Abstract
The upland rice crop system located within Brazilian savannas and Amazon Rainforest is the largest rainfed rice growing area in Latin America. To develop and release higher yield and adapted cultivars for this large region, the upland rice breeders need to conduct multiple-location trials aiming to model the genotype × location (G × L) and evaluate the germplasm yield adaptability. Here we hypothesize that regional patterns of G × L across this extensive region can be modeled by integrating factorial regression models with a geographic information system (GIS). Two sets of advanced yield trials from different germplasm pool were used in this study. From GIS tools, we collect and process geographic covariates and produce thematic maps of yield adaptability. One advantage of the methodology is that adaptability can be dissected into genotypic-sensibility coefficients related to the reaction norm for the geographic gradient. Then, breeders can discriminate different types of adaptability over a region, such as responsiveness for elevation, longitudinal or latitudinal adaptation, identifying possible ideotypes to solve current adaptation gaps for target regions. We observed that about of 53–59% of the G × L effects are due to predictable geographic-related factors. However, the upland rice germplasm is better adapted to higher elevations (> 700 m), which may indicate limitations in cultivar development because these regions do not represent the current upland rice growing region. We suggest to exploit geographic-related factors by increasing breeding efforts for northern and western Brazil environments located at lower elevations (< 300 m) and Equador’s near latitudes (2° S–2° N).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genotype-by-environment interaction (G × E) is a biological and statistical phenomenon present in breeding decision making on cultivar development (Malosetti et al. 2013). Its effect is due to the differential response of genotypes in relation to environmental variations, thus integrating the ecophysiological aspects resulting from each genotype reaction to several environmental effects (e.g., elevation and air temperature). As genotypes have different reaction norm, there are situations where G × E interaction is manifested in different degrees of impact, which in many cases result in a limitation for decision making on cultivar recommendation over multi-environment trial (MET) analysis. As regards their effects, G × E interaction patterns may be non-crossover (quantitative variations without changes in genotype ranking), crossover (qualitative variations with ranking of genotypes across environments) or null G × E interaction, where only genetic (G) and environmental (E) effects define the phenotypic mean (Y), i.e., following the Wilhelm Johannsen’s additive main effects model: Y = G + E (Lynch and Walsh 1998).
In situations where G × E interaction is present, its effect may result in depletion, e.g., negative effects when G + G×E < G; or capitalization, e.g., positive effects, when G + G×E > G. Thus, two fronts of research for germplasm improvement and cultivar development are desirable: (1) by searching for strategies to diagnose the adaptability of genotypes in order to identify situations where capitalization is desirable (e.g., Yan et al., 2007); (2) identification of regions or group of environments where there is a predominance in the capitalization of G × E interaction effects (e.g., Löffler et al., 2005; Chenu et al., 2011). For these purposes, several statistical-based methods have been used, such as regression models of phenotypic means on an environmental index (Finlay and Wilkinson 1963), mixed-effects factor analytic models (Piepho 1998; Smith et al. 2014; Smith and Cullis 2018), and linear-bilinear models, such as site-regression model on GGE-biplot analysis (see Yan and Tinker 2006). The linear-bilinear models are useful for biplot analysis, summarizing patterns of similarity between genotypes or environments (Gabriel 1971). However, biplot analysis is more descriptive and explanatory, without not much predictive ability, and it must be applied with caution once that it is unsuitable for hypothesis testing and insufficient to explain complex structures of G × E interaction (Yang et al. 2009). Biplot analysis is also graphically limited in represent genotypes spatial adaptive patterns, which are restricted only to environments within the available MET analysis.
Another perspective relies on the use of factorial regression models (FR, Denis 1988) that incorporate additional information into the MET analysis. FR models consist in a linear regression focused on dissect the non-additive effects of the G × E matrix into predictable and unpredictable genotypic responses. The predictable genotypic responses are defined by the genotypic sensitivity to certain environmental covariate (e.g., rainfall, air temperature, solar radiation) or geographic position (e.g., latitude, longitude and altitude) and have a biological meaning that can understand the environmental drivers of the G × E interaction patterns (e.g., Voltas et al. 1999; Epinat-Le Signor et al. 2001; Ortiz et al. 2007; Verhulst et al. 2011) or use them into an accurate predictive model (Ly et al. 2018; Millet et al. 2019).
The analysis of G × E interaction based on FR models has proven useful for cultivar testing analysis in several crops worldwide, such as maize (Crossa et al. 1999; Epinat-Le Signor et al. 2001; Magari et al. 1997; Romay et al. 2010; Millet et al. 2019; Crossa et al. 1999; Epinat-Le Signor et al. 2001; Magari et al. 1997; Millet et al. 2019; Romay et al. 2010), potato (Baril et al. 1995), tomato (Ortiz et al. 2007), wheat (Baril 1992; Reynolds et al. 2004; Verhulst et al. 2011; Heslot et al. 2014), melon (de Nunes 2011) and sugarcane (Ramburan et al. 2011, 2012). In these studies, the use of FR has contributed to a better understanding of the ecophysiological drivers linked to the G × E interaction. However, FR models can also contribute to the prediction of phenotypes under new environmental conditions (Ly et al. 2018; Millet et al. 2019), especially integrated with whole-genome regressions in genomics-assisted prediction-based breeding platforms (Heslot et al. 2014; Jarquín et al. 2014; Morais Júnior et al. 2018).
In this study, we applied FR models as spatial trend interpolation integrated with a geographical information system (GIS), aiming to provide geographic yield adaptability diagnosis over the target region of the upland rice breeding program in Brazil. GIS tools can be useful for cultivar targeting because of the widely available databases of environmental data and facilitate the implementation of thematic maps for several purposes (Annicchiarico 2002; White et al. 2002; Hyman et al. 2013; Xu 2016; Annicchiarico 2002; Hyman et al. 2013; White et al. 2002; Xu 2016). As upland rice breeding target population of environments (TPE) in Brazil is geographically extensive and heterogeneous (Colombari Filho et al. 2013; Heinemann et al. 2015, 2019), this can be an alternative to represent the spatial adaptation of the target germplasm.
