Abstract.
Rice double-haploid (DH) lines of an indica and japonica cross were grown at nine different locations across four countries in Asia. Genotype-by-environment (G × E) interaction analysis for 11 growth- and grain yield-related traits in nine locations was estimated by AMMI analysis. Maximum G × E interaction was exhibited for fertility percentage number of spikelets and grain yield. Plant height was least affected by environment, and the AMMI model explained a total of 76.2% of the interaction effect. Mean environment was computed by averaging the nine environments and subsequently analyzed with other environments to map quantitative trait loci (QTL). QTL controlling the 11 traits were detected by interval analysis using mapmaker/qtl. A threshold LOD of ≥3.20 was used to identify significant QTL. A total of 126 QTL were identified for the 11 traits across nine locations. Thirty-four QTL common in more than one environment were identified on ten chromosomes. A maximum of 44 QTL were detected for panicle length, and the maximum number of common QTL were detected for days to heading detected. A single locus for plant height (RZ730-RG810) had QTL common in all ten environments, confirming AMMI results that QTL for plant height were affected the least by environment, indicating the stability of the trait. Two QTL were detected for grain yield and 19 for thousand-grain weight in all DH lines. The number of QTL per trait per location ranged from zero to four. Clustering of the QTL for different traits at the same marker intervals was observed for plant height, panicle number, panicle length and spikelet number suggesting that pleiotropism and or tight linkage of different traits could be the possible reason for the congruence of several QTL. The many QTL detected by the same marker interval across environments indicate that QTL for most traits are stable and not essentially affected by environmental factors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The phenotype of an individual is affected both by genotype (G) and environment (E). Most agronomically significant characters are inherited quantitatively and are known to be affected by environmental factors. Selection based on the phenotype would be difficult for such difficult traits. In breeding programs, it is often difficult to manipulate such traits, since several inter-componential characters indirectly control them. With the advent of molecular marker techniques as well as the availability of saturated DNA marker maps it is now possible to identify and locate loci (genes) controlling complex traits like grain yield and its contributing traits. The availability of saturated molecular map (Causse et al. 1994; Kurata et al. 1994) has made it possible to elucidate the inheritance pattern of both Mendelian and quantitative trait loci (QTL). While the mapping of QTL traits has been reported by several workers, there are not many reports on the identification of the QTL in one mapping population across several environments. Most of the investigations have identified QTL either in two or three environments (Paterson et al. 1991; Stuber et al. 1992; Hayes et al. 1993; Zhuang et al. 1997; Shailaja Hittalmani et al. 2002) or used more than one population in same location (Lin et al. 1995) or one population in single environment (Wang et al. 1994; Champoux et al. 1995; Courtois et al. 1995; Li et al. 1995; Xiao et al. 1996; Hemamalini et al. 2000).
The present study was conducted with the doubled haploid (DH) lines of a IR64/Azucena rice cross in nine environments in Asia and in the 'Mean environment' computed. QTL controlling 11 growth and yield traits in rice were identified together with common QTL across different environments. G × E interaction of traits was also observed. The study was mainly aimed to detect QTL that are stable across environments and find out their phenotypic contribution to trait for the use in selecting the right type of plant material and help the breeding program to be more focused. Identifying QTL for traits that are stably expressed across diverse environments could also help in the possibility of using closely associated markers in marker-assisted-selection (MAS) for quantitative traits. Use of markers to select for quantitative traits is not common because of the skepticism that most QTL are not stable due to the environmental influence.
Materials and methods
Plant material
One hundred and twenty-five rice (Oryza sativa L.) lines of a DH-population developed from a cross between IR64, an indica variety adapted to an irrigated condition, and Azucena, an upland traditional japonica variety (Guiderdoni et al. 1992) developed in France and maintained at IRRI, Philippines, were used for the experiment. IR64 is a semidwarf, heavy-tillering, high-yielding and widely grown variety in rice-growing regions of Asia, while Azucena is a tall, sparse-tillering, low-yielding and long-grained aromatic variety from the Philippines.
Experimental locations
The experiment was carried out in nine different environments in four rice-growing countries in South and South-East Asia differing in a wide range of agro-climatic conditions (Tables 1, 2). The latitude varied from 12°NS in UAS, India to 30°NS in CNRRI in China. The longitude varied between 75°W and 122°E.
