSR (AG)10 and (CT)10 hybridization. Thus, in line HT9, both SSRs hybridized on the chromosome pairs 5A, 3B, 4B, 5B, and 6B (Fig. 1c, e; Online Resource 3). In plants of lines HT28 and HT31, the SSR (AG)10 and (CT)10 probes hybridized on the chromosome pairs 5A, 3B, and 6B (Fig. 1a, d; Online Resource 2).
The polymorphisms in number and location of the SSR probes (AG)10 and (CT)10 within and among the three HT lines demonstrated genomic restructuring, particularly in plants HT9-15 and HT31-20, after comparison with the parental species (Table 1; Fig. 1c; Online Resource 2). The observations of two SSR (AG)10 hybridization signals in one single chromosome of one HT9-15 metaphase cell (Fig. 1c) and five (CT)10 signals in one metaphase cell of the HT31-20 plant (Online Resource 2) resulted from structural rearrangements that were evidenced after FISH with H. chilense genomic DNA and pTa71 probes (Fig. 2d, e). The chromosome of plant HT9-15 that showed two (AG)10 signals (one per arm) consisted of der(5A/6HchL) (Fig. 3j). The normal wheat 5A chromosome presents the SSR (AG)10 signal in the pericentromeric region of the 5AS arm, as verified by Cabo et al. (2014a). The der(5A) chromosome of plant HT9-15 resulted from an inverted pericentric duplication of the SSR (AG)10 region, probably due to the involvement of RTN activity. A similar rearrangement was reported for SSR (AG)10 in one 6B wheat-origin chromosome of the newly synthesized hexaploid tritordeum line HT27 (Cabo et al. 2014a). In the same metaphase cell of plant HT9-15, one additional derivative chromosome was identified as being der(6Hch/4A) (Figs. 2d, 3a). Thus, the der(5A) chromosome recombined with 6HchL on the terminal region of its long arm (Figs. 1c, 2d, 3j; Online Resource 3), resulted from the simultaneous presence of the der(6Hch/4A) chromosome (Figs. 2d, 3a).
The metaphase cell of plant HT31-20 with five SSR (CT)10 signals (Online Resource 2) presented two derivative chromosomes, der(4HchLinv.6BL) and the reciprocal der(4HchSinv.6BS) with the deletion of the satellite region (Fig. 3k, l; Online Resource 3).
The remaining structural rearrangements included spontaneous intergenomic translocations mostly involving the nucleolar chromosomes 6BS and 6HchS, which were detected by FISH using simultaneously H. chilense genomic DNA and pTa71 as probes (Figs. 2, 3). It was surprising to verify the occurrence of high numbers of derivative chromosomes in two plants of these highly inbred HT lines. However, if we consider that the evolution of allopolyploids is still poorly understood and that several authors have reinforced the need of additional studies, we might suggest that different types of genomic reshuffling take place throughout the generations that follow polyploidization. In addition, the detection of genomic restructuring depends on the resolution of the techniques used and of the genomic targets under study. The first generations of recently synthesized allopolyploids seem to undergo sequence and chromosomal rearrangements (Ozkan et al. 2001; Bento et al. 2008, 2010; Cabo et al. 2014a, b, c). The present study demonstrated that after multiple generations of self-fertilization, such rearrangements also could occur.
Cross hybridization could accelerate the evolution of repetitive DNA sequences (McClintock 1978). In this study, major structural chromosomal rearrangements were detected, such as the following: deletion of a satellite region of one 6B chromosome in all HT31 metaphase cells and in one HT9-15 metaphase cell; duplications of the SSR (AG)10 region and of the 6B rDNA locus in wheat-origin chromosomes; pericentric inversions of the SSR (AG)10 region and of the 6Hch rDNA locus; and several spontaneous intergenomic reciprocal and nonreciprocal translocations (Figs. 2, 3). Most of the observed structural rearrangements involved one rDNA locus (Fig. 3). Actually, the involvement of rDNA loci in structural rearrangements has been commonly reported (Badaeva et al. 2007; Lan and Albert 2011; Singh and Barman 2013; Rosato et al. 2015; among others), suggesting these genomic regions as hotspots of recombination and chromosome breakage, as proposed by Lan and Albert (2011), due to their centromeric or subtelomeric location. Considering the present study, the fragility of the rDNA loci followed by recombination induced the formation of several structural rearrangements. Such events may arise from the stress caused by hybridization and polyploidization within a newly nascent allopolyploid, delaying the homogenization of the rDNA sequences by concerted evolution and the stabilization of the divergent genomes. We also intend to focus that the detection of particular chromosomal rearrangements are restricted by the used probes but we do not discard the occurrence of additional ones involving other classes of repetitive DNA or recombination between chromosomes of the A- and B-genomes of wheat. In this study, we optioned to characterize the three advanced HT lines with the genomic probe of H. chilense, the rDNA probe pTa71 and SSR probes, based on our previous research which also reflected the satellite region as an intensive spot of recombination. However, the use of the same probes or molecular tools could not be predictable when studying allopolyploids because our first prediction to study advanced lines of tritordeum was to evidence a highly stable genome due to the high number of self-fertilization generations. Surprisingly, we detected several rearrangements and in higher number than in newly formed HT lines.
During the analysis of derivatives of wheat–rye hybrids, the loss of terminal sites for the synthetic Oligo-pSc119.2 probe in 1B and 6B chromosome and structural variation in the 5A chromosome that showed a new intercalary site for Oligo-pSc119.2 were reported (Tang et al. 2014b). These authors also described additional structural variations in the wheat-origin chromosomes 6A, 2B, 7B, 1D, 3D and 7D. Comparing our results with those reported by Tang et al. (2014b), we could conclude that the deletion of satellite regions of chromosomes 1B and 6B could be common in allopolyploids due to the fragility of the secondary constrictions, resulting in detection of detached satellites, rDNA loci positioned at the telomeres, or division of one rDNA locus per two rearranged chromosomes (Figs. 1d, h, 2b, c, e). The high involvement of the nucleolar chromosomes 1B, 6B, and 6Hch and of their rDNA loci in the structural rearrangements described here corroborated previous assumptions about the deletion and expansion of tandem repetitive sequences as a result of RTN activity (Alkhimova et al. 1999, 2004; Cabo et al. 2014a; Tang et al. 2014b). The wheat chromosome 1B was affected the most by rearrangements in the studies of Badaeva et al. (2007).