Upland rice in Brazil is grown in a large TPE, that englobes sites with lower elevation (< 100 m above sea level), considerate hotter regions at lower equatorial latitudes (2° S to 1° N) and sites with higher elevation (> 700 m above sea level). In this TPE, there are sites whose drought-stress typology is limiting rice yields and also regions whose rainfall is excessive, as in the Amazon basin (Heinemann et al. 2019). There is still enormous variation in the physical-water and chemical properties of the soils between these regions (Cooper et al. 2005; Buol 2010). It has been also observed that the modern type cultivars have exhibit differential drought sensitive responses, which can be one of the causes that higher impact of genotype × location (G × L) interaction patterns in the multi-environment trials (MET) in the last 30-years (Colombari Filho et al. 2013; Heinemann et al. 2019).
Front to these above discussed, there is also a strategic gap related to cultivar development decisions in upland rice breeding program. How breeders can enhance adaptation diagnosis when they use a limited sample of the TPE (e.g., the MET framework) and not account regional variations in genotypic responses? i.e., only within MET specific variations. Even that breeders which adopted strategies as directing cultivar recommendation by a single State (Colombari Filho et al. 2013) or stratified the entire TPE by drought-patterns (Heinemann and Sentelhas 2011; Heinemann et al. 2015), the complex of geographic-related factors implicating into regional-spatial patterns still resulted in higher G × L effects. Thereby, this study aims to provide a new strategy for adaptation analysis based on a GIS linked to FR model in order to capitalize this geographic-related factors into a single analysis. We evaluated the merit of the use of the genotypic coefficients obtained from the FR model using simple geographic covariates (latitude, longitude and elevation) to describe the spatial region norm (or regional adaptation, denoted here as Ad) of the genotypes for an entire target region. Next, the predictive ability of the adjusted models in replacing a single missing location conditions were also evaluated in terms of accuracy (correlation between true G + G × L and predicted Ad values) and selection coincidence (the rank coincidence of the 5 top best genotypes by location). We expect to provide a friendly-user framework that will assist breeders in the diagnosis of adaptability of pre-commercial cultivars, as well as to support their decision making regarding the direction of research efforts front to the regional patterns not commonly visualized by the conventional statistical approaches of MET analysis.
Materials and methods
Study region
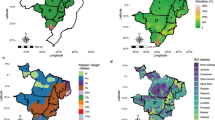
This study focused into the main target population of environments (TPE) from the national rice breeding program of the Brazilian Agricultural Research Corporation (Embrapa). The TPE region encompasses 90% of the upland rice production in Brazil (837,687 ton in 2017, IBGE—http://www.sidra.ibge.gov.br/bda/), and represents two climates according to Koppen–Geiger classification: Am equatorial and, As tropical savannah climate with a monomodal rainfall pattern (Alvares et al. 2013). Total precipitation ranges from 1000 to 2500 mm per year. Elevation ranges from 0 m to 1270 m. These multi-environment trial (MET) framework covered diverse agro-ecologically environments between 3° S and 16° S and 63° W to 42° W, involving seven Brazilian states: Goiás (GO), Mato Grosso (MT), Tocantins (TO), Pará (PA), Piauí (PI), Maranhão (MA) and Rondônia (RO) (Fig. 1a; Supplementary Table 1).
Geographic distribution of the field trial locations in Brazil, South America. a target region involving seven Brazilian states and different elevation levels above see; b geographic coordinates of the evaluated locals within the target region for MET1 and MET2 datasets (see Supplementary Table 1 for more details)
Nationwide yield trials
As a proof-of-concept, we used two independent datasets from upland rice breeding program, referred here as MET1 and MET2 (Fig. 1b; Supplementary Table 1). MET1 consists of 16 elite-lines and 4 cultivars (total of 20 genotypes) cultivated across 15 locations in rainfed season of 2004/2005 and MET2, 19 elite-lines and 4 cultivars (total of 23 genotypes) cultivated across 14 locations in rainfed season of 2012/2013. For both datasets, the experimental design was a randomized complete block with four replications. Each experimental plot consists of four rows of 5,0 m spaced of 0.3 m, with seeding density of 60 seeds m−1. Crop management are the same in both datasets (MET1 and MET2): total of 100 kg/ha of nitrogen, being 20% in the sowing, 40% 20 days after sowing and 40% 45 after sowing; pos-emergent herbicide for weeds and insecticide for caterpillar and bug; A single-trial model was previously fitted to provide a mean yield value for each genotype and environment. Adjusted means for each genotype at each trial (\(\widehat{y}_{i} = \mu + \widehat{g}_{i}\)) were obtained from ordinary least squares (OLS) following: \(y_{ik} = \mu + b_{k} + g_{i} + e_{ik}\), where \(y_{ik}\) is the vector of observed grain yield values for the genotype i-th (i = 1, 2,.., p) at block k-th (k = 1, 2, 3 and 4); \(g_{i}\) is the vector of genotype effects, and \(e_{ik}\) is the vector of residuals of the single-trial analysis, with \(e_{ik} \sim N\left( {0,I\sigma^{2} } \right)\). Only trials with experimental quality merit were inserted into the following analysis. Then, a joint analysis involving all locations was conducted to assess the magnitude and significance of the genotype by location (G × L) interaction for each dataset, following:
where \(y_{ij}\) is the vector of adjusted grain yield values for the genotype i-th (i = 1, 2,…, p) at location j-th (j = 1, 2,…, q); \(g_{i}\) is the vector of genotype effects, \(l_{ij}\) is the vector of location effects, \(gl_{ij}\) is the vector of genotype × location interaction (G × L); and \(e_{ij}\) is the vector of residuals of the joint analysis, with \(e_{ij} \sim N\left( {0,I\sigma^{2} } \right)\). From Eq. 1, the G × L effects were estimated by ordinary least squares (OLS): \((\widehat{g}l)_{ij} = Y_{ij} - \overline{Y}_{i.} - \overline{Y}_{.j} + \overline{Y}_{..}\), where: \(Y_{ij}\) is the mean of the i-th genotype for the j-th location; \(\overline{Y}_{i.}\) is the mean of the i-th genotype for any location; \(\overline{Y}_{.j}\) is the mean of the j-th location for any genotype; and \(\overline{Y}_{..}\) is the overall mean. The residual variance was tested using Bartlett’s test (1937) and the degrees of freedom associated with residual variances and G × L interaction was adjusted following Cochran (1954), due to the heterogeneity detected between the mean square errors computed in our previous statistical analysis.