Details of the field experiments
The DH population along with the parental lines was raised in a randomized complete block design (RCBD) at each location. The experimental conditions prevalent at the different environments, including number of replications, spacing, number of DH lines used, are indicated in Table 2. Twenty-five-day-old seedlings were transplanted to the main field under irrigated conditions. The selected observations were recorded at the appropriate crop growth stage during pre- and post-harvest stages. The various phenotypic observations were recorded on ten randomly selected plants per genotype and parents as per the guidelines of the Standard Evaluation System (IRRI 1988). The average of these ten plants was computed and used for analysis. The average of nine locations computed constituted the 'Mean environment' (tenth location). The traits measured for detecting QTL and their abbreviations used in the paper are indicated in Table 3.
Data were not available for the traits BMS and HID at IRRI94, and PEN at CNNRI. Data on parents was not available at CNNRI on NOS, at IRRI95 on HDD and HID, at RRI on HDD, and at SUL on BMS, FRP, HID, NOS, PEN, and TGW.
AMMI analysis for estimating G × E interaction
The 11 selected traits in nine locations were subjected to AMMI analysis (Gauch 1992) to estimate G × E interaction and the extent of interaction explained by the extraneous factors. This explains the G × E through multiplicative terms and can also improve the precision of the estimation.
QTL mapping
The chromosome map of IR64 × Azucena developed earlier (Huang et al. 1994) using 135 lines consisting of 175 polymorphic markers including 146 restriction fragment length polymorphisms, (RFLP) three isozymes, 14 randomly amplified polymorphic DNA (RAPDs) and 12 cloned genes was used. To this map 85 new markers comprising of 76 Simple sequence repeats (SSRs) (Ricegenes 2000) and two RAPDs were added. The overall map length of this population is approximately 1,822 cM (Temnykh et al. 2000). Distribution of IR64 and Azucena alleles for each marker was roughly symmetrical around 0.5, suggesting no overall bias toward either parent.
QTL mapping was carried out by interval analysis with mapmaker/qtl (Lander and Botstein 1989; Lincoln et al. 1993). The threshold LOD of 2.00 was used, considering the number of locations, to reduce type-I error and identify suggestive QTL for detecting QTL that are common across locations, as the map is not a highly saturated one. However, only those QTL with LOD above 3.2 were treated as significant. This ensures a genome-wide significance of P > 0.05 and chromosome-wide significance of P > 0.004 (Van Ooijen 1999). Chromosome-wise significance at P > 0.01 corresponds to an LOD of 2.8. This relaxed LOD was used to infer suggestive linkage if the QTL also appeared in another environment. QTL for various traits in the ten environments – nine locations and 'Mean environment' – were detected and mapped on the rice chromosomes.
Results
Trait performances
The performance of the DH lines and their parents IR64 and Azucena in ten environments for 11 traits is tabulated in Table 4. An approximate normal distribution was observed for phenotypic performance of the traits in all environments. A wide variation in the performance of the DH lines for all traits except HDD was observed in all environments. However, the performance of the parents varied considerably for BMS, HID, NOP, PEN and PHT, while the magnitude of variation was less for HDD, PLT and TGW. Parents differed in performance for traits BMS and YLD at a few locations but were largely similar in the remaining locations. Transgressive segregants in either direction were found for all of the traits measured. Maximum variation was observed for PHT, PEN, NOS and FRP, while PLT, HDD and HID had the least.
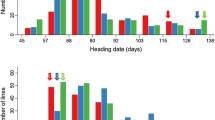
The mean performance of traits indicates the effect of environment on phenotype. PHT, PLT, HDD, and TGW were least affected by environment and had a fairly uniform environmental index, as reflected by their location mean values. PEN, YLD, BMS and NOP were the most affected by environment. The YLD of the DH lines and parents in the ten environments is depicted in Fig. 1.
G × E using AMMI analysis
Genotypic variability, phenotypic variability and the interaction effects were estimated for the 11 traits (Table 5). The AMMI model dissected the interaction component in three sects and explained the extent of interaction. HDD, PHT and TGW had the least G × E interaction, while FRP, NOS and YLD had higher G × E interaction. Of the total G × E interaction effect, the AMMI model explained 85.63% for HID, 76.19% for PHT and 71.65% for HDD and NOP. This model explained at least 58% of the total interaction effects for the traits observed (Table 5).
