Abstract
Heading date and plant height are important determinants for plant growth rate. In this study, simple sequence repeat markers were used to tag quantitative trait loci (QTL) using a recombinant inbred line mapping population derived from two important breeding parents, genetic stock Kaybonnetlpa1-1 and indica cultivar Zhe733, using data collected under field and greenhouse conditions. Interval mapping, composite interval mapping, and multiple interval mapping were performed to map QTL for heading date and plant height, and to identify epistatic interactions between the QTL. qHD3.1 on chromosome 3 from KBNTlpa1-1 had the largest effect on heading date contributing an average of 28.4 % of the total phenotypic variation. qHD7.1, 7.2, and 8.1 also had a significant contribution to heading date from Zhe733 averaging 8.1, 12.8, and 12.8 % of the phenotypic variance, respectively, and there was a positive additive-by-additive epistatic interaction between qHD7.1 and qHD8.1. QTL, qPHT1.1 and qPHT3.1, for plant height were detected on chromosomes 1 and 3, respectively. qPHT1.1 contributed the largest effect representing 38.2 % of the total phenotypic variation. Comparison of the QTL identified in our study with previous results revealed that the chromosomal locations for QTL coincided closely with positions reported previously in other rice populations worldwide, suggesting that these QTL have coevolved and become domesticated. The tightly linked SSR markers that flank these QTL should be desirable for tagging heading date and plant height genes and facilitating their incorporation into advanced breeding lines using marker assisted selection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heading date and plant height are two of the most important traits associated with yield potential in rice and they are controlled by quantitative trait loci (QTL). Appropriate heading date and plant height are therefore prerequisites for developing high-yielding varieties. Operationally, traditional selection based on phenotype for heading date and plant height relies on field plot experiments during crop growing seasons. In breeding programs, this is often problematic when natural and human resources are limited. With the advent of molecular marker techniques as well as the availability of saturated molecular maps and rice genome sequence information, it is now possible to identify and localize genes controlling complex traits for heading date and plant height.
For heading date, studies have been reported in various rice populations (Li et al. 1995; Yano et al. 1997; Takahashi et al. 2001; Lin et al. 2002; Monna et al. 2002; Lin et al. 2011). Two QTL for heading date, Qhd3a on chromosome 3 and Qhd8a on chromosome 8, were found in the population of Lemont/Teqing (Li et al. 1995) and Ghd8 was recently cloned (Yan et al. 2011). Fifteen QTL conditioning heading date, Hd1–Hd3a and Hd3b–Hd14, have been reported from a cross between a japonica variety Nipponbare and an indica variety Kasalath (Yano et al. 1997; Lin et al. 1998; Takahashi et al. 2001; Lin et al. 2002; Monna et al. 2002) and mapped on chromosomes 2, 3, 4, 6, 7, 8, 10, and 12. Of these Hd1, Hd3a, and Hd6 have been successfully cloned in rice (Yano et al. 2000; Takahashi et al. 2001; Kojima et al. 2002). In addition, two other heading date QTL, Ehd1 and Ghd7, have been cloned (Doi et al. 2004; Xue et al. 2008). It is also well known that several of these QTL interact epistatically including Hd2 with Ghd7 and Hd5 with Hd1 (Lin et al. 2003; Shibaya et al. 2011). In addition, it has been shown that Ghd7 affects expression of Ehd1 and Hd3a (Xue et al. 2008).
A number of studies have been performed to determine QTL for plant height in rice. The semi-dwarf gene sd-1 was first characterized, and subsequently cloned (Hedden 2003). In addition, several studies have reported QTL controlling plant height in rice (Li et al. 1995; Huang et al. 1996; Ashikari et al. 1999; Yu et al. 2002; He et al. 2005). Huang et al. (1996) identified 13 QTL for plant height using five rice populations. Ashikari et al. (1999) cloned a gene for the gibberellins-insensitive dwarf mutation in Dwarf 1 (D1) encoding the α-subunit of the GTP-binding protein. More recently, ten QTL for plant height on nine of the 12 chromosomes have been identified by using single segment substitution lines (He et al. 2005). Heading date gene Ghd7 has also been reported to increase plant height when overexpressed under long-day conditions (Xue et al. 2008).