Several spontaneous translocations involving wheat chromosomes of the B genome have been described (Belay and Merker 1998; Carvalho et al. 2009; Cabo et al. 2014a; Tahmasebi et al. 2015, among others). Badaeva et al. (2007) reported that wheat chromosomes from the B genome are more frequently involved in translocations, followed by chromosomes of the A and D genomes, and explained that within an allopolyploid nucleus, one genome seems to have greater potential for structural chromosomal evolution than the others presumably exposed to the same selection pressures. Perhaps due to the trend of wheat chromosomes of the B genome to evolve quickly and more intensely than those of the A and D genomes, the donor species of the B genome remains to be found.
Mosaicism and male sterility in plant HT9-15
Unequal chromosome division is frequent in the mitosis of allopolyploids (Sachs 1952; Gernand et al. 2005; Hua et al. 2006; Tu et al. 2009; Tang et al. 2012; Fu et al. 2013). Tritordeum has an aneuploidy rate ranging from 4 to 23 % (Padilla and Martin 1986).
Plant HT9-15 showed a high number of structural rearrangements and numerical irregularities, such as the presentation of few metaphase cells with aneuploid chromosome numbers of 2n = 40 and 2n = 41. Given that most of the cells exhibited the expected chromosome number of 2n = 42, the individual was considered a mosaic (Fig. 2a–d). The aneuploid metaphases detected in the HT9-15 plant probably arose from abnormal chromosome pairing in the previous meiosis derived from the high occurrence of intergenomic translocations–putative intragenomic translocations not detected with the probes used and/or other structural rearrangements. Such features avoid the diploid-like meiotic behavior that is characteristic of well-established amphiploids. In addition, the regular homologous chromosome pairing is also compromised by the absence of some chromosomes in the aneuploid cells, resulting in a vicious cycle that ended with the male sterility of this plant. Chromosome recombination also could happen during mitosis (Peterhans et al. 1990; LaFave and Sekelsky 2009; Lee et al. 2009; Rosa et al. 2013; Richter et al. 2014). The lack of one homologous chromosome during both meiotic and mitotic divisions hampers genomic repair mechanisms of DNA damage (Comai 2000). In the aneuploid metaphase cells of plant HT9-15, one or two chromosomes will lack their homologous partner to use for repair, which means that rearranged chromosomes will be repaired by other mechanisms with low fidelity or that they will not be repaired at all, producing DNA errors, deletions, and other irregularities. Furthermore, several studies showed that biotic and abiotic stress may increase somatic homologous recombination in plants (Ries et al. 2000; Lucht et al. 2002; Kovalchuk et al. 2003; Boyko et al. 2005; Molinier et al. 2005). Actually, for some stresses, the high frequency of somatic recombination can be high even within subsequent non-stressed plant generations (Molinier et al. 2006).
Previous morphological and yield characterizations (Lima-Brito 1998) and meiotic pairing analysis performed in these tritordeum lines showed regular meiosis and fertility (Carvalho 2004). However, when crossed with other cereals, spontaneous intergenomic translocations were observed. This feature was verified in one F1 multigeneric hybrid plant of triticale × HT9 (genomic constitution AABBRHch) that presented the translocation 7BS/7RL (Carvalho et al. 2009).
In addition to the stress induced by allopolyploidization, the tritordeum lines studied here also could have been exposed to some particular abiotic stress during their growth. The repair mechanisms could be different from cell to cell and may depend on the cell type, explaining the mosaicism. Another possible explanation for mosaicism and rearrangements is the activation of mobile elements, widely described as a response to allopolyploidization (Zhao et al. 1998; Hanson et al. 2000; Chen et al. 2007) and frequently associated with stress situations (Wessler 1996; Grandbastien 1998).
The sterility of plant HT9-15 also could be caused by the pericentric inversion of the 6Hch rDNA locus in homozygous and heterozygous conditions (Figs. 1f, 2c, 3g). The 6HchS arm is a carrier of restoration fertility genes, and its homozygous or heterozygous addition in male-sterile alloplasmic bread wheat lines containing cytoplasm of H. chilense line H1 restores the lines fertility (Martín et al. 2008). Our data support that chromosome 6HchS also has an important role in the fertility of hexaploid tritordeum.
Plant HT31-20 only showed one metaphase cell with structural rearrangements as well as the normal chromosome number (2n = 42), and it was fertile.
Probable explanations for the detected rearrangements
Globally, hexaploid tritordeum lines are fertile and meiotically stable (Lima-Brito 1998; Carvalho 2004). Among the three HT advanced lines studied here, we considered HT28 the most genetically stable. Nonetheless, in this study we detected high genomic instability in two plants, HT9-15 and HT31-20, as well as one particular structural variation in all plants of line HT31, namely the heteromorphism of the chromosome pair 6B due to deletion of the satellite region of one chromosome 6B. Even so, the present results should not constitute a disadvantage in the acceptability of tritordeum as an alternative crop. We only advise that advanced hexaploid HT lines should be cytogenetically and/or molecularly monitored before their use.