QTL detection by interval analysis
The interval analysis detected a total of 292 QTL above an LOD of 2.0, of which 126 were significant (above LOD 3.2) (Table 6), for the 11 traits across the ten environments, with an average of 1.15 per trait per location. The maximum number of QTL were identified for PLT (20) and NOP (20) and a minimum for FRP (1) and YLD (2). Mean environment detected maximum QTL (20); the minimum (2) was at the UAS location in India.
Table 7 shows the QTL for various traits along with marker interval of peak LOD, number of locations in which QTL appeared, additive effect and direction of the QTL, percentage variation explained and the peak LOD. In total, 34 QTL were detected for 11 traits that appeared in at least two environments. The additive effect of a QTL was consistent in its direction when detected in more than one environment, but its magnitude varied (Table 7).
Biomass (BMS)
A total of Six QTL significantly affecting BMS were detected. Three were confined to one environment, while other three were identified in more than one environment albeit at lower LOD. The highest number of QTL (2) were identified in SUL and Mean environment, while none were detected at CNNRI, IRRI95 and PAU. Of the six QTL, four had a negative influence.
Fertility percentage (FRP)
One significant QTL for grain fertility at CNNRI was detected. This QTL also appeared at location RRI although at a lower LOD of 2.24.
Heading date (HDD)
A total of ten significant QTL with five distinct QTL were detected, of which two were detected in only one environment and other three appeared in more than one environment. A maximum of two QTL were identified in IARI and Mean environment. None were detected at IRRI95 and SUL. The region between PGMS0.7-RG477 on chromosome 7 showed QTL in seven environments. One suggestive QTL between RM257-RZ228 was also inferred. Interestingly, QTL in the interval RG190-RG908 appeared in four locations with the LOD ranging between 2.07 and 2.50. These two regions had a negative influence on trait.
Harvest index (HID)
A total of 13 significant QTL, eight of which were distinct, were found to affect HID and detected across ten environments, with four QTL being confined to one environment and the others detected in two to four environments. Maximum QTL were detected in Mean environment (3) and the least at IARI and UAS in India. Two QTL, one each on chromosome 1 (RZ801-RG331) and chromosome 3 (RZ574-RZ284) were consistent in four environments and could explain up to 20% variation. However, the former was exerting negative influence and latter positive influence for harvest index. Three suggestive QTL were also inferred on chromosomes 7 and 8.
Number of panicles (NOP)
A total of 20 significant QTL, with six being distinct, were detected for NOP. The number of QTL at any location varied from zero to three. QTL located between markers RZ730 and RG810 on chromosome 1 explained 15.10–27.70% of the total phenotypic variation, and its position was consistent across eight environments. Another QTL was consistently mapped between markers RG163 and RG214 on chromosome 4 across six environments and accounted for 18.0–25.70% of the total phenotypic variation. Another QTL for panicle number on chromosome 12 (RM235-RG181) consistent across five environment explained variation ranging from 15.0% to 19.10%. Interestingly, all QTL were exerting negative influence trait, contributing to a lower number of panicles from the female parent.
Number of spikelets (NOS)
A total of 12 significant QTL with four being distinct were detected. All of the QTL were located on chromosome 4. The number of QTL at a location ranged from zero to three. Of the QTL identified, the one between markers RG163-RG214 was detected in eight environments and accounted for at least 10.60% variation. In 'Mean environment' it could explain 41.10% of the total phenotypic variation. This locus enhanced NOS by 24.97 per Azucena allele in SUL in China. Two suggestive QTL were also inferred on chromosome 1 and 7.
Panicle exsertion (PEN)
A total of eight QTL, three of which were distinct, that significantly affected panicle exertion were located. The highest number of QTL (3) was detected in PAU and none in IRRI95, IARI, SUE and UAS. The QTL located between RZ730 and RZ810 on chromosome 1 was consistent in six environments and explained variation ranging from 12.90% to 23.00% of the total phenotypic variation. Another QTL for PEN consistently mapped between the markers RG190 and RG908 on chromosome 4 across four environments. The third QTL appeared in two environments. The latter two had a negative impact on the trait.