The indica cultivar Zhe733 (PI 629016) and genetic stock Kaybonnetlpa1-1 (KBNTlpa1-1, PI 632282) have been important resources for rice breeding for improved yield, nutrient quality, and disease resistance in the USA and worldwide (Rutger and Tai 2005). Zhe733 and KBNTlpa1-1 are known to possess distinct phenotypic differences for plant height and heading dates; therefore, they are ideal for introducing different QTL. It is important to note that Zhe733 is a high yielding and early maturing semi-dwarf cultivar that can be grown in the southern USA, and KBNTlpa1-1 is a mutant of Kaybonnet that has lower phytic acid content than its parent resulting in increased mineral nutritional value. A mapping population (K/Z) of the cross of KBNTlpa1-1 and Zhe733with a linkage map was previously constructed and used for mapping QTL for resistance to rice sheath blight and blast disease (Liu et al. 2008; Lee et al. 2009; Jia and Liu 2011). However, QTL for heading date and plant height present in K/Z population and their potential importance in US rice breeding have not been determined. Thus, the objective of this study was to identify QTL for heading date and plant height in KBNTlpa1-1 and Zhe733 using the K/Z mapping population grown in the rice fields and in a greenhouse.
Materials and methods
Plant materials, experimental design, and phenotyping
The indica semi-dwarf cultivar Zhe733 and the genetic stock KBNTlpa1-1 were used as the parents in this study. KBNTlpa1-1 is the first low-phytate mutant induced in the rice cultivar Kaybonnet by γ-irradiation (Bryant et al. 2005). A total of 255 F10-11 recombinant inbred lines (RILs) were used for determining the chromosomal locations of QTL for heading date and plant height. The F10–11 RILs together with two parents were transplanted into the field at the University of Arkansas Rice Research and Extension Center, Stuttgart, AR, USA in the 2002, 2003, and 2004 rice-growing seasons. Each RIL was randomly assigned to a plot of 0.46 m2. Data were collected from the field plots for days to heading (days from planting to 50 % heading) and plant height (distance, cm, from soil surface to panicle tip before harvesting). The average of the data for heading date and plant height from each year was used for QTL mapping. In order to detect QTL under highly controlled environmental conditions the same population was grown in a greenhouse in 2011. The greenhouse experiment was designed as random complete block with three replications, one set of replicates per bench. Each pot was filled with sterilized local clay soil and 3–5 seeds per line were sown. One plant per pot was maintained until heading. Data collected from sowing date, heading date, and plant height for each pot and each line; the average from the three replications was used for QTL analysis.
Simple sequence repeat (SSR) analysis
One hundred and eighteen SSR markers in 255 F10–11 RILs were analyzed for mapping QTL for heading date and plant height in the K/Z population. PCRs were performed as previously described by Liu et al. (2008), except the reaction volume was changed to 25 μL. Amplified products were diluted between 40 and 2000×, and 2 μl of the diluted product was added to 9 μL of formamide containing ROX-labeled size standard. PCR products from three primer pairs with different size ranges and fluorophore labels were combined to determine the sizes of SSR alleles. The reaction was run on an ABI Prism 3700 DNA Analyzer according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA, USA). Fragment sizing and SSR marker genotype analysis were performed using GeneScan® and Geneotyper® software Version 3.7 (Applied Biosystems, Foster City, CA, USA).