Factorial regression (FR)
Factorial regression (FR) is a technique applied to dissect G × E interaction effects into predictable genotypic responses and residual (unpredictable) effects (van Eeuwijk et al. 1996). The predictable responses are given by the estimative of genotypic sensitivity to certain environmental (e.g., rainfall, air temperature, solar radiation) or geographic position (e.g., latitude, longitude and elevation). For this reason, FR models are a technique to link envirotyping (environmental + typing Xu (2016)) approaches to adaptability models unrevealing the main environmental drivers of the G × E for a target germplasm over a target region. FR-based analysis combines predictive power and exploratory analytical tools, which is useful to guide cultivar development strategies. Here, we applied FR models as a spatial trend interpolation in GIS framework and as exploratory tool helping to visualize regional adaptation trends in two sources of the upland rice germplasm (MET1 and MET2). From the G × L interaction matrix obtained in Eq. 1, we applied the FR using environmental covariates as showed below:
where \(b_{\text{ik}}\) is the coefficient of regression that describes the particular sensitivity of the i-th genotype to the k-th environmental covariate (k = 1, 2,… v); \(z_{\text{kj}}\) is the scaled value for mean = 0 and variance = 1 associated with k-th environmental covariate in the j-th location; \(u_{ij}\) is the residual effects not captured by the variables selected in G × L interaction modeling. Thus, the complete model (Eq. 1), in its original version and after decomposition (Eq. 2), becomes:
where \(y_{ij} - \overline{y}_{.j}\) can be viewed as the relative yield adaptability for the i-th genotype at the j-th location free of local-specific unpredictable environmental effects \(y_{ij} - (\overline{y}_{.j} + u_{ij} )\). Assuming that is expected to model predictable genotypic effects and responses to an environmental gradient, the expected yield adaptability (or regional adaptation, \(Ad_{ij}\)) for a specific location is now obtained (adapted from Martins 2004 master’s degree thesis):
The model presented in (Eq. 4) can be assumed as the reaction norm model environmental-centered, and modeling the expected cultivar adaptability for any new location. In other words, it is a spatial interpolation obtained from (Eq 1.) enriched by geographic information. From now on in this study, we call the genotypic value \(g_{i}\) which is also known as the mean genotypic value (MGV) assuming a position of a genotypic-specific intercept.
FR with geographic information systems (GIS)
The FR model was incorporated into GIS tools as a global spatial trend interpolator of yield adaptability. Thus, predictions are made for different points in a spatial grid for a target region, resulting in the possibility of producing adaptability maps. In this study, we used latitude, longitude and elevation information as geographic covariates in order to model spatial trends of phenotypic plasticity among the genotypes. The elevation data were obtained by the SRTM file (available in: https://lta.cr.usgs.gov/SRTM) consisting of a grid of 0.16° × 0.16°, based on datum WGS84. The reference coordinates were assumed as the observed at the evaluation trials location. The measurements of the covariables given were centered on the mean, with variance 1 and mean 0, that is, \(z\sim N\left( {0,1} \right)\). The Eq. 4 is then updated as:
where \(\widehat{b}_{1}\), \(\widehat{b}_{2}\) and \(\widehat{b}_{3}\) are de genotypic sensibility coefficients of the i-th genotypes for the effects of latitude (kg ha−1 unit of latitude degree−1), longitude (kg ha−1 unit of longitude degree−1) and elevation (kg ha−1 m−1 above sea level), assuming each unit of latitude degree equal of unit of longitude degree = 0.16° = ud.
Mapping consistence statistics
A cross-validation scheme was used to verify the FR-model predictive ability to reproduce spatial trends within MET and for new locations. The scheme used a bootstrap approached based on 10,000 random samplings, which generated a training population by resampling 10% of the genotypes (~ 2 genotypes) and leaving one environment (location) out. Then, we can verify the model consistence by using the training population set to predict the genotypes and environments removed from the analysis. At each boot were computed the genotypic sensibility coefficients, the MGV and the contribution of each geographic covariable to the sum of squares of each regression (by genotype). The predictive ability was estimated by the Pearson’s moment correlation (r) between the observed (\(g_{i} + gl_{ij}\)) and predicted (\(Ad_{ij} )\) values.
We also compared the consistency of the FR-model in reproducing the selection performed in each trial as a criterion for recommending new cultivars per state. For this, we compared the selection coincidence of the top 5 pre-commercial cultivars, in each test, for each boot, based on the magnitude of the observed and predicted effects for each location within each state.
Yield adaptability maps
Finally, yield adaptability maps were generated using the genotypic sensitivity coefficients obtained by the mean of the coefficients computed in the aforementioned 1000-time bootstrap stage. Each prediction was performed under the pixel of 0.16° × 0.16°, approximately an area of 17 km × 17 km, a total of 21,1147 pixels (~ 21 M pixels) and 6,682,160 km2 area. The expected values of yield adaptability E(\(Ad_{ij}\)) for each pixel were grouped in classes of 500 kg ha−1 empirically delimited and colored with warm (negative adaptability) and cold (positive adaptability) colors. This approach was designed to facilitate genotypic discrimination through visual interplay. In fact, for a new cultivar to be released, the breeding program must specify which state the new cultivar is adapted to the Brazilian Ministry of Agriculture, Cattle and Supplying (MAPA). Therefore, diagnostic procedures for cultivar targeting and the spatial interpolator consistency were computed following the territorial limits of each state in the target region.
Software and data availability
All statistical process and GIS tools, in this study, were carried out using the R platform 3.5.0 (R Core Team, 2018). In this study, the R-codes and dataset applied were compiled into a new R package named frGIS. The R package and the database is freely available at GitHub at https://github.com/gcostaneto/frGIS.