Plant height (PHT)
A total of 15 significant QTL influencing plant height were detected in ten environments, of which three QTL were distinct. The number of QTL detected in an environment varied from one to two. Of these QTL identified, the one between markers RZ730 and RZ810 on chromosome number 1 was detected in all ten environments and explained at least 25.20% of the total phenotypic variation. At IRRI94, this locus could explain 62.20% of the variation in PHT. This is the map position of gene sd-1 (Huang et al. 1994; Cho et al. 1998). This is a good example of major genes also harboring QTL (Beavis et al. 1991). Another QTL between RZ284 and RZ574 on chromosome 3 accounted for more than 11% of the total phenotypic variation in six environments. At this particular locus, each allele of Azucena, the taller parent, could reduce PHT by at least 7 cm. A fourth QTL on chromosome 7 was inferred as it appeared in four locations, although at a lower LOD. Although Azucena is the taller parent, of the four QTL detected, only one locus on chromosome 1 could enhance height.
Panicle length (PLT)
A total of 20 significant QTL, six of which were distinct, were found to influence panicle length across ten environments. The number of QTL found varied from zero to four. Five QTL were common in more than one environment. QTL between RG418A and RG910 on chromosome 3 was identified in nine environments and accounted for 9.90% to 19.70% of the total phenotypic variation. One locus located between markers RZ730 and RZ810 on chromosome 1 was consistent across nine environments and explained at least 15.60% of the total variation with a maximum of 37.10% at IRRI94. Another two QTL, one each on chromosome 3 (RZ574-RZ284) and chromosome 10 (RG257-RG241) were detected in five environments and explained at least 12% of the variation. Two suggestive QTL were inferred on chromosome 6 and 10. Of these QTL, five were acting negatively and other three positively, indicating the effect of male and female parental alleles contribution for manipulation in the phenotype.
1000 grain weight (TGW)
Nineteen QTL significantly affecting TGW were detected with nine of these being distinct. The number of QTL detected at each location ranged from four (CNNRI) to none (UAS, IARI). Of these QTL identified, one between markers RG134 and RZ500 on chromosome 10 was detected in five environments and explained 25.10% of total phenotypic variation in RRI. One QTL on chromosome 1 was found to enhance TGW by at least 1.16 g per Azucena allele and explained more than 12.20% of variation in six environments. Another QTL on chromosome 3 (RZ574-RZ284) caused a reduction in TGW by more than 1 g per Azucena allele and explained a minimum variation of 9.80% in six environments. Three QTL were limited to a single environment. One QTL on chromosome 6 was inferred based on suggestive linkage.
Grain yield (YLD)
A single significant QTL influencing YLD was detected each in SUE and the Mean environment. The QTL located between RG91 and RG449 on chromosome 4 exerted a positive influence, explaining a minimum of 11.40% of the total variation, and its position was consistent across three environments. One QTL on chromosome 10 appeared only at RRI. Another QTL between RZ12 and RM201 on chromosome 9 was inferred, based on suggestive linkage. This was detected in two environments and exhibited a negative influence, contributing to the lower grain yield.
Congregation of QTL
QTL affecting different traits are shown in Fig. 2. Different QTL affecting HDD, HID, NOP, PHT, PEN, PLT and TGW were clustered together between markers RZ730 and RZ810 on chromosome 1 in six, ten, nine, two, one, eight and one of the environments respectively. Similarly, region RZ801-RG331 could explain variance for TGW, BMS and HID in six, two and four environments respectively. Common QTL for both PLT and TGW were detected on chromosome 6 (RG433-Cat-1) in three environments. Chromosomes 2, 5 and 11 were devoid of any QTL. On chromosome 3, the region flanked by markers RZ574 and RZ284 detected common QTL for PHT, PLT, TGW and HID consistently in six, five, six and four environments. Another locus between RZ284 – pRD10A showed common QTL for BMS, HID and PHT in two, one and six environments. On chromosome 4, the locus between RG190 – RG908 explained variance for HDD, BMS, PEN, HID in four, two, four and one of the environment. Similarly region between RG91 – RG449 could influence YLD and HID in three and one of the environments, respectively. The interval RG449 – RG788 showed a QTL for FRP and NOS in two and one of the environments, respectively. The region flanked by RG163 – RG214 possessed QTL for PLT, NOP and NOS in two, six and eight environments respectively. QTL for HID, PLT and PHT were detected on chromosome 7 between markers RG769 and RG511 in two, two and four environments. Similarly, the interval RG477 – PGMS0.7 also explained variance for HDD and BMS in seven and two environments, respectively. The region RG257 – RG241 on chromosome 10 showed common QTL for YLD and PLT in one and five environments, respectively.