Construction of linkage map and QTL analysis
A linkage map of 118 polymorphic SSR markers was constructed using JoinMap® 4 (Van Ooijen 2006). A genetic map with cumulative cM distances was generated using Kosambi’s mapping function from 255 RILs of ‘Zhe733’ × ‘KBNTlpa1-1’. QTL analysis was performed using the phenotypic and genotypic data of heading date and plant height from 255 F10–11 RILs. Interval mapping, composite interval mapping (CIM), and multiple interval mapping (MIM) were performed to identify SSR markers associated with heading date and plant height using the software Windows QTL Cartographer 2.5 (Wang et al. 2007). The value of additive genetic effect and phenotypic variation explained by each putative QTL were obtained from the program concurrently. QTL positions on chromosomes were determined using a logarithm of odds (LOD) threshold of 3.2–3.5 for plant height and heading date for field data and 2.9 for heading date and 3.0 for plant height for greenhouse data, as determined by performing 1,000 permutations, for an experiment-wise significance level of 0.05 using CIM. QTL were named following the gene nomenclature system for rice (McCouch 2008).
Results
Phenotypic variation for heading date and plant height
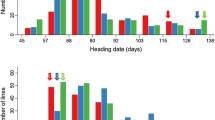
The overall days to heading for the F10–11 RIL population from KBNTlpa1-1 and Zhe733 was 66–110 days (Fig. 1). The difference of heading date between parents was approximately 20 days with KBNTlpa1-1 having a heading date of 95 ± 5.7 days and Zhe733 having a heading date of 75 ± 5.7 days. The plant height of KBNTlpa1-1 (104 cm) and Zhe733 (90 cm) differed by approximately 14 cm while the height of F10–11 RIL individuals varied from 78 to 170 cm (Fig. 1). A number of RILs with increased plant height phenotype was observed in the F10–11 population. In the RIL population, heading date and plant height showed a two-peak distribution with a large transgressive segregation over the two parents (Fig. 1), indicating quantitative inheritance.
Adjusted means frequency distributions for heading date (a) and plant height (b) of 255 F10–11 RIL population from the cross “KBNTlpa1-1 and Zhe733”. Plant height was assessed for 2 years (2002 and 2003) and heading date was assessed for 3 years (2002, 2003, and 2004) in the field plot located at Stuttgart, Arkansas. Arrows indicate the adjusted mean values of the parents, Zhe733 and KBNTlpa1-1
QTL for heading date
Using 118 SSR markers in the K/Z population, CIM and MIM detected six QTL, qHD3.1, qHD7.1, qHD7.2, qHD8.1, qHD8.2, and qHD10.1, for heading date (LOD of 2.9–32.8) from field data collected in 2002, 2003, and 2004 and greenhouse data collected in 2011 (Fig. 2). Only QTL identified in three years data for heading date and two years data for plant height were considered valid. The first QTL, qHD3.1's peak location was 3.8 cM on chromosome 3 between RM132 and RM231 and provided the largest phenotypic effect and contributed an average of 28.4 % of total variation for heading date. Nine known QTL were located near qHD3.1 (Fig. 2). Two other QTL, qHD7.2 and qHD8.1, on chromosomes 7 and 8, respectively, provided the second largest effects both contributing an average of 12.8 % of the phenotypic variation. qHD8.2 on chromosome 8 and qHD10.1 on chromosome 10 contributed the least phenotypic variation with 5.4 and 7.4 %, respectively (Table 1). The estimated additive genetic effects resulted in an average decrease of heading date by 1.7–4.2 days (Table 1). The earliness allele at qHD3.1 was contributed by KBNTlpa1-1 and alleles at qHD7.1, qHD7.2, qHD8.1, and qHD8.2 were contributed by Zhe733 for the earliness of heading date. qHD10.1 had different effects in the field and greenhouse with the KBNTlpa1-1 allele conferring earliness in the greenhouse and the Zhe733 allele conferring earliness in the field. The location of qHD10.1 was slightly different between the greenhouse and field data, possibly indicating that these are two different QTL. A positive additive-by-additive epistatic interaction was identified between qHD7.1 and qHD8.1, indicating that the Zhe733 alleles at both loci together will reduce heading date by an additional 2 days bringing a total of 6 day reduction.