Results
Predictable spatial patterns
For both datasets the genotype × location (G × L) interaction effects were highly significant (see Table 1, as * significance at 5% for F-test). The largest variation in the main effects was due to location, accounting for 61% (MET1) and 76% of the sum of squares—SS (MET2, 2012/2013). However, G × L interaction were also an important source of variation, accounted for 32% (MET1, 2004/2005) and 18% of the SS (MET2, 2012/2013), being more than four (× 4) and three (× 3) times, the variance explained by the genotypic main effects in MET1 (7%) and MET2 (6%), respectively.
Table 1 shown the mean values for SS and mean squares (MS) obtained from 1000-bootstrap. This approach enabled to calculate the theoretical parametric mean fraction of contribution of each geographic covariate for the G + G×L SS under different multiple-environment arrangements. For simulated condition, a high variation of the G + G × L fraction explained by the geographic covariates was observed. For MET1, the fraction of the G + G × L variance explained by latitude (11 ± 10%), longitude (10 ± 12%) and elevation (32 ± 12%) allowed to identify up to 53% of repeatable spatial patterns. In MET2, this value reached up to 59%, but with different covariates contribution, being latitude (26 ± 13%) and longitude (16 ± 8%) showed higher contribution than elevation (17 ± 15%).
The observed G + G × L effects and the captured regional patterns are showed in Fig. 2a, b, respectively, for each dataset (MET1 and MET2). As expected for an extensive and environmentally complex region, the relative positioning of the genotypes summarized over locations changes considerably. The observed lower correlation between trials in both datasets indicates that crossover patterns of G × L interaction are predominant in the upland rice growing region (Supplementary Figure 1). This phenomenon implicates into an unpredictable biological response and an obstacle for cultivar targeting (Annicchiarico 2002; Kang 2002), especially for extensive regions such as upland rice in Brazil. In these Fig. 2a, b, the genotypes were ordered vertically in descending order of magnitude G + G × L and Ad. It is possible to observe that a great part of the genotype ordering is due to the spatial patterns, since the same arrangement in Fig. 2a, b has been maintained.
Graphical summary of adjusted means of each genotype (Y-axis) at each location (X-axis), expressed in kg ha−1,for datasets MET1 and MET2. a full observed genotype plus genotype by location interaction effects (G + G × L) patterns from the ordinary least squares (OLS) estimations; b regional adaptation trends captured by factorial regression with geographic covariates related to latitude, longitude and elevation. The values were ordered on the X and Y axis
Spatial trends consistence
The predictive ability and the selection coincidence were measures applied to verify the consistency of the spatial interpolation in predicting the ordering of genotypes in environments within the MET structure and in new sites by state (Fig. 3). Predictive ability values were computed by the Pearson moment correlation (r, given in percent %) between the values of G + G × L and Ad for scenarios within the MET and new locations (Fig. 3a). Then, were observed a mean (± standard deviation) r values for MET1 and MET2 equal to 50 ± 10 and 67 ± 10%, in the within MET scenario; and − 10 ± 17; and 23 ± 1% for prediction of new sites not included in the MET structure.
Statistics of spatial interpolation consistency by State. a predictive ability based on the Pearson’s correlation between observed G + G×L and predicted Ad values; b percentage of selection coincidence between the field trial phenotypic selection using magnitude values of G + G×L and Ad. The bars represent average values of 1000-boots, with their respective standard deviations. Acronyms GO, MA, MT, PA, PI, RO and TO refers to the States of Goiás, Maranhão, Mato Grosso, Pará, Piauí, Rondônia and Tocantins
The r values depended on the State, dataset, and scenario. For MET conditions, the highest r values was achieved in Goiás (GO) in MET2 (97 ± 1%), Tocantins (TO) in MET1 (75 ± 1%), Piauí (PI) in MET2 (75 ± 1%) and Pará (PA) in MET2 (75 ± 1%), respectively. We found that the breeding nursery (Santo Antonio de Goiás, STG) has a low correlation with most sites in the experimental rice test network (see Supplementary Figure 1). For this reason, the removal of the STG assays, the only assay in the state of Goiás in MET2, did not significantly impair the predictive capacity of the models. However, the forecast of new locations within the states did not follow the same trend. For new sites, the r values ranged from − 21 ± 28% (Pará State at MET1) to 57 ± 1% (Goiás Sate at MET2). In MET1, in addition to the r State with negative values, we also observed an inability to predict new sites in MA, GO, TO and PI. In MET1, this cause may be due to the greater adaptation of the genotypes to the warmer and lower latitude regions, such as PA and Northern MT. However, in MET2 the lowest r values were observed in Rondônia (RO) with 13 ± 29%, MA (14 ± 1%) and PI (16 ± 1%).In this case, the current germplasm is more adapted to regions of higher elevation and latitude, such as those found in the nursery of the upland rice breeding program located in STG.
The selection coincidence (SC, %) of the top-5 best genotypes per location was applied as a criterion of consistency in the prediction of the higher yield cultivars by State (Fig. 3b). SC values varied from 13 ± 15% (PI, MET1) to 89 ± 9% (GO, MET2), with an overall mean of 49 ± 15% for conditions within MET; and 2 ± 5% (PI, MET2) up to 51 ± 17% (MT, MET1), with an overall mean of 23 ± 14% when predicting new locations. The most consistent state based on SC criterion was Mato Grosso (MT), with SC values equal to 62 ± 18% (within MET) and 50 ± 18% (new locations) in MET1; and 51 ± 10% (within MET) and 43 ± 20% (new locations) in MET2. The state of Pará (PA), which has a large territorial extension as MT, showed values of SC equal to 64 ± 11% (within MET) and 42 ± 16% (new locations) in MET2.