Discussion
Grain yield in rice is a complex trait dependent on various growth and component traits. Yield-contributing traits such as NOP, NOS, TGW influence the yield directly and are affected by environment. A number of QTL loci are reported to control the growth and yield components. A number of quantitative traits such as the ones we observed show a high magnitude of G × E interaction. G × E interaction is a challenge to plant breeders and has been shown to reduce the progress of the quantitative traits from selection. G × E interactions are vital in expression of the QTL effect. The present investigation was undertaken to estimate the G × E interaction on the growth traits, grain yield and yield-contributing traits and map them in the DH population of indica × japonica rice cross in nine selected locations across four Asian countries. The trial sites varied significantly with respect to geographical locations and provided varied environmental conditions. The experimental conditions, planting dates and seasons varied in the different countries and also within locations at IRRI, Philippines. In addition to these nine locations, a tenth environment, 'Mean environment', was computed by averaging over all nine locations. This virtual environment reduces the variance due to error and increases the precision of QTL environment (Knapp and Bridges 1990). A few studies in maize have reported this to be an efficient method (Veldboom and Lee 1996a, b; Austin and Lee 1998). In this study, 'Mean environment' could detect 35 out of 55 common QTL and four new QTL.
In a given genotype, genes acting in both positive and negative direction would control expression of any quantitative trait. Plant breeding is directed towards accumulating favorable genes/alleles for trait by exerting selection. Identifying the undesirable/desirable alleles at different loci could efficiently and conveniently do this task. Molecular marker technology is potential enough to provide us such information. In this population, the contribution of negative alleles from Azucena ranged from none (NOS) to seven (TGW). Thus, the performance of any trait was affected by presence of positive or negative alleles at a QTL. This gives us ample scope to exercise selection for desirable alleles at genotype or molecular level.
The G × E interaction analyses by the AMMI statistical model provided a more advanced tool in dissecting the interaction effects and as well explained the extent of interaction effect. The genotypic and G × E effects estimated by AMMI analysis based on phenotypic performance were consistent with the presence of QTL. Traits with a higher genotypic effect were governed by QTL explaining higher variance as observed for PHT, PLT and TGW. Similarly, BMS and YLD having lower genotypic effects had QTL explaining lower variance. For traits with a high environment effect (>57%), like BMS, YLD and HDD, few QTL could be detected. For traits with a low environment influence (<18%), like PHT and PLT, there were more QTL. FRP and YLD having a high G × E interaction had fewer QTL. On the other hand, PHT, HDD, TGW having a low G × E interaction, had more QTL.
Quantitative traits show a range of sensitivities to environment. QTL of such traits could be detected only in one/two environments. In the present study, QTL of traits such as FRP and YLD with a high environment main effect could be detected only in two or three environments. Furthermore, consistent with this observation, those genomic regions of quantitative traits with the least genotype environment interaction and high genotypic main effects would express the same way across different environments. QTL of traits such as PHT and PLT with low G × E interaction and high genotypic main effect were detected across nine environments. PHT and PLT were the only traits having common QTL in almost all environments, strongly indicating the stability of QTL in different ecosystems because of the minimum influence of environmental factors. These are the traits that were found to be stable. NOS, NOP were equally stable, with one QTL for each trait present in eight environments.
Among the various common QTL identified, many explained phenotypic variation larger than 30%. Some of the QTL were located in interval RG163-RG214 on chromosome 4 for NOS, RG348-RM231 on chromosome 3 for HDD and RZ730-RZ810 on chromosome 1 for PHT and PLT. In these cases, one would assume that it would be a major gene rather than a QTL. Consistent with prior observations (Huang et al. 1994), the presence of major gene for PHT would be assumed rather than a QTL in the region RZ730-RZ810 on chromosome 1. This locus affected all length components like PHT, PLT, and PEN. Thus, the gene affecting cell elongation may be the candidate gene at this locus to be dissected.
Phenotypically correlated traits are known to map together (Albert et al. 1991; Paterson et al. 1991; Lebreton et al. 1995; Shashidhar et al. 1999; Shailaja Hittalmani et al. 2002). Molecular marker technology is capable of identifying close relationships and would help in discerning between pleiotrophy and tight linkage or overlapping genes. The magnitude and direction of influence of these loci on the different phenotypes will bear heavily on the utility of such loci in selection for simultaneous improvement of these traits. PHT, PLT, PEN are strongly positively correlated among themselves and with BMS, HDD, TGW. Traits like PHT, PLT, PEN, HDD and TGW were negatively correlated with NOP, HID. This relationship is brought out by congregation of QTL at same locus as could be seen on chromosome 1, 3, 4, 6, and 7.