Molecular genetic maps of QTL controlling plant height and heading date in 255 F10–11 RIL population from the cross “KBNTlpa1-1 and Zhe733”. Six QTL for heading dates and two QTL for plant height were detected in this study. Previously identified QTL from different mapping populations are listed next to each QTL. The significance threshold of the logarithm of odds (LOD) was between 2.9 and 3.5 for the detection of putative QTL based on the threshold established by 1,000 permutations
The chromosomal positions of qHD3.1 and qHD7.1 were highly similar between field data and greenhouse data. Three QTL, qHD7.2, qHD8.1, and qHD8.2, were only detected in the field experiment. qHD10.1’s position was slightly different between the field and greenhouse. Similar additive effect was observed for qHD3.1 and qHD7.1; however, opposite additive effect was observed for qHD10.1 (Tables 1 and 2).
QTL for plant height
Two QTL, qPHT1.1 and qPHT3.1, for plant height were identified on chromosomes 1 and 3 (Fig. 2), respectively. qPHT1.1 on chromosome 1 between flanking SSR markers, RM315 and RM431, had the largest effect contributing about 38.2 % of total phenotypic variation for plant height (LOD = 34.9) with the Zhe733 allele, conferring a reduction of 11.8 cm in height. Fourteen known QTL, including semi-dwarf gene sd-1, were located near qPHT1.1 (Fig. 2). qPHT3.1 explained 7.2 % of phenotypic variation (LOD = 5.9) and caused a reduction of 7.8 cm in plant height. Five known QTL were located near qPHT3.1 (Fig. 2). The height-reducing allele from qPHT1.1 was contributed by parent Zhe733 whereas qPHT3.1 was contributed by KBNTlpa1-1 (Table 1). No epistasis was observed between these two QTL.
All plant height QTL identified in the field were also identified in our greenhouse experiment (Table 2). The chromosomal positions of qPHT1.1 and qPHT3.1 were highly similar between each year of field and greenhouse data. Similar additive effects were observed for qPHT1.1 and qPHT3.1.
Discussion
Both heading date and plant height are major determinants of the growth rate of rice and are known to be influenced by the environment. The present study identified six heading date QTL and two plant height QTL in KBNTlpa1-1 and Zhe733 using phenotypical data from controlled (greenhouse) and uncontrolled (multiyear field) conditions. Most of the QTL identified in the field study were also identified in a replicated greenhouse study. These QTL correlate with previously reported heading date and plant height QTL in rice (Table 3; Fig. 2). It has been reported that most QTL for domestication related traits are clustered in chromosomal blocks and the position of these clusters are consistent with those reported in different populations (Lee et al. 2005). Therefore, it is difficult to conclude whether the QTL identified in our study are the same QTL as those previously identified in other studies. Furthermore, to our knowledge, this is the first study to compare QTL of plant height and heading date for rice under both greenhouse and field conditions. This advancement is important because it may encourage researchers worldwide to identify useful QTL involved in plant height and heading date under controlled environmental conditions.