Geographic drivers of yield adaptability
The impact of geographic effects on the adaptability of cultivars in terms of genotypic responsiveness (genotype sensitivity coefficients, \(\widehat{b}_{k}\)) and their participation in the sum of square of the G × L interaction plus genetic effect (G) are summarized in Figs. 4a and 4b. Differently from regression coefficients, such as that of Finlay and Wilkinson (1963), the plasticity of the cultivars was detailed in responsiveness to latitude (\(\widehat{b}_{1}\)), longitude (\(\widehat{b}_{2} )\) and elevation (\(\widehat{b}_{3} )\) plus a mean genetic effect (MVG, kg ha−1). For MET1, there is a high effect of elevation for most genotypes, which tended to respond positively to the increase in elevation (from 3.2 kg ha−1 m−1 above sea level for genotype G04 and to 0 kg ha−1 m−1 for G12 genotype). The fraction of the G + G×L variance explained by this effect reached up to 54% in G13 and 61% in G08. However, some genotypes showed a greater response to other effects, such as latitude (35% in G01, with a coefficient equal to − 101.7 kg ha−1 ud −1) and longitude (41% in G12, with coefficient equal to − 43.7 kg ha−1 ud−1).
Impact of the geographic effects on the upland rice yield adaptability for datasets MET1 and MET2. a Genotypic sensibility coefficients for geographic covariates and mean genotypic effect (MGV) obtained from 1000-boos; b fraction of the G + GL SS explained by the geographic covariates for each genotype
Yield adaptability maps for cultivar development support
In the yield adaptability maps (Fig. 5) for the MET1 and MET2 datasets, negative values in red represents the genotype not adapted to the target pixel region. Examples of the performance of the genotypes for each dataset were highlighted in Figs. 5a (MET1) and 5b (MET2) and the others not-selected are showed in the Supplementary Fig. 2. Adaptability has been identified point to point and is quantified as mean values, e.g., + 500 kg ha−1 is equivalent to affirming that the genotype will capture + 500 kg ha−1 above the average of the evaluated germplasm at the same MET conditions. For example, genotype G01 has adaptability to the northern region of Brazil (Pará state, PA), but is not adapted to the central region (Goiás state, GO). The genotype G26 has adaptation to the environments of the Goiás and southern Tocantins sates, but showed little adapted to the northern regions (northwest of Mato Grosso and Pará).
Yield stability can also be viewed spatially by the adaptability variability classes over the target region. Genotypes G02 and G14, for example, are highly stable because they have the same adaptability throughout the region. This can also be characterized as a broad regional adaptability. However, the G25, G26 and G28 genotypes are highly adapted to the higher latitude regions in the state of Goiás, such adaptability is not repeatable throughout the target region, resulting in non-adaptation to the northern region, characterized by lower elevations (from 0 to 100 m a.s.l) and higher latitudes (from 0° to 5° S). Therefore, they are fewer stable genotypes to an extensive target region, but with specific adaptability to conditions of higher elevation (from 500 to 800 m a.s.l) and lower latitudes (from 13° S to 20° S). These concepts can be associated with the type I stability (Lin and Binns 1991). The ideotype sought is that one occupying the wider area, has higher performance (adaptability) and lower variation in the adaptability classes (agronomic stability). Based on the criteria listed above, the ideal genotypes are G14 (broad + high average adaptation in the northern region) and G28 (wide adaptation in the western + medium–high regions).
Discussion
Factorial regression with GIS as a complementary model in advanced yield tests
FR model is currently used to associate non-genetic additional information from environmental variables (e.g., elevation) with phenotypic observations or G × E interaction effects from MET analysis (Denis 1988). Traditionally, this approach focused in exploratory analysis for recovery some useful biological or agronomic pattern which can explain G × E interaction drivers. More details about the different FR models and their association with traditional linear-bilinear models (e.g., AMMI) can be found in van Eeuwijk et al. (1996). Martins (2004) provide the first report of using FR models to visualize spatial patterns of G × E interaction. In this study, we focus on the potential of using this methodology, associated with the computational resampling techniques (e.g., bootstraping) and geographic information systems (GIS), aiming not only to understand the patterns but also to explore regional yield plasticity trends for cultivar adaptability diagnosis purposes.
The use of GIS for spatial diagnostic mapping of the G × L interaction can contribute to the cultivar targeting and spatial diagnosis of agroecological zones (Hyman et al. 2013). However, the integration with FR models facilitates both the diagnosis of adaptation zones of the genotypes and a better understanding of the spatial causes of phenotypic plasticity. Here we seek to expand the interpretation concept of the G × E interaction via FR not testing the significance of the covariables but rather verifying the participation of the same explaining the variance of the effects G + G × L, i.e., the effects that denote the genetic responsiveness to the environmental variation. Thereby, we use here a concept of reaction norm applied to the explanation of regional adaptation.
Nevertheless, the methodology was also efficient as an exploratory method, especially when we sought some biological meaning for the environmental covariates in explaining the G × L interaction. In the past we see that the search for the identification of effect covariates has sought an empirical understanding of the causes of G × E interaction in diverse crops (Epinat-Le Signor et al. 2001; Ortiz et al. 2007; de Nunes 2011; Verhulst et al. 2011). A central question when using FR models is the choice of covariables for description of G × E interaction. Thermal-related co-variates, such as accumulated degree-days in diverse development stages, or simple co-variates such as average air temperature were used with success in the past (Baril et al. 1995; Magari et al. 1997; Voltas et al. 2005; Ortiz et al. 2007). These covariates, in combination with hydrological factors (e.g., evapotranspiration, accumulated precipitation) explained from 44% up to 91% of G × E interaction SS. Here, we were able to explain up to 59% of G + G × L SS using only the three simple geographic co-variates, latitude, longitude and elevation.
Ecophysiological interpretation of the G × L interaction spatial trends
As the first step in developing a future high-density GIS-based envirotyping framework to support cultivar testing, we evaluate here the merit of the use of simple geographic covariates (latitude, longitude and elevation) in representing latent regional-specific environmental patterns and how the genotypes capitalize the consequent G × E interaction effects from these patterns. Thus, we were able to find exploitable ecophysiological interpretations from these geographic effects. The geographic covariates studied here represented the spatial variability present in a target population of environments (TPE), such as local-specific factors (e.g., soil type variations across locations) and possible biotic relations, such as the differential response of crops in growth and development and also their interaction into pathosystems differentially respond to geographic variation.