Genealogy of the advanced HT lines and Gc genes
The advanced lines HT9, HT28 and HT31 were produced during the 1990’s and the compilation of data reported by Rubiales et al. (1991), Chauhan and Singh (1997) and Ballesteros et al. (2005) allowed us to trace some of the crosses performed between primary and secondary tritordeums till their production. The production and improvement of these tritordeum lines were performed during years with other goals of study, such as the transference of traits from the wheat D- genome into tritordeum in order to improve its baking properties or resistance to some diseases. At the present time, we were not able to achieve every single line involved in the genealogy of the advanced HT lines for a complete study of all intermediate generations. However, we previously characterized plants of lines HT9, HT28 and HT31 at the yield, morphological and cytogenetic levels (Lima-Brito 1998; Carvalho 2004). These cytogenetic studies were performed in mitotic and meiotic chromosome spreads. Plants from HT28 line showed a more regular meiosis than those of lines HT9 and HT31, where univalents and multivalents, mainly from H. chilense-origin, but also of wheat-origin, were found (Carvalho 2004). Besides, the HT9 plants differed in their morphology relative to plants of the other lines and were less productive. However, no intergenomic translocations were seen at mitosis or meiosis of those plants (Lima-Brito 1998; Carvalho 2004). Nonetheless, when we used HT9 as male parent in interspecific crosses with bread and durum wheat, and with triticale, some of the F1 hybrids showed intergenomic translocations (Carvalho et al. 2009); neocentromeres (Carvalho et al. 2008); or other structural rearrangements involving rDNA loci (Carvalho 2004). The HT plants used in this study are descendants of those that previously showed regular mitosis and meiosis, and absence of intergenomic translocations or other rearrangements. We also studied the HT22 line which is an intermediate material in the production of these advanced lines. This tritordeum line showed molecular rearrangements (Cabo et al. 2014b, c) and no structural rearrangements were detected after characterization with the same techniques used here. Thus, the intermediate plant material studied before (Lima-Brito 1998; Carvalho 2004; Cabo et al. 2014b, c) showed less instability than that found in plants HT9-15 and HT31-20.
According to the information reported by Rubiales et al. (1991), Chauhan and Singh (1997) and Ballesteros et al. (2005), the production of advanced HT lines with interesting agronomic traits of H. chilense origin required crosses with octoploid tritordeum plants. Lines HT9 and HT31 resulted from a cross between a hexaploid tritordeum and an octoploid one (line HT18; AABBDDHchHch; 2n = 56), which could explain the genomic instability detected in the plants HT9-15 and HT31-20, whereas line HT28 derived from the cross of two hexaploid HT lines (HchHchAABB; 2n = 42). These crossing schemes might be responsible for the variability among the three advanced lines.
Regarding the several structural rearrangements observed in plants HT9-15 and HT31-20, we also hypothesized the probable introduction of gametocidal (Gc) genes into tritordeum through homoeologous recombination with gametocidal chromosomes from the D-genome of the octoploid tritordeum (line HT18). These genes or factors cause chromosomal breaks, translocations, and other irregularities at different developmental stages (Endo 1988; Tsujimoto and Noda 1990; King and Laurie 1993; Nasuda et al. 1998; Joshi et al. 2013). Triticum speltoides (Tausch) Gren. ex Richter (syn. Aegilops speltoides Tausch) is the most acceptable donor species of the wheat B genome and Triticum tauschii (syn. Aegilops squarrosa, Aegilops tauschii) as the donor species of the D genome (Feldman et al. 1995; Bálint et al. 2000; Feldman 2001). The Gc genes could be introduced into bread and durum wheat through interspecific hybridization and backcrossing with related Aegilops species (Endo 1990; Nasuda et al. 1998). The Gc genes are strong distorters of chromosome segregation that affect plant fertility through differential functioning of the gametes (Nasuda et al. 1998). The 6S chromosome of T. speltoides was referred to as a gametocidal chromosome since it carries Gc genes (Tsujimoto and Tsunewaki 1983, 1984; Kota and Dvorak 1998; Nasuda et al. 1998; See 2007). The monosomic or disomic substitution of chromosome 6S of T. speltoides by chromosome 6B of bread wheat cultivar ‘Chinese Spring’ induced chromosomal rearrangements, such as deletions and translocations, but the rearrangements were not verified in plants without 6S (Kota and Dvorak 1998). Overall, we could hypothesize the earlier recombination of these chromosomes during wheat evolution, the consequent introduction of Gc genes into the wheat-origin 6B (or 6D) chromosome, and the activation of these factors under stress as those induced by allopolyploidization. Thus, the involvement of species from the genus Aegilops (carrier of Gc genes) and of octoploid HT plants in the genealogy of lines HT9 and HT31 could justify the introduction of such genes to their genomes and explain the occurrence of the chromosomal rearrangements reported here.
Colchicine treatment
Chemical agents have been pointed out as responsible for induction of structural variations, such as 5-azacytidine, which was previously ascribed as an inductor of neocentromeres in wheat-origin chromosomes of one F1 multigeneric triticale × HT9 hybrid (AABBRHch) (Carvalho et al. 2008). This multigeneric hybrid also presented a wheat-origin chromosome with three constrictions (polycentric chromosome)—the tandem duplication of its rDNA locus and normal centromere (Carvalho et al. 2008), resembling der(6B) detected in Fig. 2b.
The primary tritordeum plants involved in the genealogy of lines HT9, HT28, and HT31 resulted from chromosome doubling of F1 interspecific hybrids H. chilense × durum wheat and of F1 H. chilense × bread wheat (Rubiales et al. 1991; Chauhan and Singh 1997; Ballesteros et al. 2005). The chromosome doubling was induced by colchicine, which is a potent chemical mutagen that induces polyploidy and chromosomal mutants (Ahirmar and Verma 2015). The consequences of using colchicine for the production of synthetic allopolyploids, especially regarding sequence elimination, have been questioned previously. Nonetheless, the studies developed by Feldman et al. (1997) and Liu et al. (1997, 1998a, b) revealed that sequence elimination occurs in both natural and synthetic allopolyploids (Chen and Chen 2007), discarding the hypothesis of colchicine treatment as an inductor of sequence elimination. However, the involvement of this chemical in the induction of other structural rearrangements could not be discarded completely.