Many of the QTL reported by Yano et al. (1997) were located in regions similar to the heading date QTL identified in this study (Table 3; Fig. 2). The most effective heading date QTL, qHD3.1, accounted for 21 % of total phenotypic variation and was found near Hd9 on chromosome 3 (Lin et al. 2002). As shown in Table 3, the map location of two other QTL, qHD7.1 and qHD7.2, are similar to the location of Hd2 and Hd4 on chromosome 7 (Lin et al. 2000; Yano et al. 1997). qHD8.1 was also detected near Hd5 on chromosome 8 (Lin et al. 2002; Yano et al. 1997). qHD8.2 was located near qDTH-8 previously observed in an Oryza glaberrima by Oryza sativa cross in which the favorable allele came from O. glaberrima (Suh et al. 2005). Our results indicate that O. sativa alleles can possibly be used to reduce heading; however the magnitude of effect is not as high as that of observed in O. glaberrima. qHD10.1 was located near Ehd-1 (Doi et al. 2004). Additionally, the conserved chromosomal locations on heading date QTL may suggest that the QTL may have undergone human selection to diversify the heading date of rice during crop domestication or the early stage of cultivation. Understanding the molecular basis involved in this epistasis would be a foundational advancement in regulation of plant growth rate. Additive-by-additive epistatic interaction was observed between qHD7.1 and qHD8.1. The presence of both Zhe733 alleles can reduce heading date by 6 days. An epistatic interaction in this region has been observed in a cross of a weedy rice accession with a domesticated pure line of O. sativa (Gu and Foley 2007). This is the first time such an interaction was identified in a cross involving two domesticated O. sativa varieties. Molecular basis of epistatic interaction of these two QTL needs to be elucidated in the future. The development of target amplification polymorphism (TRAP) marker (Hu and Vick 2003) at qHD7.1 and qHD8.1 should help to delimit the genomic regions and shed more light on the molecular basis of epistatic interactions.
The sum of the additive effects of the plant height QTL identified in our study was similar to the differences in plant height between KBNTlpa1-1 (japonica) and Zhe733 (indica). However, as shown in Fig. 1, a large phenotypic variation of RIL individuals was observed in the frequency distributions of plant height. A number of RILs were much taller (>30 cm) than KBNTlpa1-1 (104 cm), indicating the complexity of genetic regulation of plant height. Even though two plant height QTL were identified between KBNTlpa1-1 and Zhe733, a single QTL locus may contain multiple alleles for plant height. In rice, it has been reported that dozens of genes play a role in regulating plant height (Sakamoto and Matsuoka 2004). Any possible combination of genes associated with plant height can directly affect metabolic pathways for plant development. Phenotypic gain without genetic contribution may suggest that transgressive variation observed for plant height may also be due to epigenetic regulation. Indeed, several nucleotide differences nearby height controlling gene, OsBAK1, were predicted to alter the gene’s functions (Li et al. 2009). Continued examination of these changes that affect the binding of transcription factors resulting in altered gene expression should provide a better understanding of epigenetic regulation.
The two subspecies of cultivated rice, japonica and indica, have different evolutionary histories and their potentially independent domestication has resulted in distinctly different genomes (He et al. 2011). Pleiotropic effects on both heading date and plant height have been reported in the population of Lemont and Teqing (Li et al. 1995). All QTL for heading date in Lemont and Teqing populations were mapped to similar locations as plant height QTL. In the present study, heading date and plant height did not correlate with each other. Indeed changes in biochemical and physiological states can affect and promote gene expression profiles that are associated with plant height. Whether or not there are any pleiotropic effects on these two traits is difficult to predict, it is known that genes controlling heading date are associated with the transition of rice from the vegetative phase to the reproductive phase (Yan et al. 2011). To determine any pleiotropic roles in regulating multiple traits in rice, the gene needs to be cloned and functionally characterized.
qPHT1.1 and qPHT3.1 are located in the same genomic region as other known QTL reported in previous studies (Table 3). Two other knownQTL, Ph3 and ph3-2, are located close to qPHT3.1 (Yan et al. 1998). The semi-dwarf gene, sd-1, of rice was found on chromosome 1 at the location of 38.7 Mb (Monna et al. 2002; Spielmeyer et al. 2002). One of the plant height QTL, qPHT1.1, lies near this location. The SSR marker, RM431, closely linked to qPHT1.1, was found on chromosome 1 at 39.2 Mb and this marker is 0.5 Mb away from sd-1. Zhe733 is semi-dwarf and the majority of semi-dwarf varieties contain sd-1 (Spielmeyer et al. 2002). Zhe733's pedigree has IR24, which contains sd-1 (Khush and Gomez 1985; Monna et al. 2002), indicating that sd-1 may be one of the factors controlling height variations in Zhe733.