Latitude and longitude showed statistical importance as spatial modulators in modeling G × L patterns, as observed by Hyman et al. (2013). Latitude is a covariate related to the radiation balance in the atmosphere, as well as day length and heat intensity received in the canopy (Allen et al. 1998). Elevation is a covariable inversely related to the air temperature and atmospheric pressure (Allen et al. 1998). Regions with higher altitudes tend to have lower temperatures and higher daily range in air temperature. For these conditions, there are reports of increased susceptibility to blast disease (Magnaporthe grisea), considered the main disease in rice worldwide (Raboin et al. 2016). This is due to the formation of dew and mild temperatures, favorable conditions for the development of the fungus (Kato 1974; Kim, 1994; dos Santos et al. 2011). For upland rice in Brazil, the pathogen races are very region-specific, exhibit higher diversity of races according to the regional climate, soil and management practices (da Silva et al. 2012). Therefore, latitude and longitude variations may also express the regional diversity of the breed of these pathogens (Filippi et al. 2002), among others, resulting in differential pathogen-host interactions throughout a state or region.
Regional variations in soil properties can also be observed (Cooper et al. 2005; Buol 2010). Lower regions close to rivers tend to have soils of lower depth of A-horizon, having higher levels of sand than of clay and higher elevation regions are characterized by the predominance of Oxisols (Cooper et al. 2005; Heinemann et al. 2019). Therefore, the geographic variables are indirect descriptors of geo-spatial conditions of relief implying in the dynamics of the agricultural systems. Drought-stress is one of the most important yield-limiting factors of rainfed crops such as upland rice (Heinemann et al. 2015). There is a spatial variation among drought-stress in Brazil, which combined with the lower resilience of the modern type cultivars (Heinemann et al. 2019) have resulted into differential patterns of G × L interaction among states. In this context, in our study, the combined effect of latitude, longitude and elevation may represent the latent effect of regional drought typologies among states.
Regional trends reveal germplasm adaptation shifts
Although genotypes of the two datasets are from to distinct phases of the upland rice breeding program (Colombari Filho et al. 2013), it is possible to observe similar trends in their regional crop adaptation for some covariates. We observed that in MET1 the genotypes responded positively to the increase of elevation and negatively to the increase of longitude and latitude. Therefore, there are indications that genotypes were more adaptable to regions with higher elevations and lower latitudes and longitudes, such as those presented near the nursery of the breeding program in Goiás state (see STG location in Fig. 1). However, for MET2, the increase in elevation is also beneficial to capitalize G × L interactions, but the genotypes tended to present better performance in regions with higher longitudes, such as western regions of the state of Mato Grosso.
There are also indications of the existence of significant differences related to the G × L interaction patterns due to latitude effects. In a study conducted using data from 30-year yield trials, Colombari Filho et al. (2013) observed that there are notable differences in G × L interaction patterns between and within states. We can conclude that regional differences within the states may be associated with the differential response of cultivars to variations such as altitude, longitude and elevation, especially in states with a large territorial area, such as Mato Grosso and Pará. Considering that the two datasets are from different time of the breeding program, it is possible to infer that these differences may be related to the selection process directed to specific environments, as in northern and western of Mato Grosso state. From the last 15-years, the breeding program strategy to select genotypes have been changed. In this context, it’s possible that shift variations among datasets may reveal the impact of this breeding strategies into upland rice adaptation. Heinemann et al. (2019) using a long-term diagnosis demonstrated that drought-resilience has been decreased in the last 30 years. However, this study was based only in 3 representative genotypes, which their agronomy performances were highly distinct among them. Here we used an entire set of the elite-germplasm, which allows us to visualize more accurately the plastic shifts between (years) and within (G × L interactions) the two sets. Further studies involving more years of MET are needed to prove this hypothesis.
Adaptability Island around breeding program nursery
The geographic representation of yield adaptability highlights the existence of an “adaptability island” around to the breeding program nursery (located Santo Antonio de Goiás, GO State, latitude: 16.47 S; longitude: 49.28 W; elevation: 800 m a.s.l.). On the other hand, there is a difficulty to select better-adapted genotypes for the northern states (e.g., Pará, Maranhão, and Piauí). Similar trends in upland rice cropping area in Brazil were achieved by Heinemann et al. (2019) using cropping modelling system (CMS) and GIS tools. These authors, based on CMS approaches identified shifts in germplasm adaptation for drought stress in the last 30 years. However, they observed that regions near to the breeding program nursery has different drought-stress patterns than the rest of the entire TPE. For this reason, the modern type cultivars developed by the breeding focused on higher yield tend to be less resilient than the cultivars of the 80 s and 90 s. Here, we observed a quantitative increased in yield adaptability for the Goiás states from the years of 2004/2005 (MET1) to 2012/2013 (MET2) (Fig. 5).
In MET1, the maximum Ad values observe were equal to + 2000 kg ha−1. However, in MET2, it was observed values reaching + 3000 kg ha−1, denoting that selection gains for grain yield were achieved for higher elevations such as at breeding nursery conditions. Thereby, we also observed an increased in not adapted regions (red colors in the map) at equatorial and lower elevation regions when compared the germplasms from 2004/2005 to 2012/2013. By combining this information with our results, we hypothesize that the breeding strategies may lead to increased yield and adaptation to higher elevations of ‘Cerrado’ biome, which represents a fewer proportion of the entire TPE. To improve adaptation for equatorial Amazon regions, which has lower elevations, efforts in evaluating and selecting for grain yield must be direct to the regions at northern Mato Grosso State, Rondônia and Pará States.
Perspectives of GIS-based virtual screening for yield adaptability
Our results suggests a step-forward in cultivar testing over extensive regions, highlighting that geographic coordinates can be used to indicate some exploitable spatial trends related to G × L interaction. For early yield performance evaluations conducted at regional multiple-environment trials (with higher number of genotypes and lower number of locations), the FR-models using other environmental covariates can accommodate also molecular markers or pedigree information, exploiting relationship patterns of reaction norm (Heslot et al. 2014; Ly et al. 2018; Millet et al. 2019). However, when these models are embedded with GIS databases, the methodology present here can be used to predict and exploit the latent spatial trends of yield adaptability. In addition, there is also a possibility that incorporate several GIS databases (e.g., WorlClim, http://www.worldclim.org or NASA Power, http://power.larc.nasa.gov/) in order to provide a high-density envirotyping framework both for cultivar testing that germplasm adaptation diagnosis.