Genomic reshuffling evidenced by molecular markers
REMAP, IRAP, and ISSR are feasible tools for DNA fingerprinting (Kalendar et al. 1999; Carvalho et al. 2005) but also for detecting genomic restructuring in allopolyploids (Bento et al. 2008, 2010; Cabo et al. 2014b). In this study, we also were able to verify that the iPBS markers developed by Kalendar et al. (2010) were also suitable for detection of molecular rearrangements in tritordeum relative to their parental species. According to Bento et al. (2008), rearranged markers are those of parental origin (in one or both parents) that were not transmitted to the allopolyploid (lost parental bands or genetic loss) as well as bands that are exclusively amplified in the allopolyploid (novel bands or genetic gain) being absent from both parents. The rearranged bands are considered polyploidization-induced rearrangements (Bento et al. 2008; Cabo et al. 2014b).
REMAP, IRAP, ISSR and iPBS markers revealed the loss of parental bands in the three advanced HT lines by comparison with the molecular patterns of the parental species, H. chilense and durum wheat (Table 2), indicating genomic restructuring.
Overall, most of the polymorphic markers inherited by the HT lines had a wheat origin (Table 2). These results indicated non-random patterns of gain and loss of bands of wheat and H. chilense origin, respectively, since the same was described in newly synthesized HT lines (Cabo et al. 2014b). Similar results were also reported for triticale and wheat–rye addition lines relative to the preferential loss of bands of parental rye genome origin (Bento et al. 2008, 2010). Our data revealed that the number of IRAP bands of H. chilense origin inherited by the HT individuals was slightly higher (18) to those of wheat origin (14) (Table 2). Cabo et al. (2014b) reported the inheritance of almost double the IRAP bands of H. chilense origin (51) comparatively to those of wheat origin (27) in two newly formed lines of tritordeum. Fragment loss of RTNs seems to occur at very early stages (F1 and S1), and the allopolyploids then remain relatively static in this respect (Han et al. 2003). This assumption could explain the differences between the present results obtained in advanced HT lines and those described by Cabo et al. (2014b) for newly formed tritordeum. Additionally, in newly formed hexaploid tritordeum lines, it is more frequent to detect novel bands (Cabo et al. 2014b, c), probably as a result of hybridization of the divergent genomes. Since in this study none of the three highly inbred HT lines showed novel bands, we might suggest a non-random pattern of band elimination in the following generations after polyploidization. Such an elimination pattern could provide a physical basis for reducing the homoeologous chromosome pairing and genome redundancy, improving the fitness and establishment of allopolyploids in nature, benefiting their diploid-like behavior and subsequent fertility (Feldman et al. 1997; Ozkan 2000; Feldman and Levy 2005; Ma and Gustafson 2005). Despite the report of fast elimination of highly repetitive DNA sequences in newly formed allopolyploids of the tribe Triticeae (Ozkan et al. 2001), the present results allowed us to suggest a continuous elimination of repetitive sequences through multiple generations of self-fertilization after polyploidization.
Through the years, tritordeum lines have been selected in early generations for the following criteria: fertility, earliness, and resistance to wheat and H. chilense diseases (Ballesteros et al. 2005). However, similarly to the morphological aspect of tritordeum that mostly resembles the wheat parent, also at the genomic level, some interesting agronomic traits of H. chilense origin were unable to be expressed, being inhibited by the presence of wheat genes (Ballesteros et al. 2005).
Overall the molecular approaches revealed genomic restructuring by the occurrence of non-random loss of parental bands.
Even if we have not detected novel REMAP, IRAP, ISSR or iPBS bands, the cytogenetic approaches allowed the detection of a novel SSR-rich region (AG)10 on the long arm of a 5A chromosome as a result of duplication followed by pericentric inversion and recombination with the 6HchL arm (Fig. 3h). Excluding the intergenomic translocation, a similar rearrangement involving the SSR (AG)10 region was also detected in the newly formed HT27 line (Cabo et al. 2014a). In both cases, we consider the structural rearrangements involving the SSR (AG)10 as a result of RTN activity. The occurrence of the novel SSR-rich region on the 5AL arm of plant HT9-15 is in accordance with the molecular data, since most of the REMAPs and ISSRs inherited by the three advanced HT lines had wheat origin.
Conclusions
In this study, the cytogenetic and molecular approaches proved to be suitable for the determination of genomic reshuffling in advanced lines of hexaploid tritordeum.
Whenever possible, cytogenetic data should be complemented and/or confirmed by molecular results and vice versa. Such approach requires the choice of suitable marker systems that would target the same genomic regions as the chromosomal probes. Most of the ISSRs and REMAPs inherited by the HT plants had wheat origin, as demonstrated by the ND-FISH experiments performed with SSR probes, and confirmed after discrimination of the parental genomes by FISH. On the other hand, a non-random elimination of REMAP and ISSR regions of H. chilense origin was verified, which could be assigned as the most prevalent rearrangement induced by polyploidization in tritordeum. The number of IRAP bands of H. chilense origin inherited by tritordeum was slightly higher than those of wheat origin, suggesting that RTN sequences might stabilize faster than SSR-rich regions in this amphiploid.
The present results were somewhat similar to those obtained previously by our group in newly synthesized HT lines. Actually, this study revealed that genomic reshuffling in tritordeum is not restricted to the first generation after polyploidization. Instead, it seems to be ongoing, as it should be for the evolutionary processes to ensure adaptation ability. Furthermore, our study demonstrates the need of monitoring tritordeum in the process of selection of improved lines.