Marker assisted selection (MAS) is a powerful tool to accelerate conventional rice breeding. Despite abundant DNA markers for heading date, flowering time, and plant height reported in rice (Hittalmani et al. 2003; Yu et al. 2002; Septiningsih et al. 2003; Thomson et al. 2003; Yan et al. 1998; Yano et al. 2001; Moncada et al. 2001; You et al. 2006), few breeding programs heavily depended on the use of MAS for selection for traits other than disease resistance (Bertrand et al. 2008). Possible reasons to explain the low impact of MAS in rice improvement are (i) QTL effects vary depending on genetic background or are influenced by environmental conditions, (ii) accuracy of QTL mapping studies, and (iii) poor integration of molecular genetics and conventional breeding (Bertrand et al. 2008). Among these reasons, reliability of QTL mapping is the most critical step to the success of MAS.
For accurate QTL mapping, reliable phenotypic data using multiple replications, environments, and confirmation of QTL results in independent populations are critical. QTL governing heading date and plant height identified in the present study are also conserved across many different mapping populations used by other research groups. The major QTL of these two traits can now be detected under controlled greenhouse conditions suggesting that major QTL have consistent effects across environmental conditions. This finding suggests that the major QTL may have been strongly selected during rice domestication. Nevertheless, SSR markers tagging QTL controlling heading date and plant height identified in our study should be an additional valuable molecular tool for breeding for early flowering semi-dwarf rice possibly resulting in substantial yield improvements.
Abbreviations
- CIM:
-
Composite interval mapping
- IM:
-
Interval mapping
- KBNTlpa1-1:
-
Kaybonnetlpa1-1
- LOD:
-
Logarithm of odds
- MAS:
-
Marker assisted selection
- MIM:
-
Multiple interval mapping
- QTL:
-
Quantitative trait loci
- RIL:
-
Recombinant inbred line
- SSR:
-
Simple sequence repeat
References
Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A (1999) Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the a-subunit of GTP-binding protein. Proc Natl Acad Sci USA 96:10284–10289
Bertrand C, Collard Y, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Phil Trans R Soc B 363:557–572
Bryant RJ, Dorsch JA, Peterson KL, Rutger JN, Raboy V (2005) Phosphorus and mineral concentration in whole grain and milled low phytic acid (lpa) 1-1 rice. Cereal Chem 82:517–522
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes & Dev 18:926-936
Gu X-Y, Foley ME (2007) Epistatic interactions of three loci regulate flowering time under short and long daylengths in a backcross population of rice. Theor Appl Genet 114:745–754
He P, Li J, Zheng X, Shen L, Lu C, Chen Y, Zhu L (2001) Comparison of molecular linkage maps and agronomic trait loci between DH and RIL populations derived from the same rice cross. Crop Sci 41:1240–1246
He F, Xi Z, Zeng R, Akshay T, Zhang G (2005) Identification of QTL for plant height and its components by using single segment substitution lines in rice (Oryza sativa). Rice Sci 12:151–156
He Z, Zhai W, Wen H, Tang T, Wang Y, Lu X, Greenberg AJ, Hudson RR, Wu C, Shi S (2011) Two evolutionary histories in the genome of rice: the roles of domestication genes. PLoS Genet 7(6):e1002100. doi:10.1371/journal.pgen.1002100
Hedden P (2003) The genes of the green revolution. Trends Genet 19:5–9
Hemamalini GS, Shashidhar HE, Hittalmani S (2000) Molecular marker assisted tagging of morphological and physiological traits under two contrasting moisture regimes at peak vegetative stage in rice (Oryza sativa L.). Euphytica 112:69–78