The GIS-based FR model approach can improve an in silico screening for yield adaptability, enabled an optimized effort in further MET trials of advanced phases of breeding programs. Similar approaches are also in current application, such as the use of CMS (Messina et al. 2018), single covariate effects incorporated into whole-genome regressions (Ly et al. 2018) and genomic-based FR approaches combining genotype-specific index (Millet et al. 2019). Here, we demonstrated similar evidence of increased predictive ability and capability to screen target germplasm for an entire breeding region. We also demonstrated that shifts in breeding strategies may promote differential sensibility responses, as observed from the germplasm of 2004/2005 (MET1) to the germplasm of 2012/2013 (MET2), when the effects of latitude have been changed and the specific adaptation for higher elevations remains the main gap for improving wide adaptation genotypes. We suggest that early efforts in crossing, selecting and evaluating genotypes are conducted at MET framework, and if possible, to incorporate some environmental and genomic information in order to allow the training of FR models for spatial prediction as an in silico early screening for adaptation.
References
Allen RG, Pereira LS, Raes D, Smith M (1998) Crop evapotranspiration: guidelines for computing crop water requirements. FAO Irrigation and drainage paper 56/Food and Agriculture Organization of the United Nations
Alvares CA, Stape JL, Sentelhas PC et al (2013) Köppen’s climate classification map for Brazil. Meteorol Zeitschrift 22:711–728. https://doi.org/10.1127/0941-2948/2013/0507
Annicchiarico P (2002) Genotype × environment interactions: challenges and opportunities for plant breeding and cultivar recommendations. Food and AgricultureOrganisation of the United Nations, Rome
Baril CP (1992) Factor regression for interpreting genotype-environment interaction in bread-wheat trials. Theor Appl Genet 83:1022–1026
Baril CP, Denis J-B, Wustman R, Van Eeuwijk FA (1995) Analysing genotype by environment interaction in Dutch potato variety trials using factorial regression. Euphytica 84:23–29
Buol SW (2010) Soils and agriculture in central-west and north Brazil. Sci Agric 66:697–707. https://doi.org/10.1590/s0103-90162009000500016
Chenu K, Cooper M, Hammer GL et al (2011) Environment characterization as an aid to wheat improvement: interpreting genotype–environment interactions by modelling water-deficit patterns in North-Eastern Australia. J Exp Bot 62:1743–1755
Cochran WG (1954) The combination of estimates from different experiments. Biometrics 10:101–129
Colombari Filho JM, de Resende MDV, de Morais OP et al (2013) Upland rice breeding in Brazil: a simultaneous genotypic evaluation of stability, adaptability and grain yield. Euphytica 192:117–129. https://doi.org/10.1007/s10681-013-0922-2
Cooper M, Mendes LMS, Silva WLC, Sparovek G (2005) A national soil profile database for brazil available to international scientists. Soil Sci Soc Am J 69:649. https://doi.org/10.2136/sssaj2004.0140
Crossa J, Vargas M, Van Eeuwijk FA et al (1999) Interpreting genotype × environment interaction in tropical maize using linked molecular markers and environmental covariables. Theor Appl Genet 99:611–625
da Silva GB, de Araújo LG, da Lobo VL et al (2012) Use of local rice cultivars as additional differentials to identify pathotypes of Pyricularia oryzae. Bragantia 70:860–868. https://doi.org/10.1590/s0006-87052011000400019
de Nunes GH (2011) Influência de variáveis ambientais sobre a interação genótipos x ambientes em meloeiro. Rev Bras Fruttic 33:1194–1199. https://doi.org/10.1590/S0100-29452011000400018
Denis JB (1988) Two way analysis using covarites. Statistics (Ber) 19:123–132
dos Santos GR, Chagas JFR, Tavares AT et al (2011) Danos causados por doenças fúngicas no arroz cultivado em várzeas no Sul do Estado do Tocantins. Bragantia 70:869–875. https://doi.org/10.1590/S0006-87052011000400020
Epinat-Le Signor C, Dousse S, Lorgeou J et al (2001) Interpretation of genotype x environment interactions for early maize hybrids over 12 years. Crop Sci 41:663–669. https://doi.org/10.2135/cropsci2001.413663x
Filippi MC, Prabhu AS, De Faria JC (2002) Genetic diversity and virulence pattern in field populations. Race 1:1681–1688
Finlay KW, Wilkinson GN (1963) The analysis of adaptation in a plant breeding programme. J Agric Res 14:742–754
Gabriel KR (1971) The biplot graphic display of matrices with application to principal component analysis. Biometrika. https://doi.org/10.1093/biomet/58.3.453
Heinemann AB, Sentelhas PC (2011) Environmental group identification for upland rice production in central Brazil. Sci Agríc 68:540–547
Heinemann AB, Barrios-Perez C, Ramirez-Villegas J et al (2015) Variation and impact of drought-stress patterns across upland rice target population of environments in Brazil. J Exp Bot 126:1–14
Heinemann AB, Ramirez-Villegas J, Rebolledo MC et al (2019) Upland rice breeding led to increased drought sensitivity in Brazil. F Crop Res. https://doi.org/10.1016/j.fcr.2018.11.009
Heslot N, Akdemir D, Sorrells ME, Jannink J-L (2014) Integrating environmental covariates and crop modeling into the genomic selection framework to predict genotype by environment interactions. Theor Appl Genet 127:463–480
Hyman G, Hodson D, Jones P (2013) Spatial analysis to support geographic targeting of genotypes to environments. Front Physiol 4:1–13. https://doi.org/10.3389/fphys.2013.00040
Jarquín D, Crossa J, Lacaze X et al (2014) A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor Appl Genet 127:595–607. https://doi.org/10.1007/s00122-013-2243-1
Kang MS (2002) Quantitative genetics, genomics and plant breeding, 1st edn. Cabi Publishing, Wallingford
Kato H (1974) Epidemiology of rice blast disease. Rev Plant Prot Res 7:1–20
Kim CK (1994) Blast management in high input, high yield potential, temperate rice ecossystems. In: Zeigler RS, Leong SA, Teng PS (eds) Rice blast disease. CAB International, Wallingford, pp 451–464