References
Ahirmar R, Verma RC (2015) Colchicine induced asynaptic chromosomal behaviour at meiosis in Allium cepa L. The Nucleus 58:47–51
Alkhimova AG, Heslop-Harrison JS, Shchapova AI, Vershinin AV (1999) Rye chromosome variability in wheat-rye addition and substitution lines. Chromosome Res 7:205–212
Alkhimova OG, Mazurok NA, Potapova TA, Zakian SM, Heslop-Harrison JS et al. (2004) Diversity patterns of the tandem repeats organization in rye chromosomes. Chromosoma 113:42–52
Alvarez JB, Ballesteros J, Sillero JA, Martin LM (1992) Tritordeum: a new crop of potential importance in the food industry. Hereditas 116:193–197
Atienza SG, Ballesteros J, Martín A, Hornero-Mendez D (2007) Genetic variability of carotenoid concentration and degree of esterification among tritordeum (×Tritordeum Ascherson et Graebner) and durum wheat accessions. J Agric Food Chem 55:4244–4251
Badaeva ED, Dedkova OS, Gay G, Pukhalskyi VA, Zelenin AV, Bernard S, Bernard M (2007) Chromosomal rearrangements in wheat: their types and distribution. Genome 50:907–926
Bálint AF, Kovács G, Sutka J (2000) Origin and taxonomy of wheat in the light of recent research. Acta Agron Hung 48:301–313
Ballesteros J, Ramirez MC, Martínez C, Atienza SG, Martín A (2005) Registration of HT621, a high carotenoid content tritordeum germplasm line. Crop Sci 45:2662–2663
Beckmann JS, Weber JL (1992) Survey of human and rat microsatellites. Genomics 12:627–631
Belay G, Merker A (1998) Cytogenetics analysis of a spontaneous 5B/6B translocation in tetraploid wheat landraces from Ethiopia, and implications for breeding. Plant Breed 117:537–542
Bento M, Pereira HS, Rocheta M, Gustafson P, Viegas W, Silva M (2008) Polyploidization as a retraction force in plant genome evolution: sequence rearrangements in Triticale. PLoS ONE 3:e1402. doi:10.1371/journal.pone.0001402
Bento M, Gustafson P, Viegas W, Silva M (2010) Genome merger: from sequence rearrangements in triticale to their elimination in wheat-rye addition lines. Theor Appl Genet 121:489–497
Boyko A, Filkowski J, Kovalchuk I (2005) Homologous recombination in plants is temperature and day-length dependent. Mutat Res 572:73–83
Cabo S, Carvalho A, Martín A, Lima-Brito J (2014a) Structural rearrangements detected in newly-formed hexaploid tritordeum after three sequential FISH experiments with repetitive DNA sequences. J Genet 93:183–188
Cabo S, Carvalho A, Rocha L, Martín A, Lima-Brito J (2014b) IRAP, REMAP and ISSR fingerprinting in newly formed hexaploid tritordeum (×Tritordeum Ascherson et Graebner) and respective parental species. Plant Mol Biol Rep 32:761–770
Cabo S, Ferreira L, Carvalho A, Martins-Lopes P, Martín A, Lima-Brito JE (2014c) Potential of Start Codon Targeted (SCoT) markers for DNA fingerprinting of newly synthesized tritordeums and their respective parents. J Appl Genet 55:307–312
Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D, Waugh R (2000) Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156:847–853
Carvalho AIF (2004) Emparelhamento meiótico e DNA fingerprint em anfiplóides e híbridos interespecíficos da tribo Triticeae. MCS dissertation, University of Tras-os-Montes and Alto Douro, Vila Real
Carvalho A, Matos M, Lima-Brito J, Guedes-Pinto H, Benito C (2005) DNA fingerprint of F1 interspecific hybrids from the Triticeae tribe using ISSRs. Euphytica 143:93–99
Carvalho A, Guedes-Pinto H, Heslop-Harrison JS, Lima-Brito J (2008) Wheat neocentromeres found in F1 triticale × tritordeum hybrids (AABBRHch) after 5-azacytidine treatment. Plant Mol Biol Rep 26:46–52
Carvalho A, Martín A, Heslop-Harrison JS, Guedes-Pinto H, Lima-Brito L (2009) Identification of the spontaneous 7BS/7RL intergenomic translocation in one F1 multigeneric hybrid from the Triticeae tribe. Plant Breed 128:105–108
Carvalho A, Guedes-Pinto H, Lima-Brito J (2013) Polymorphism of the simple sequence repeat (AAC)5 in the nucleolar chromosomes of Old Portuguese wheat cultivars. J Genet 92:583–586
Charles M, Belcram H, Just J, Huneau C, Viollet A, Couloux A, Segurens B, Carter M, Huteau V, Coriton O, Appels R, Samain S, Chalhoub B (2008) Dynamics and differential proliferation of transposable elements during the evolution of the B and A genomes of wheat. Genetics 180:1071–1086
Chauhan RS, Singh BM (1997) Resistance to Karnal bunt in Hordeum chilense and its amphiploids with Triticum species. Euphytica 96:327–330
Chen LZ, Chen JF (2007) Allopolyploid-induced sequence elimination. Genes Genom Genet 1:113–117
Chen L, Lou Q, Zhuang Y, Chen J, Zhang X, Wolukau JN (2007) Cytological diploidization and rapid genome changes of the newly synthesized allotetraploids Cucumis × hytivus. Planta 225:603–614
Comai L (2000) Genetic and epigenetic interactions in allopolyploid plants. Plant Mol Biol 43:387–399
Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, Byers B (2000) Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12:1551–1567
Cuadrado A, Jouve N (2007) The non-random distribution of long clusters of all possible classes of trinucleotide repeats in barley chromosomes. Chromosome Res 15:711–770
Cuadrado A, Jouve N (2010) Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 119:495–503
Cuadrado A, Schwarzacher T (1998) The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma 107:587–594
Cuadrado A, Schwarzacher T, Jouve N (2000) Identification of different chromatin classes in wheat using in situ hybridization with simple sequence repeat oligonucleotides. Theor Appl Genet 101:711–717