The congruence of the QTL loci on the chromosome for various traits may be due to either linkage or pleiotropism. This signifies the plural selection efficiency by selecting markers closely associated with these traits. Since the direction of the additive effect of the QTL was also in the same direction, selection if exerted would be very effective.
In the present study using a DH population for detecting QTL across ten environments, QTL were detected to be stable across all environments for PHT and for all other traits except BMS and FRP, in at least three environments, indicating the stability of QTL for some of the traits observed. This is an encouraging result where QTL markers could be fine-mapped and made use of for detecting even the complex traits like grain yield or its contributing traits. Considering the geographic distinctiveness of the locations selected, the commonality of loci detected indicates the broad-based environment independent activity/expression of the gene(s) in question. Loci with less consistent expression can be used for selection at specific locations. QTL, which function consistently over a range of environments, are preferred for plant breeding and are most likely useful candidates for MAS programs with immediate application or after fine mapping for use in wider locations. Location-specific QTL can be used for selection at specific locations or to develop better genotype with different environment specific QTL. The skepticism prevailing regarding the stability of QTL can be set aside for the common QTL identified and plant breeder is now in position to venture into the practice of MAS for the QTL trait.
References
Albert BSB, Edwards MD, Stuber CW (1991) Isoenzymatic identification of quantitative trait loci in crosses of elite maize inbreds. Crop Sci 31:267–274
Austin DF, Lee M (1998) Detection of quantitative trait loci for grain yield and yield components in maize across generations in stress and nonstress environments. Crop Sci 38:1296–1308
Beavis WD, Grant D, Albertsen M, Fincher M (1991) Quantitative trait loci for plant height in four maize populations and their association with qualitative genetic loci. Theor Appl Genet 83:141–145
Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald PC, Harrington SE, Second G, McCouch SR, Tanksley SD (1994) Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138:1251–1274
Champoux MC, Wang G, Sarkarung S, Mackill DJ, O'Toole JC, Huang N, McCouch SR (1995) Locating genes associated with root morphology and drought avoidance in rice via linkage to molecular markers. Theor Appl Genet 90:969–981
Cho YG, McCouch SR, Kuiper M, Kang M, Pot J, Groenen JTM, Eun MY (1998) Integrated map of AFLP, SSLP and RFLP markers using a recombinant inbred population of rice (Oryza sativa L.). Theor Appl Genet 97:370–380
Courtois B, Huang N, Guiderdoni E (1995) RFLP mapping of genes controlling yield components and plant height in an indica × japonica DH population. In: IRRI (ed) Fragile lives in fragile ecosystems. Proc Int Rice Res Conf IRRI, Los Banos, Philippines, pp 963–976
Gauch HG (1992) Model selection and validation for yield trials with interaction. Biometrics 44:705–712
Guiderdoni E, Gallinato E, Luistro J, Vergara G (1992) Anther culture of tropical japonica/indica hybrids of rice (Oryza sativa L.). Euphytica 62:219–224
Hayes PM, Liu BH, Knapp SJ, Chen F, Jones B, Blake T, Franckowiak J, Rasmusson D, Sorells M, Ullrich SE, Wesenberg D, Kleinhofs A (1993) Quantitative trait locus effects and environmental interaction in a sample of North American barley germplasm. Theor Appl Genet 87:392–401
Hemamalini GS, Shashidhar HE, Shailaja Hittalmani (2000) Molecular marker assisted tagging of morphological and physiological traits under two contrasting moisture regimes at peak vegetative stage in rice (Oryza sativa L.). Euphytica 112:69–78
Huang N, McCouch SR, Mew T, Parco A, Guiderdoni E (1994) Development of an RFLP map from a doubled haploid population in rice. Rice Genet Newslett 11:134–137
International Rice Research Institute (1988) Standard evaluation system for rice, 3rd edn, IRRI, Los Banos, Philippines
Knapp SJ, Bridges WC (1990) Using molecular markers to estimate quantitative trait locus parameters; power and genetic variances for un-replicated and replicated progeny. Genetics 126:769–777