Lin CS, Binns MR (1991) Genetic properties of four types of stability parameter. Theor Appl Genet 82:505–509. https://doi.org/10.1007/BF00588606
Löffler CM, Wei J, Fast T et al (2005) Classification of maize environments using crop simulation and geographic information systems. Crop Sci 45:1708–1716. https://doi.org/10.2135/cropsci2004.0370
Ly D, Huet S, Gauffreteau A et al (2018) Whole-genome prediction of reaction norms to environmental stress in bread wheat (Triticum aestivum L.) by genomic random regression. F Crop Res 216:32–41. https://doi.org/10.1016/j.fcr.2017.08.020
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits, 1st edn. Sinauer Associates, Sunderland, Massachussets
Magari R, Kang MS, Zhang Y (1997) Genotype by environment interaction for ear moisture loss rate in corn. Crop Sci 37:774–779
Malosetti M, Ribaut JM, van Eeuwijk FA (2013) The statistical analysis of multi-environment data: modeling genotype-by-environment interaction and its genetic basis. Front Physiol. https://doi.org/10.3389/fphys.2013.00044
Martins AS (2004) Aplicação de sistema de informações geográficas no estudo da interação genótipos com ambientes. Master's dissertation, Universidade Federal de Goiás, Goiás, Brazil
Messina CD, Technow F, Tang T et al (2018) Leveraging biological insight and environmental variation to improve phenotypic prediction: Integrating crop growth models (CGM) with whole genome prediction (WGP). Eur J Agron. https://doi.org/10.1016/j.eja.2018.01.007
Millet EJ, Kruijer W, Coupel-Ledru A, et al (2019) Genomic prediction of maize yield across European environmental conditions. Nat Genet 51:952–956. https://doi.org/10.1038/s41588-019-0414-y
Morais Júnior OP, Duarte JB, Breseghello F et al (2018) Single-step reaction norm models for genomic prediction in multienvironment recurrent selection trials. Crop Sci 58:592–607. https://doi.org/10.2135/cropsci2017.06.0366
Ortiz R, Crossa J, Vargas M, Izquierdo J (2007) Studying the effect of environmental variables on the genotype × environment interaction of tomato. Euphytica 153:119–134
Piepho H-P (1998) Empirical best linear unbiased prediction in cultivar trials using factor-analytic variance-covariance structures. Theor Appl Genet 97:195–201
Raboin LM, Ballini E, Tharreau D et al (2016) Association mapping of resistance to rice blast in upland field conditions. Rice 9:1–12. https://doi.org/10.1186/s12284-016-0131-4
Ramburan S, Zhou M, Labuschagne M (2011) Interpretation of genotype × environment interactions of sugarcane: identifying significant environmental factors. F Crop Res 124:392–399
Ramburan S, Zhou M, Labuschagne M (2012) Integrating empirical and analytical approaches to investigate genotype × environment interactions in sugarcane. Crop Sci 52:2153–2165. https://doi.org/10.2135/cropsci2012.02.0128
Reynolds MP, Trethowan R, Crossa J et al (2004) Physiological factors associated with genotype by environment interaction in wheat. F Crops Res 75:253
Romay MC, Malvar RA, Campo L et al (2010) Climatic and genotypic effects for grain yield in maize under stress conditions. Crop Sci 50:51–58
Smith AB, Cullis BR (2018) Plant breeding selection tools built on factor analytic mixed models for multi-environment trial data. Euphytica 214:1–19. https://doi.org/10.1007/s10681-018-2220-5
Smith AB, Ganesalingam A, Kuchel H, Cullis BR (2014) Factor analytic mixed models for the provision of grower information from national crop variety testing programs. Theor Appl Genet 128:55–72
van Eeuwijk FA, Denis JB, Kang MS (1996) Incorporating additional information on genotypes and environments in models for two-way genotype by environment tables. In: Kang MS, Gauch HG (eds) Genotype-by-environment interction. CRC Press, New York, pp 15–49
Verhulst N, Sayre KD, Vargas M et al (2011) Wheat yield and tillage–straw management system × year interaction explained by climatic co-variables for an irrigated bed planting system in northwestern Mexico. F Crop Res 124:347–356
Voltas J, Van Eeuwijk FA, Araus JL et al (1999) Integrating statistical and ecophysiological analyses of genotype by environment interaction for grain filling of barley II: grain growth. F Crop Res 62:75–84
Voltas J, Lopez-Corcoles H, Borras G (2005) Use of biplot analysis and factorial regression for the investigation of superior genotypes in multi-environment trials. Eur J Agron 22:309–324
White JW, Corbett JD, Dobermann A (2002) Insufficient geographic characterization and analysis in the planning, execution and dissemination of agronomic research? F Crop Res. https://doi.org/10.1016/S0378-4290(02)00041-2
Xu Y (2016) Envirotyping for deciphering environmental impacts on crop plants. Theor Appl Genet 129:653–673. https://doi.org/10.1007/s00122-016-2691-5
Yan W, Tinker NA (2006) Biplot analysis of multi-environment trial data: principles and applications. Can J Plant Sci 86:623–645
Yan W, Tinker NA (2011) Biplot analysis of multi-environment trial data: principles and applications. Can J Plant Sci. https://doi.org/10.4141/p05-169
Yan W, Kang MS, Ma B et al (2007) GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop Sci 47:643–653
Yang RC, Crossa J, Cornelius PL, Burgueño J (2009) Biplot analysis of genotype x environment interaction: proceed with caution. Crop Sci 49:1564–1576
Acknowledgements
The authors would like to thank the entire rice breeding team of Embrapa, especially the research assistants and field workers who performed the field trials and collected the data used in this study. We also wish to thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) for granting scholarships to the GMFCN and OPMJ.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Costa-Neto, G.M.F., Morais Júnior, O.P., Heinemann, A.B. et al. A novel GIS-based tool to reveal spatial trends in reaction norm: upland rice case study. Euphytica 216, 37 (2020). https://doi.org/10.1007/s10681-020-2573-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-020-2573-4