Cuadrado A, Cardoso M, Jouve N (2008a) Increasing the physical markers of wheat chromosomes using SSRs as FISH probes. Genome 51:809–815
Cuadrado A, Cardoso M, Jouve N (2008b) Physical organization of the simple sequence repeats (SSRs) in Triticeae: structural, functional and evolutionary implications. Cytogenet Genome Res 120:210–219
Cuadrado A, Golczyk H, Jouve N (2009) A novel, simple and rapid nondenaturing FISH (ND-FISH) technique for the detection of plants telomeres—potential used and possible target structures detected. Chromosome Res 17:755–762
Delgado A, Carvalho A, Martín AC, Martín A, Lima-Brito J (2016) Use of the synthetic Oligo-pTa535 and Oligo-pAs1 probes for identification of Hordeum chilense-origin chromosomes in hexaploid tritordeum. Genet Resour Crop Evol 63(6):945–951
Depeiges A, Goubely C, Lenoir A, Cocherel S, Picard G, Raynal M, Grellet F, Delseny M (1995) Identification of the most represented repeated motifs in Arabidopsis thaliana microsatellite loci. Theor Appl Genet 91:160–168
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Endo TR (1988) Induction of chromosomal structural changes by a chromosome of Aegilops cylindrical L. in common wheat. J Hered 79:366–370
Endo TR (1990) Gametocidal chromosomes and their induction of chromosome mutations in wheat. Jpn J Genet 65:135–152
Fedoroff N (2000) Transposons and genome evolution in plants. Proc Natl Acad Sci USA 97:7002–7007
Feldman M (2001) The origin of cultivated wheat. In: Bonjean AP, Angus WJ (eds) The world wheat book. A history of wheat breeding. Lavoisier Tech & Doc, Paris, pp 3–56
Feldman M, Levy AA (2005) Allopolyploidy: a shaping force in the evolution of wheat genomes. Cytogenet Genome Res 109:250–258
Feldman M, Lupton FGH, Miller TE (1995) Wheats. In: Smartt J, Simmonds NW (eds) Evolution of crop plants. Longman Scientific and Technical, Harlow, pp 185–192
Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM (1997) Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147:1381–1387
Fu S, Yang M, Fei Y, Tan F, Ren Z, Yan B, Zhang H, Tang Z (2013) Alterations and abnormal mitosis of wheat chromosomes induced by wheat-rye monosomic addition lines. PLoS ONE 8(7):e70483. doi:10.1371/journal.pone.0070483
Fu S, Chen L, Wang Y, Li M, Yang Z, Qiu L, Yan B, Ren Z, Tang Z (2015) Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci Rep 5:10552
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885
Gernand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S et al (2005) Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell 17:2431–2438
Grandbastien M-A (1998) Activation of plant retrotransposons under stress conditions. Trends Plant Sci 3:181–187
Han FP, Fedak G, Ouellet T, Liu B (2003) Rapid genomic changes in interspecific and intergeneric hybrids and allopolyploids of Triticeae. Genome 46:716–723
Hanson RE, Islam-Faridi MN, Crane CF, Zwick MS, Czeschin DG, Wendel JF, McKnight TD, Price HJ (2000) Ty1-copia-retrotransposon behaviour in a polyploid cotton. Chromosome Res 8:73–76
Heslop-Harrison JS, Harrison GE, Leitch IJ (1992) Reprobing of DNA:DNA in situ hybridization preparations. Trends Genet 8:372–373
Hua YW, Liu M, Li ZY (2006) Parental genome separation and elimination of cells and chromosomes revealed by AFLP and GISH analyses in a Brassica carinata × Orychophragmus violaceus cross. Ann Bot 97:993–998
Jiang J, Friebe B, Gill BS (1994) Chromosome painting of Amigo wheat. Theor Appl Genet 89:811–813
Joshi GP, Endo TR, Nasuda S (2013) PCR and sequence analysis of barley chromosome 2H subjected to the gametocidal action of chromosome 2C. Theor Appl Genet 126:2381–2390
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman A (1999) IRAP and REMAP: two retrotransposon-base DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, Schulman AH (2008) Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci USA 105:5833–5838
Kalendar R, Antonius K, Smýkal P, Schulman AH (2010) iPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor Appl Genet 121:1419–1430
Katti MV, Ranjekar PK, Gupta VS (2001) Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol 18:1161–1167
King IP, Laurie DA (1993) Chromosome damage in early embryo and endosperm development in crosses involving the preferentially transmitted 4S1chromosome of Aegilops sharonensis. Heredity 70:52–59
Koba T, Takumi S, Shimada T (1997) Isolation, identification and characterization of disomic and translocated barley chromosome addition lines of common wheat. Euphytica 96:289–296
Komuro S, Endo R, Shikata K, Kato A (2013) Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56:131–137
Kota RS, Dvorak J (1998) Genomic instability in wheat induced by chromosome 6BS of Triticum speltoides. Genetics 120:1085–1094
Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, Hohn B (2003) Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423:760–762
LaFave MC, Sekelsky J (2009) Mitotic recombination: why? when? how? where? PLoS Genet 5(3):e1000411. doi:10.1371/journal.pgen.1000411
Lagercrantz U, Ellegren H, Andersson L (1993) The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res 21:1111–1115
Lan T, Albert VA (2011) Dynamic distribution patterns of ribosomal DNA and chromosomal evolution in Paphiopedilum, a lady’s slipper orchid. BMC Plant Biol 11:126. http://www.biomedcentral.com/1471-2229/11/126. 12 Sept 2011
Lee PS, Greenwell PW, Dominska M, Gawel M, Hamilton M, Petes TD (2009) A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet 5(3):e1000410. doi:10.1371/journal.pgen.1000410
Leitch IJ, Bennett MD (1997) Polyploidy in angiosperms. Trends Plant Sci 2:470–476