Kurata N, Nagamura Y, Yamamoto A, Harushima Y, Sue N, Wu J, Antonnio BA, Shomura A, Shimizu T, lin SY, Inoue T, Fukuda A, Shimona T, Kuboki Y, Momma Y, Umehara Y, Yano M, Sasaki T, Minobe Y (1994) A 300 Kb interval genetic map of rice including 883 expressed sequence. Natl Genet 8:365–372
Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Lebreton C, Lazic-Jancic V, Steed A, Pekic S, Quarrie SA (1995) Identification of QTL for drought responses in maize and their use in testing causal relationships between traits. J Exp Bot 46:853–865
Li Z, Pinson SRM, Stansel JW, Park WD (1995) Identification of quantitative trait loci (QTL) for heading date and plant height in cultivated rice (Oryza sativa L.). Theor Appl Genet 91:920–927
Lin H-X, Qian H-R, Zhuang J-Y, Lu J, Min S-K, Xiong Z-M, Huang N, Zheng K-L (1995) RFLP mapping of QTL for yield and related characters in rice (Oryza sativa L.) Theor Appl Genet 92:920–927
Lincoln SE, Daly MJ, Lander ES (1993) Mapping genes controlling quantitative traits with mapmaker/qtl1.1: a tutorial and reference manual. Whitehead Institute for Biomedical Research technical report, 3rd edn, Cambridge, Mass
McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newslett 14:11–13
Paterson AH, Damon S, Hewitt JD, Zamir D, Rabinowitch HD, Lincoln SE, Tanksley SD (1991) Mendelian factors underlying quantitative traits in tomato: comparison across species, generations and environments. Genetics 127:181–197
Ricegenes (2000) http://genome.cornell.edu/
Shailaja Hittalmani, Shashidhar HE, Prashanth G. Bagali, Ning Huang, Sidhu JS, Singh VP, Khush GS (2002) Molecular mapping of quantitative trait loci for plant growth, yield and yield related traits across three diverse locations in a doubled haploid rice population. Euphytica 125:207–214
Shashidhar HE, Hemamalini GS, Hittalmani S (1999) Molecular marker-assisted tagging of morphological and physiological traits at the peak vegetative stage: two contrasting moisture regimes. In: Ito O, O'Toole, JC, Hardy B (eds) Genetic improvement of rice for water-limited environments. IRRI, Los Banos, Manila, Philippines, pp 239–256
Stuber CW, Lincoln SE, Wolf DW, Helenjaris T, Lander ES (1992) Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics 132:823–839
Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet 100:697–712
Van Ooijen JW (1999) LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83:613–624
Veldboom LR, Lee M (1996a) Genetic mapping of quantitative trait loci in maize in stress and nonstress environments: 1.Grain yield and yield components. Crop Sci 36:1310–1319
Veldboom LR, Lee M (1996b) Genetic mapping of quantitative trait loci in maize in stress and nonstress environments. II. Plant height and flowering. Crop Sci 36:1320–1327
Wang G, Mackill DJ, Bonman JM, McCouch SR, Champoux MC, Nelson RJ (1994) RFLP mapping of genes conferring complete and partial resistance to blast in a durable resistant rice cultivar. Genetics 136:1421–1434
Xiao J, Li J, Yuan L, Tanksley SD (1996) Identification of QTL affecting traits of agronomic importance in a recombinant inbred population derived from a sub-specific rice cross. Theor Appl Genet 92:230–244
Zhuang JY, Lin HX, Lu J, Qian HR, Hittalmani S, Huang N, Zheng KL (1997) Analysis of QTL × Environment interaction for yield components and plant height in rice. Theor Appl Genet 95:799–808
Acknowledgements.
We thank the Asian Rice Biotechnology Network and the Rockefeller Foundation, USA for the financial support (RF98001/698and 671 to SH). We are also grateful to all who have indirectly helped in conducting the trials at different locations.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H.C. Becker
The order of authors is in accordance with their contribution towards conducting trials, generating data, data analysis and preparation of the manuscript
Rights and permissions
About this article
Cite this article
Hittalmani, S., Huang, N., Courtois, B. et al. Identification of QTL for growth- and grain yield-related traits in rice across nine locations of Asia. Theor Appl Genet 107, 679–690 (2003). https://doi.org/10.1007/s00122-003-1269-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1269-1