Hittalmani S, Huang N, Courtois B, Venuprasad R, Shashidhar HE, Zhuang J-Y, Zheng K-L, Liu G-F, Wang G-C, Sidhu JS, Srivantaneeyakul S, Singh VP, Bagali PG, Prasanna HC, McLaren G, Khush GS (2003) Identification of QTL for growth- and grain yield-related traits in rice across nine locations of Asia. Theor Appl Genet 107:679–690
Hu J, Vick BA (2003) Target region amplification polymorphism: a novel marker technique for plant genotyping. Plant Mol Biol Rep 21:289–294
Huang N, Courtois B, Khush GS, Lin HX, Wang GL, Wu P, Zheng KL (1996) Association of quantitative trait loci for plant height with major dwarfing genes in rice. Heredity 77:130–137
Jia Y, Liu G (2011) Mapping quantitative trait loci for resistance to rice blast. Phytopathology 101:176–187
Khush GS, Gomez KA (1985) Parentage of IRRI crosses IR1-IR50000. The International Rice Research Institute, Manila
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabadopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43:1096–1105
Lee S-J, Oh CS, Suh JP, McCouch SR, Ahn SN (2005) Identification of QTL for domestication-related and agronomic traits in an Oryza sativa x O. rufipogon BC1F7 population. Plant Breed 124:209–219
Lee S, Wamishe Y, Jia Y, Liu G, Jia MH (2009) Identification of two major resistance genes against race IE-1 k of Magnaporthe oryzae in the indica rice cultivar Zhe733. Mol Breed 24:127–134
Li Z, Pinson SRM, Stansel JW, Park WD (1995) Identification of quantitative trait loci (QTL) for heading date and plant height in cultivated rice (Oryza sativa L.). Theor Appl Genet 91:374–381
Li ZK, Yu SB, Lafitte HR, Huang N, Courtois B, Hittalmani S, Vijayakumar CH, Liu GF, Wang GC, Shashidhar HE, Zhuang JY, Zheng KL, Singh VP, Sidhu JS, Srivantaneeyakul S, Khush GS (2003) QTL x environment interactions in rice. I. Heading date and plant height. Theor Appl Genet 108:141–153
Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K (2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7:791–806
Lin SY, Sasaki T, Yano M (1998) Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor Appl Genet 96:997–1003
Lin H, Yamamoto T, Sasaki T, Yano M (2000) Characterization and detection of epistatic interactions of 3 QTL, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines. Theor Appl Genet 101:1021–1028
Lin H, Ashikari M, Yamanouchi U, Sasaki T, Yano M (2002) Identification and characterization of a quantitative trait locus, Hd9, controlling heading date in rice. Breed Sci 52:35–41
Lin H, Liang ZW, Sasaki T, Yano M (2003) Fine mapping and characterization of quantitative trait loci Hd4 and Hd5 controlling heading date in rice. Breed Sci 53:51–59
Lin Y-R, Wu S-C, Chen S-E, Tseng T-H, Chen C-S, Kuo S-C, Wu H-P, Hsing Y-IC (2011) Mapping of quantitative trait loci for plant height and heading date in two inter-subspecific crosses of rice and comparison across Oryza genus. Botanical Studies 52:1–14
Liu G, Bernhardt JL, Jia MH, Wamishe YA, Jia Y (2008) Molecular characterization of the recombinant inbred line population derived from a japonica-indica rice cross. Euphytica 159:73–82
Lu C, Shen L, Tan Z, Xu Y, He P, Chen Y, Zhu L (1997) Comparative mapping of QTL for agronomic traits of rice across environments using a doubled-haploid population. Theor Appl Genet 94:145–150
McCouch SR (2008) Gene nomenclature system for rice. Rice 1:72–84
Mei HW, Luo LJ, Ying CS, Wang YP, Yu XQ, Guo LB, Paterson AH, Li ZK (2003) Gene actions of QTL affecting several agronomic traits resolved in a recombinant inbred rice population and two testcross populations. Theor Appl Genet 107:89–101
Moncada P, Martinez CP, Borrero J, Chatel M, Gauch H, Guimaraes E, Tohme J, McCouch SR (2001) Quantitative trait loci for yield and yield components in an Oryza sativa X Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet 102:41–42
Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y (2002) Positional cloning of rice semi-dwarfing gene, sd-1: rice “Green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res 9:11–17
Rutger JN, Tai T (2005) Registration of K/Z mapping population of rice. Crop Sci 45:2671–2672
Sakamoto T, Matsuoka M (2004) Generating high-yielding varieties by genetic manipulation of plant architecture. Curr Opin Biotechnol 15:144–147
Septiningsih EM, Prasetiyono J, Lubis E, Tai TH, Tjubaryat T, Moeljopawiro S, McCouch SR (2003) Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor Appl Genet 107:1419–1432
Shibaya T, Nonoue Y, Ono N, Yamanouchi U, Hori K, Yano M (2011) Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions. Theor Appl Genet 123:1133–1143
Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20 oxidase gene. Proc Natl Acad Sci USA 99:9043–9048
Suh J-P, Ahn S-N, Cho Y-C, Kang K-H, Choi I-S, Kim Y-G, Suh H-S, Hong H-C (2005) Mapping of QTL for yield traits using an advanced backcross population from a cross between Oryza sativa and O. glaberrima. Korean J Breed 37:214–220
Takahashi Y, Shomura A, Sasaki T, Yano M (2001) Hd6, a rice quantitative locus involved in photoperiod sensitivity, encodes the ß subunit of protein kinase CK2. Proc Natl Acad Sci USA 98:7922–7927
Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME, Lobos KB, Xu Y, Martinez CP, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493
Van Ooijen JW (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart. Accessed 22 July 2011
Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40:761–767
Yan J, Zhu J, He C, Benmoussa M, Wu P (1998) Molecular dissection of developmental behavior of plant height in rice (Oryza sativa L.). Genetics 150:1257–1265
Yan WH, Wang P, Chen H-X, Zhou H-J, Li Q-P, Wang C-R, Ding Z-H, Zhang Y-S, Yu S-B, Xing Y-Z, Zhang Q-F (2011) A major QTL, Ghd8, plays pleotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant 4:319–330
Yano M, HarushimaY Nagamura Y, Kurata N, Minobe Y, Sasaki T (1997) Identification of quantitative trait loci controlling heading date of rice using a high-density linkage map. Theor Appl Genet 95:1025–1032
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2483
Yano M, Kojima S, Takahashi Y, Lin HX, Sasaki T (2001) Genetic control of flowering time in rice, a short-day plant. Plant Physiol 127:1425–1429
You A, Lu X, Jin H, Ren X, Liu K, Yang G, Yang H, Zhu L, He G (2006) Identification of quantitative trait loci across recombinant inbred lines and testcross populations for traits of agronomic importance in rice. Genetics 172:1287–1300
Yu SB, Li JX, Xu CG, Tan YF, Li XH, Zhang Q (2002) Identification of quantitative trait loci and epistatic interactions for plant height and heading date in rice. Theor Appl Genet 104:619–625
Acknowledgments
The authors thank the Arkansas Rice Research and Promotion Board for financial support; Michael Lin, Tracy Bianco, Ellen McWhirter, Alan Sites, Tony Beaty, and Dr. Joseph Kepiro for excellent technical support; and Lorie Bernhardt and Dr. J. Neil Rutger of Genetic Stock-Oryza (GSOR) collection of Dale Bumpers National Rice Research Center for providing the RIL population of the cross of KBNTlpa1-1 and Zhe733. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Additional information
Seonghee Lee and Melisa H. Jia contributed equally to this report.
Rights and permissions
About this article
Cite this article
Lee, S., Jia, M.H., Jia, Y. et al. Tagging quantitative trait loci for heading date and plant height in important breeding parents of rice (Oryza sativa). Euphytica 197, 191–200 (2014). https://doi.org/10.1007/s10681-013-1051-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-013-1051-7