Lima-Brito J (1998) Estudos citogenéticos em híbridos multigenéricos de trigo, centeio e cevada. Ph.D. thesis, University of Tras-os-Montes and Alto Douro, Vila Real
Liu B, Segal G, Vega JM, Feldman M, Abbo S (1997) Isolation and characterization of chromosome-specific sequences from a chromosome arm genomic library of common wheat. Plant J 11:959–965
Liu B, Vega JM, Feldman M (1998a) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41:535–542
Liu B, Vega JM, Segal G, Abbo S, Rodova M, Feldman M (1998b) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy noncoding DNA sequences. Genome 41:272–277
Lucht JM, Mauch-Mani B, Steiner HY, Metraux JP, Ryals J, Hohn B (2002) Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat Genet 30:311–314
Lukens LN, Pires JC, Leon E, Vogelzang R, Oslach L et al (2006) Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol 140:336–348
Ma XF, Gustafson JP (2005) Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenet Genome Res 109:236–249
Ma XF, Gustafson JP (2006) Timing and rate of genome variation in triticale following allopolyploidization. Genome 49:950–958
Madlung A, Tyagi A, Watson B, Jiang HM, Kagochi T et al. (2005) Genomic changes in synthetic Arabidopsis polyploids. Plant J 41:221–230
Martín A (1988) Tritordeum: the first ten years. Rachis 7:2–15
Martín A, Sánchez-Monge Laguna E (1982) Cytology and morphology of the amphiploid Hordeum chilense × Triticum turgidum conv. durum. Euphytica 31:261–267
Martín A, Martínez-Araque C, Rubiales D, Ballesteros J (1996) Tritordeum: triticale’s new brother cereal. In: Guedes-Pinto H, Darvey N, Carnide VP (eds) Triticale: today and tomorrow. Kluwer, London, pp 57–72
Martín A, Martín LM, Cabrera A, Ramirez MC, Giménez MJ, Rubiales D, Hernandez P, Ballesteros J (1998) The potential of Hordeum chilense in breeding Triticeae species. In: Jaradat AA (ed) Triticeae III. Science Publish, Washington, pp 377–386
Martín A, Alvarez JB, Martín LM, Barro F, Ballesteros J (1999) The development of tritordeum: a novel cereal for food processing. J Cereal Sci 30:85–95
Martín AC, Atienza SG, Ramírez MC, Barro F, Martín A (2008) Male fertility restoration of wheat in Hordeum chilense cytoplasm is associated with 6HchS chromosome addition. Aust J Agric Res 59:206–213
McClintock B (1978) Mechanisms that rapidly reorganize the [maize] genome. Stadler Genet Sym 10:25–48
Mellado-Ortega E, Atienza SG, Hornero-Méndez D (2015) Carotenoid evolution during postharvest storage of durum wheat (Triticum turgidum conv. durum) and tritordeum (×Tritordeum Ascherson et Graebner) grains. J Cereal Sci 62:134–142
Metzgar D, Bytof J, Wills C (2000) Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res 10:72–80
Millán T, Martín A, de Haro A (1988) Field trial of tritordeum. Cereal Res Commun 16:31–38
Molinier J, Oakeley EJ, Niederhauser O, Kovalchuk I, Hohn B (2005) Dynamic response of plant genome to ultraviolet radiation and other genotoxic stresses. Mutat Res 571:235–247
Molinier J, Ries G, Zipfel C, Hohn B (2006) Transgeneration memory of stress in plants. Nature 442:1046–1049
Molnár-Láng M, Linc G, Friebe BR, Sutka J (2000) Detection of wheat-barley translocations by genomic in situ hybridization in derivatives of hybrids multiplied in vitro. Euphytica 112:117–123
Mukai Y, Nakahara Y, Yamamoto M (1993) Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 36:489–494
Nasuda S, Friebe B, Gill BS (1998) Gametocidal Genes induce chromosome breakage in the interphase prior to the mitotic cell division of the male gametophyte in wheat. Genetics 149:1115–1124
Ozkan H (2000) Genomic changes in newly synthesized amphiploids of Aegilops and Triticum. Ph.D. Thesis, University of Cukurova, Adana, Turkey
Ozkan H, Levy A, Feldman M (2001) Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13:1735–1747
Ozkan H, Tuna M, Arumuganathan K (2003) Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) group. J Heredity 94(3):260–264
Padilla JA, Martin A (1986) Aneuploidy in hexaploid tritordeum. Cereal Res Commun 14:341–346
Pavia I, Carvalho A, Rocha L, Gaspar MJ, Lima-Brito J (2014) Physical location of SSR regions and cytogenetics instabilities in Pinus sylvestris chromosomes revealed by ND-FISH. J Genet 93:567–571
Peterhans A, Schlüpmann H, Basse C, Paszkowski J (1990) Intrachromosomal recombination in plants. EMBO J 9:3437–3445
Richter KS, Kleinow T, Jeske H (2014) Somatic homologous recombination in plants is promoted by a geminivirus in a tissue-selective manner. Virology 452–453:287–296
Ries G, Heller W, Puchta H, Sandermann H, Seidlitz HK, Hohn B (2000) Elevated UV-B radiation reduces genome stability in plants. Nature 406:98–101
Rosa M, von Harder M, Cigliano RA, Schlögelhofer P, Scheid OM (2013) The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination and meiosis. Plant Cell 25:1990–2001
Rosato M, Moreno-Saiz JC, Galián JA, Rosseló JA (2015) Evolutionary site-number changes of ribosomal DNA loci during speciation: complex scenarios of ancestral and more recent polyploid events. AoB Plants 7:135. doi:10.1093/aobpla/plv135
Rubiales D, Ballesteros J, Martín A (1991) The reaction of ×Tritordeum and its Triticum spp. and Hordeum chilense parents to rust diseases. Euphytica 54:75–81
Sachs L (1952) Chromosome mosaics in experimental amphiploids in the Triticeae. Heredity 6:157–170
Salina EA, Ozkan H, Feldman M, Shumny VK (2000) Subtelomeric repeat reorganization in synthesized amphiploids of wheat. In: Proceedings of the international conference on biodiversity and dynamics of systems in north Eurasia, Novosibirsk, Russia, pp 102–105
Scheid OM, Jakovleva L, Afsar K, Maluszynska J, Paszkowski J (1996) A change in ploidy can modify epigenetic silencing. Proc Natl Acad Sci USA 93:7114–7119
Schmidt T, Boblenz K, Metzlaff M, Kaemmer D, Weising K, Kahl G (1993) DNA fingerprinting in sugar beet (Beta vulgaris)—identification of double-haploid breeding lines. Theor Appl Genet 85:653–657
Schwarzacher T, Heslop-Harrison JS (2000) Practical in situ hybridization. BIOS Scientific Publishers Limited, Oxford, UK, ISBN 185996138 X
Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS (1989) In situ localization of parental genomes in a wild hybrid. Ann Bot 64:315–324
See DR (2007) Genomic targeting and mapping of a gametocidal gene in wheat. MCS dissertation, Kansas State University
Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA (2001) Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant J 13:1749–1759
Singh M, Barman AS (2013) Chromosome breakages associated with 45S ribosomal DNA sequences in spotted snakehead fish Channa punctatus. Mol Biol Rep 40:723–729
Song K, Lu P, Tang K, Osborn TC (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Trends Ecol Evol 9:348–352
Subramanian S, Mishra RK, Singh L (2003) Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol 4:R13. doi:10.1186/gb-2003-4-2-r13
Tahmasebi S, Heidari B, Paknivat H, Dadkhodaie A (2015) Consequences of 1BL/1RS translocation on agronomic and physiological traits in wheat. Cereal Res Commun. doi:10.1556/0806.43.2015.016
Tang ZX, Fu SL, Yan BJ, Zhang HQ, Ren ZL (2012) Unequal chromosome division and inter-genomic translocation occurred in somatic cells of wheat-rye allopolyploid. J Plant Res 125:283–290
Tang Z, Li M, Chen L, Wang Y, Ren Z et al. (2014a) New types of wheat chromosomal structural variations in derivatives of wheat-rye hybrids. PLoS ONE 9(10):e110282. doi:10.1371/journal.pone.0110282
Tang Z, Yang Z, Fu S (2014b) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet 55:313–318
Tautz D, Schlötterer C (1994) Simple sequences. Curr Opin Genet Dev 4:832–837
Teo CH, Tan SH, Ho CL, Faridah QZ, Othman YR, Heslop-Harrison JS, Kalendar R, Schulman AH (2005) Genome constitution and classification using retrotransposon-based markers in the orphan crop banana. J Plant Biol 48:96–105
Toth G, Gaspari Z, Jurka J (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10:967–981
Tsujimoto H, Noda K (1990) Mutation of five marker genes in wheat by gametocidal gene of Ae. speltoides, Gc1a. Wheat Inf Serv 71:6–9
Tsujimoto H, Tsunewaki K (1983) Genetic analyses on a gametocidal gene originated from Aegilops aucheri. In: Proceedings of the 6th international wheat genetics symposium, Kyoto, Japan, pp 1077–1081
Tsujimoto H, Tsunewaki K (1984) Gametocidal genes in wheat and its relatives. I. Genetic analysis in common wheat of a gametocidal gene derived from Aegilops speltoides. Can J Genet Cytol 26:78–84
Tu YQ, Sun J, Ge XH, Li ZY (2009) Chromosome elimination, addition and introgression in intertribal partial hybrids between Brassica rapa and Isatis indigotica. Ann Bot 103:1039–1048
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis of frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7:537–546
Villegas D, Casadesús J, Atienza SG, Martos V, Maalouf F, Karam F, Aranjuelo I, Nogués S (2010) Tritordeum, wheat and triticale yield components under multi-local mediterranean drough conditions. Field Crops Res 116:68–74
Wallace RB, Johnson MJ, Hirose T, Miyake T, Kawashima EH, Itakura K (1981) The use of synthetic oligonucleotides as hybridization probes. II. Hybridization of oligonucleotides of mixed sequence to rabbit β-globin DNA. Nucleic Acids Res 9:879–894
Wegscheider E, Benjak A, Forneck A (2009) Clonal variation in Pinot noir revealed by S-SAP involving universal retrotransposon-based sequences. Am J Enol Vitic 60:104–109
Wendel JF, Schnabel A, Seelanan T (1995) Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc Natl Acad Sci USA 92:280–284
Wessler SR (1996) Plant retrotransposons: turned on by stress. Curr Biol 6:959–961
Zhao XP, Si Y, Hanson RE, Crane CF, Price HJ, Stelly DM, Wendel JF, Paterson NH (1998) Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Res 8:479–492
Acknowledgments
This work was partially supported by the HY-WHEAT project—International Consortium (P-KBBE/AGRGPL/0002/2010), funded by the Portuguese Foundation for Science and Technology (“Fundação para a Ciência e a Tecnologia” [FCT]), the “Programa Operacional de Factores Competitividade” (COMPETE), “Quadro de Referência Estratégico Nacional 2007–2013” (QREN), and “Fundo Europeu de Desenvolvimento Regional” (FEDER) of the European Union, and also by the project AGL2013-43329-R funded by the “Ministerio de Economía y Competitividad” of Spain.
Authors’ contribution
The authors have made the following declarations regarding their contributions: Conceived and designed the experiments: JLB and AC. Performed the experiments: AD and AC. Analyzed the data: AD, AC, JLB. Contributed with plant material/reagents/analysis tools: ACM, AM, JLB. Contributed to the writing of the manuscript: all authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Andreia Delgado and Ana Carvalho have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Delgado, A., Carvalho, A., Martín, A.C. et al. Genomic reshuffling in advanced lines of hexaploid tritordeum. Genet Resour Crop Evol 64, 1331–1353 (2017). https://doi.org/10.1007/s10722-016-0439-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-016-0439-3